Abstract
Activating K-RAS mutations are the most frequent oncogenic mutations in human cancer. Numerous downstream signaling pathways have been shown to be deregulated by oncogenic K-ras. However, to date there are still no effective targeted therapies for this genetically defined subset of patients. Here we report the results of a small molecule, synthetic lethal screen using mouse embryonic fibroblasts derived from a mouse model harboring a conditional oncogenic K-rasG12D allele. Among the >50,000 compounds screened, we identified a class of drugs with selective activity against oncogenic K-ras–expressing cells. The most potent member of this class, lanperisone, acts by inducing nonapoptotic cell death in a cell cycle- and translation-independent manner. The mechanism of cell killing involves the induction of reactive oxygen species that are inefficiently scavenged in K-ras mutant cells, leading to oxidative stress and cell death. In mice, treatment with lanperisone suppresses the growth of K-ras–driven tumors without overt toxicity. Our findings establish the specific antitumor activity of lanperisone and reveal oxidative stress pathways as potential targets in Ras-mediated malignancies.
Keywords: targeted therapy, synthetic lethality
Activating mutations in the RAS oncogenes (H-, N-, and K-RAS) are found in ∼20% of all human tumors. These mutations occur most frequently in K-RAS and are predominantly missense mutations that compromise the GTPase activity of Ras. In clinical practice, K-RAS mutation defines a subset of patients for whom prognosis is generally poor and treatment options are limited (1). K-RAS mutation has been associated with resistance to cytotoxic chemotherapy and radiation therapy (2, 3). Several recent studies demonstrate that K-RAS mutation also confers resistance to newer biologic agents such as EGF receptor (EGFR)-directed therapies (4–6).
At the cellular level, mutant-activated K-ras mediates several key aspects of oncogenesis, including deregulated cell growth, evasion of apoptosis, and induction of angiogenesis. These effects result from the integrated activities of multiple Ras signaling pathways, many of which are potential targets for cancer therapy. The most intensively studied, so-called “canonical” Ras signaling pathways are the Raf/Mek/Erk cascade, involved in mitogenic signaling, and the PI3K/Akt pathway involved in cell survival. The utility of these canonical pathways as targets for the treatment of Ras-mediated cancers remains uncertain. For example, administration of the MAPK/ERK kinase (MEK) inhibitor CI-1040 to mice harboring K-rasG12D-driven lung adenocarcinomas results in a significant reduction in tumor burden, suggesting a dependency on MAPK signaling for tumor growth and survival (7). However, in a phase II clinical trial involving patients with a variety of advanced cancers, CI-1040 showed minimal antitumor activity (8). Recently, the combination of a MEK inhibitor with a PI3K inhibitor showed impressive antitumor activity in mice with K-ras–driven lung tumors (9); whether this combination is effective in killing human cell lines and tumors harboring K-RAS mutations remains to be determined.
Targeting canonical Ras signaling pathways is complicated by the fact that these pathways are also essential for the proliferation and survival of normal cell types. To target oncogenic Ras selectively, several different strategies have been taken. One of the initial strategies involved inhibition of Ras farnesylation, a posttranslational modification required for localization of Ras to the plasma membrane. Preclinical studies of farnesyltransferase inhibitors (FTIs) in transgenic mouse models overexpressing oncogenic Ras showed potent antitumor activity (10). However, in patients with solid tumors, FTIs have shown little if any clinical activity, likely due to redundancies in this pathway (11, 12).
More recent approaches to targeting oncogenic Ras have used synthetic lethal screening to identify novel anticancer agents capable of selectively killing tumor cells harboring a specific mutation. In the case of oncogenic K-ras, several compounds have already been identified, including sulfinyl cytidine derivative (SC-D) (13), erastin (for eradicator of RAS and ST-expressing cells) (14), and oncrasin-1 (for oncogenic Ras tumor-inhibiting compound 1) (15). Though erastin has been shown to induce mitochondrial dysfunction (16), and oncrasin may require protein kinase C iota (PKCι) for activity (15), the precise mechanisms by which any of these compounds selectively kill K-ras mutant cells remain uncertain. Recently, synthetic lethal screens using shRNAs targeting the kinome have identified other potential target pathways in K-ras mutant cell lines, including the noncanonical IκB kinase TBK1 involved in NF-κB signaling (17), the mitotic kinase PLK1 (18), and the serine/threonine protein kinase STK33 of unknown biological function (19).
Despite these promising developments, there are still no effective therapeutic agents or regimens in the clinic for patients with K-RAS mutant tumors. Here we report the results of a synthetic lethal chemical screen using mouse embryonic fibroblasts (MEFs) derived from mice harboring a conditional, oncogenic K-rasG12D allele. We have identified a family of compounds, represented by lanperisone (LP), with selective activity in cells harboring oncogenic K-ras. In contrast to other putative Ras inhibitors, LP, which was originally developed as a muscle relaxant (20), has already been tested in the clinic, and serves as a potential lead in targeting Ras mutant cancers safely and effectively.
Results
Identification of Tolperisone and the Related Compound Lanperisone by Synthetic Lethal Chemical Screening.
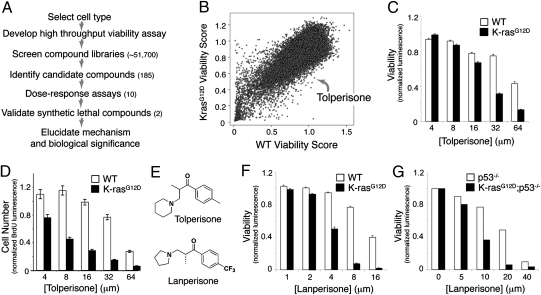
We performed a high-throughput, synthetic lethal chemical screen to identify small molecules that selectively kill MEFs expressing oncogenic K-ras (Fig. 1A). We derived K-rasG12D–expressing MEFs using two different methods. MEFs from K-rasLSL-G12D embryos were treated in vitro with a retroviral, self-excising Cre recombinase (21). Genomic PCR confirmed complete recombination of the K-rasLSL-G12D allele (Fig. S1). We also generated MEFs from K-rasLSL-G12D;Mox2-Cre compound mutants. In these mutant embryos, Cre-mediated recombination occurs in utero and efficiently activates K-rasG12D throughout the embryo proper as shown previously (22). Several lines of evidence indicate that MEFs are a valid surrogate cell type in which to model oncogenic K-ras signaling. First, MEFs expressing physiologic levels of oncogenic K-rasG12D are partially transformed (21). Second, K-rasG12D MEFs do not exhibit augmented activation of canonical Ras signaling pathways (21), similar to the early pulmonary lesions in K-rasLSL-G12D mice.
Fig. 1.
Identification of tolperisone and tolperisone-like drugs as synthetic lethal small molecule inhibitors of K-rasG12D-transformed MEFs. (A) Schematic of high-throughput small molecule library screening. (B) Representative data from primary screening. The viability score is the normalized CTG luminescent value associated with each compound. A viability score of 1.0 indicates that there is no difference in viability relative to DMSO-treated cells. (C) Viability of wild-type and K-rasG12D-expressing MEFs across a range of tolperisone concentrations, as assessed using the CTG assay. Mean ± SEM of three independent lines per genotype, each plated in triplicate. (D) Cell number after tolperisone treatment of wild-type and K-rasG12D MEFs, as assessed using a BrdU cytoblot assay. Mean ± SEM of three independent lines per genotype, each plated in triplicate. (E) Chemical structures of tolperisone and the tolperisone-like drug LP. (F and G) Dose–response experiment testing the effects of LP on cell viability in K-rasG12D (F) or K-rasG12D;p53−/− MEFs. Mean ± SEM of three independent lines per genotype, each plated in triplicate. For all dose–response experiments shown, assays were performed 48 h after compound addition.
Using an ATP-based cell viability assay (CellTiter-Glo or CTG), we screened over 50,000 compounds and identified two with potent and selective activity against K-ras mutant cells compared with wild-type controls (Fig. 1B and Fig. S2). We focused on a piperidine derivative, tolperisone, which is an orally available, centrally acting muscle relaxant used to treat painful muscle spasms. Tolperisone exhibited significant differential activity in both the cell viability assay (Fig. 1C) as well as in a second, independent assay based on BrdU incorporation (Fig. 1D). To explore structure-function relationships, we synthesized or obtained from commercial sources >10 derivatives of tolperisone (Fig. 1E and Fig. S3). Among tolperisone and its derivatives, LP showed the most potent and selective activity against K-rasG12D MEFs, with an IC50 value in the CTG viability assay of 4 μM (Fig. 1F). We also created compound mutant MEFs carrying the K-rasLSL-G12D allele in a p53-null background. K-ras was activated using the same retroviral, self-excising Cre recombinase described above. As shown in Fig. 1G, K-rasG12D;p53−/− MEFs were similarly sensitive to LP-induced killing as K-rasG12D MEFs, indicating that the death was p53 independent. Both K-rasG12D and K-rasG12D;p53−/− MEFs were used in subsequent experiments and showed similar results.
LP-Mediated Induction of Cell Death in K-ras Mutant Cells.
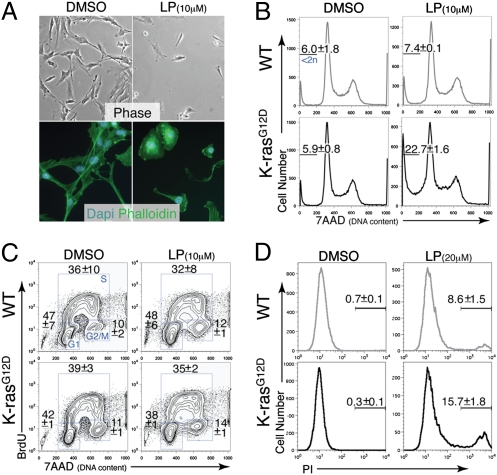
To determine the cellular basis for the decreased viability of LP-treated, K-ras mutant MEFs, we analyzed cells after short-term (6 h) exposure to LP or vehicle (DMSO). In contrast to DMSO-treated K-ras mutant MEFs, LP-treated cells showed striking morphologic changes (Fig. 2A), with the vast majority of cells becoming spherical and beginning to detach from the plate. Wild-type MEFs exposed to LP or DMSO showed minimal changes in morphology (Fig. S4A). We next performed FACS analyses on both wild-type and K-ras mutant MEFs treated with LP or DMSO for 6 h. Consistent with the morphologic observations, LP treatment of K-ras mutant, but not wild-type MEFs, induced a large population of cells with sub-2N DNA content (Fig. 2B). FACS analysis for BrdU incorporation demonstrated normal cell cycle progression of LP-treated cells (Fig. 2C). These results suggest that LP selectively kills K-rasG12D MEFs in a cell cycle-independent fashion.
Fig. 2.
Lanperisone induces cell death of oncogenic K-ras–expressing MEFs. (A) Morphological changes induced by LP treatment of K-rasG12D-expressing MEFs. Phase contrast (Upper) and Oregon Green 488 phalloidin fluorescence (Lower) are shown. (B) Enhanced cell killing of K-rasG12D-expressing MEFs compared with wild-type MEFs, as assessed by FACS for DNA content. Percentages are the mean ± SD of three independent MEF lines. (C) Cell cycle analysis of LP vs. DMSO-treated MEFs. Percentages are the mean ± SD of three independent MEF lines. (D) FACS for PI. Percentages are the mean ± SD of three independent MEF lines. For all experiments shown, K-rasG12D-expressing MEFs were derived from K-rasLSL-G12D;Mox2-Cre embryos. Cells were treated with LP or DMSO for 6 h. Similar results were observed with K-rasG12D;p53−/− MEFs.
To examine the mechanism of LP-induced cell death, we first stained cells with propidium iodide (PI) for FACS analysis. Compared with controls, K-ras mutant MEFs treated with LP for 6 h showed a significantly higher percentage of dying, PI+ cells (Fig. 2D). In separate FACS-based TUNEL-labeling experiments, LP consistently induced higher levels of TUNEL positivity in K-ras mutant compared with wild-type MEFs (Fig. S4B). To investigate whether LP induces death through apoptosis, caspase activation and the effect of the pan caspase inhibitor zVAD on LP-induced cell death were determined. Cleaved caspase 3 was variably detected by immunoblotting and flow cytometry after LP treatment of MEFs, but pretreatment with zVAD did not inhibit LP-induced death (Fig. S4C). Taken together, these results suggest that the cell death induced by LP involves DNA fragmentation, but occurs predominantly via a nonapoptotic mechanism.
Evaluation of Candidate Target Pathways Identified by Gene Expression Studies and Gene Set Enrichment Analysis.
Gene expression profiling has been widely used for the molecular analysis and classification of human cancers. More recently, gene expression profiling has also been used to elucidate the target pathways of biologically active, small molecules of unknown mechanism (23, 24). To begin to understand the mechanism by which LP selectively kills K-ras mutant MEFs, we first defined the gene expression signatures associated with LP treatment of wild-type vs. K-ras mutant MEFs. These signatures were identified by gene expression profiling of RNA from MEFs treated with LP or DMSO for 6 h, followed by comparative marker selection to identify genes that distinguish LP-treated from control profiles (24).
We next used gene set enrichment analysis (GSEA) to compare the signatures associated with LP treatment with a curated database of gene expression changes associated with a variety of biologic pathways (i.e., gene sets) (25). The LP signatures in both wild-type and K-ras mutant MEFs demonstrated marked similarity with gene sets associated with hypoxia and the expression of hypoxia-inducible factors (HIFs; Table S1). This observation led to the hypothesis that LP might induce hypoxia or a hypoxia-like state, which then sensitizes K-ras mutant MEFs to cell death. To address this hypothesis, we first performed immunoblotting of MEF lysates using HIF-specific antibodies. LP treatment was associated with induction of HIF-1, but not HIF-2, in control and K-ras mutant MEFs (Fig. S5A). To determine whether the induction of hypoxia and/or HIF-1 plays a functional role in LP-mediated cell killing, we examined the consequences of activating or inhibiting HIF in the setting of LP treatment. To activate HIF, we exposed MEFs to varying concentrations of the prolyl hydroxylase inhibitor DMOG or to hypoxia (0% O2) before treatment with LP. Neither DMOG nor hypoxia significantly enhanced cell killing by LP (Fig. S5B). Similarly, overexpression of either wild-type HIF-1 or HIF-2, or a mutant HIF-1 or HIF-2 in which the oxygen degradation domain (ODD) is deleted, did not enhance LP-mediated cell killing (Fig. S5C). Finally, overexpression of a dominant-negative form of HIF-1 and HIF-2 did not reduce LP-induced cell death (Fig. S5C). These findings suggest that though HIFs are induced by LP treatment, they do not play a functional role in the selective killing of K-ras mutant cells by LP.
Enhanced Induction of Reactive Oxygen Species by LP in K-ras Mutant Cells.
By GSEA, the LP gene expression signatures also show similarity to those associated with oxidative stress pathways (Table S1). In addition, we queried the Connectivity Map (CMAP; http://www.broadinstitute.org/cmap/) to identify known drugs that induced similar gene expression changes as did LP (23). This query yielded numerous drugs that showed strong positive (i.e., highly correlated with the LP signature) or negative (anticorrelated with the LP signature) connectivities with LP treatment. Among the strong positive connectivities were multiple drugs of different mechanism but all known to induce oxidative stress, including parthenolide (a sesquiterpene lactone),15-δ-prostaglandin J2 (an endogenous anti-inflammatory signaling molecule), lomustine (an alkylating agent), and hsp90 inhibitors such as geldanamycin (Fig. S6). Of note, these connectivities were similar in both wild-type and K-ras mutant MEFs.
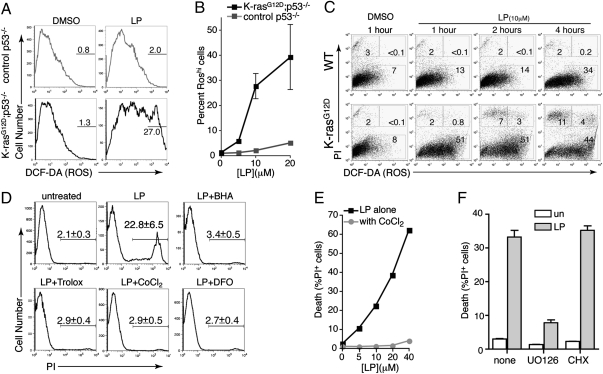
To test whether the mechanism of LP-induced cell killing does indeed involve oxidative cell death, we first measured the levels of reactive oxygen species (ROS) in wild-type and K-ras mutant MEFs. Both control and LP-treated MEFs were stained with the redox-sensitive dye DCF-DA, followed by FACS analysis to quantify intracellular ROS levels. After 6 h of exposure to LP, K-ras mutant MEFs displayed a significant surge in ROS levels compared with LP-treated wild-type MEFs (Fig. 3A). Approximately 30–40% of K-ras mutant cells showed high ROS levels, compared with 5% or less of wild-type controls (Fig. 3 A and B). Significant ROS levels were detected as early as 1 h after LP treatment in K-ras mutant MEFs, whereas wild-type MEFs treated with LP showed a more gradual accumulation of intracellular ROS over time (Fig. 3C).
Fig. 3.
Lanperisone-mediated induction of intracellular reactive oxygen species and oxidative cell death in K-ras mutant MEFs. (A) FACS of DCF-DA stained MEFs. MEFs were treated with 10 μM LP for 6 h. (B) High level ROS induction as a function of LP dose, as shown by DCF-DA staining and FACS analysis. MEFs were treated with the indicated concentrations of LP for 6 h. Values shown are mean ± SD of triplicate wells. (C) Time course of LP-mediated ROS induction and cell death, as assessed by FACS and costaining for DCF-DA and PI. Percent of cells in each gate is shown. (D) Suppression of LP-mediated cell death by pretreatment with antioxidants, as shown by FACS. K-rasG12D;p53−/− MEFs were pretreated and then exposed to 20 μM LP for 6 h. (E) Suppression of LP-induced death by cobalt chloride, even at high doses of LP. Cells were treated with LP for 6 h. Data are representative of three independent K-rasG12D MEF lines. (F) MEK-dependent, translation-independent cell killing by LP (10 μM, 6 h). Data are the mean ± SD of triplicate wells and is representative of three independent MEF lines.
To assess the functional importance of ROS induction in LP-treated K-ras mutant MEFs, we pretreated cells with a variety of different ROS scavengers, including deferoxamine (DFO), butylated hydroxyanisole (BHA), and the antioxidant trolox, a vitamin E analog. As shown in Fig. 3D, all of the ROS scavengers tested completely abolished cell killing by LP. FACS confirmed the viability of cells pretreated with ROS scavengers, and also demonstrated a variable reduction in intracellular ROS levels, depending on the ROS scavenger used (Fig. 3D and Fig. S7A). Consistent with these results, hydrogen peroxide (H2O2), a cell-permeable ROS, synergized with LP in killing K-rasG12D-expressing MEFs (Fig. S7B). In addition, cobalt chloride, which is known to compete with iron for cellular transport and for binding sites on a variety of proteins, did not significantly diminish ROS production, but potently blocked LP-induced cell death (Fig. 3E and Fig. S8). Furthermore, LP-induced cell death was also inhibited by pretreatment with the MEK1/2 inhibitor U0126 but not by the protein synthesis inhibitor cycloheximide (Fig. 3F and Fig. S8). Taken together, these results demonstrate that the selective killing of K-ras mutant cells by LP is mediated by the induction of ROS, and is both iron- and Ras/MAPK-dependent.
Antitumor Activity of LP in Vivo.
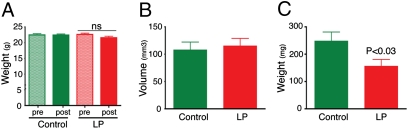
To test whether LP has activity against K-ras mutant tumors in vivo, we used compound mutant MEFs carrying the oncogenic K-ras allele in a p53-null background. Unlike K-rasG12D MEFs, K-rasG12D;p53−/− MEFs form s.c. tumors when injected into nude mice. Mice with established tumors were treated with either water or with LP at 40 mg/kg twice daily by oral gavage. The mice that received LP tolerated treatment well, with no overt toxicities and no statistically significant change in body weight (Fig. 4A). After 7 d, tumors were measured, dissected, and weighed. Control and LP-treated mice both showed similar tumor sizes at the start of treatment (Fig. 4B). However, compared with tumors from control animals, tumors from LP-treated mice were 24% smaller based on estimated tumor volumes (P = 0.16), and 37% smaller based on actual tumor weights (P < 0.03; Fig. 4C).
Fig. 4.
Lanperisone inhibits tumor growth in vivo. Fully transformed, compound mutant K-rasG12D;p53−/− MEFs were injected s.c. into the flanks of nude mice. Tumor-bearing mice were then treated with either vehicle (dH2O) or LP. (A) Body weights before and after treatment in control and LP-treated mice. Mean ± SEM is shown. ns, not significant. (B) Initial estimated volumes of s.c. tumors before treatment. Mean ± SEM is shown (n = 23/cohort). (C) Final tumor weights of s.c. tumors after 7 d of control or LP treatment. Mean ± SEM is shown (n = 23/cohort).
Discussion
Through synthetic lethal chemical screening, we have identified a class of drugs with selective activity against oncogenic K-ras–expressing cells. These drugs have been developed as centrally acting muscle relaxants, and until now have not been studied as potential anticancer agents. Here we show that LP, the most potent member of this family, induces nonapoptotic cell death in a cell cycle- and translation-independent manner. Based on gene expression and biochemical studies, the mechanism of selective cell killing appears to involve the induction of ROS, which may be either overproduced and/or inefficiently scavenged in the setting of oncogenic K-ras activation. High intracellular levels of ROS in turn appear to contribute to the selective, oxidative cell death of K-ras mutant cells.
To date, among the K-ras targeted therapies identified through synthetic lethal chemical screening, LP is structurally distinct. However, in terms of potential mechanism of action, LP seems to mostly closely resemble erastin. Erastin was first discovered in a synthetic lethal chemical screen using engineered human tumor cells derived from primary BJ fibroblasts (14). Like LP, erastin induces nonapoptotic, oxidative cell death in cell lines expressing oncogenic Ras (14, 16). Affinity purification and mass spectrometry led to the identification of mitochondrial voltage-dependent anion channels (VDACs) as putative targets of erastin (16). By binding to VDACs, erastin is believed to alter VDAC gating, leading to mitochondrial dysfunction, release of ROS, and oxidative cell death. The mechanistic basis for erastin's selectivity, however, remains poorly understood, and has been attributed to either up-regulation of VDACs (16) or higher levels of iron (26) in HRAS mutant cells.
Though the molecular target of LP and related family members is unknown, functional studies suggest that this class of compounds may also perturb voltage-gated ion channels. In dorsal root ganglion neurons, the related drug tolperisone inhibits voltage-gated sodium and calcium channels (27). This combined blockade leads to presynaptic inhibition of neurotransmitter release from primary afferent terminals, which in turn depresses the spinal reflex machinery. As a result, tolperisone-type drugs like LP have been developed to treat painful reflex muscle spasms. Whether LP interacts with voltage-gated channels in K-ras mutant cells to causes mitochondrial dysfunction remains to be determined. However, this hypothesis is tantalizing in light of the known pharmacology of LP (27), as well as the similarities in LP- and erastin-mediated cell killing. Of note, erastin also exhibited selective lethality for K-ras mutant MEFs relative to wild-type controls (Fig. S9). Additionally, in the CMAP experiments, lanperisone did show moderate connectivity with erastin (Fig. S6E).
The potential role of ROS in promoting and maintaining the oncogenic phenotype of cancer cells is well known. However, ROS and the resulting oxidative stress can also induce cellular senescence and apoptosis in certain contexts, suggesting a role for ROS-generating drugs as anticancer agents. The exact response to ROS depends on the magnitude of ROS induction, the type of ROS produced, and cell type and tissue-specific factors. A number of drugs with known ROS-generating activity have already proved therapeutically useful: arsenic trioxide, for example, is an FDA-approved treatment for patients with relapsed or refractory acute promyelocytic leukemia (APL). Numerous lines of evidence suggest that the antitumor activity of arsenic trioxide in APL is due to the production of ROS (28), possibly via arsenic-induced up-regulation of NADPH oxidase (29). Whether arsenic trioxide is also active in cancer cells harboring mutant K-ras is unknown; however, our findings with LP provide a rationale for pursuing future studies of arsenic trioxide specifically in the setting of oncogenic K-ras activation.
In cancers harboring oncogenic K-ras, the various roles of ROS have not been fully elucidated. K-ras activation itself is believed to augment ROS production and levels of oxidative stress (30). Consistent with the notion of ROS as cancer-promoting, recent work using the same MEF system has shown that ROS are required for K-ras–induced, anchorage-independent growth (31). Of note, we did not detect elevated basal levels of ROS in K-ras mutant MEFs compared with controls (Fig. 3A), raising the possibility that under steady-state conditions, ROS scavengers can counteract the elevation in ROS induced by endogenous oncogenic K-ras. Based on all of the available data, we propose that ROS may serve as a double-edged sword in K-ras mutant cancers. The oxidative stress related to oncogenic activation may help drive and maintain the transformed state, while also sensitizing cells to exogenous ROS inducers such as LP and erastin. This sensitization may result from the increased demand on intrinsic ROS-scavenging systems, shifting the redox state so that cells can no longer appropriately respond to further oxidative stress. Consequently, high levels of ROS can rapidly accumulate and trigger cell death. This model is supported by the observation that H-RAS transformation of engineered human tumor cells confers marked sensitivity to β-phenylethyl isothiocyanate (PEITC), a natural compound that effectively depletes cellular glutathione (30).
Because activating K-RAS mutations occur predominantly in human tumors of epithelial origin, the relevance of synthetic lethal interactions discovered in fibroblast cell-based systems is important to consider. Both MEFs and engineered human fibroblasts are ideally suited for synthetic lethal screens because each enables comparison of isogenic lines differing only in the expression of an oncogene. However, there are clearly intrinsic differences in the biology of fibroblast vs. epithelial cells (32), which may in part reflect differential utilization of downstream RAS effectors. In preliminary studies, we have examined the activity of LP in a variety of mouse and human cancer cell lines. Overall, lung cancer cell lines harboring oncogenic K-ras appear to be relatively resistant to LP-induced cell killing, whereas one K-ras mutant mouse sarcoma cell line is relatively sensitive to LP (Fig. S10). We speculate that intrinsic differences in the redox states of mesenchymal vs. epithelial-derived tissues may underlie the differences in sensitivity to LP. We cannot exclude the possibility of species-specific differences, because the majority of the lung cancer cell lines were of human origin, whereas the sarcoma cell line was of mouse origin.
In conclusion, we have identified the tolperisone-like drug LP as a potential targeted therapy for K-ras mutant cancers. By inducing high levels of ROS, LP appears to selectively target K-ras mutant cells by exploiting their inherent vulnerability to oxidative stress. The remarkable mechanistic similarities of LP as well as erastin underscore the potential of ROS-mediated therapies as a novel strategy to treat K-ras mutant tumors. Such mechanisms may also underlie the function of two recently identified, synthetic lethal K-ras targets, TBK1 and STK33 (17, 19). Among all of the candidate K-ras–targeted agents, LP is the only one with human safety and pharmacokinetic data. If preclinical studies of LP continue to show promise, this drug could undergo rapid clinical development and prove efficacious for K-ras mutant tumors, particularly those of mesenchymal origin.
Materials and Methods
Mouse Strains and MEFs.
The K-rasLSL-G12D strain was interbred to Mox2-Cre mice to enable recombination within the embryo proper and derivation of K-rasG12D–expressing MEFs (22). Recombination of the K-rasLSL-G12D allele was also performed in vitro using self-excising Cre recombinase as described (21). K-rasLSL-G12D mice were crossed to p53+/− mice to generate K-rasLSL-G12D;p53−/− MEFs. Mice and MEFs were either on a pure 129Sv or a mixed 129 Sv/C57BL/6 background. Early passage (P3-4) MEFs were used for all experiments.
Small Molecule Screening.
High-throughput small molecule screening was performed at the Institute for Chemical and Cellular Biology, Harvard Medical School. At least three independently derived MEF lines of each genotype (wild-type or K-rasG12D) were plated in triplicate in 384-well format at 1,500 cells/well. Compounds were added robotically the following day to a final concentration of ∼10 μM. Compounds (∼51,700) were obtained from commercial libraries (ChemDiv, Maybridge, Peakdale, and Bionet), known bioactives collections (National Institute of Neurological Disorders and Stroke, SpecPlus, Broad Institute), natural product libraries (Starr Foundation Extracts and Philippines Plant Extracts), and diversity oriented synthesis (DOS) collections from the Broad Institute. Growth inhibition was determined 48 h after compound addition using a luminescent cell viability assay (CellTiter-Glo; Promega). Viability scores were calculated for each compound within each genotype by normalizing to DMSO controls. Positives were defined as any compounds that reduced the relative viability of mutant vs. wild-type MEFs by 0.3. Positives were selected for secondary screening using a BrdU cytoblot assay as previously described (33), and for dose–response assays.
Proliferation, Cell Cycle, and Apoptosis Assays.
BrdU incorporation was determined using the BD BrdU staining kit according to the manufacturer's protocol (BD Bioscience). Fluorescein TUNEL staining was performed with the In Situ Cell Death Detection Kit according to the manufacturer's protocol (Roche). The caspase inhibitor Z-VAD-FMK was used at 25 μM (Promega). Cells were analyzed using FacScan or FacsCaliber flow cytometers (BD Bioscience) and analyzed using FlowJo Software (TreeStar).
Gene Expression Profiling and Analysis.
Three independently derived MEF lines per genotype were treated with either DMSO or LP (10 μm) for 6 h. RNA was isolated by TRIzol extraction, and labeled cRNA was hybridized to Affymetrix Mouse Genome 430A 2.0 microarrays according to the manufacturer's protocol. Expression signatures were calculated using the Comparative Marker Selection module of the GenePattern software suite, using the signal-to-noise statistic and standard settings. GSEA was performed using the GSEA module (25) of GenePattern; P values were determined by random permutation of gene sets. CMAP analysis was performed with the CMAP Web tool found at http://www.broadinstitute.org/cmap/ (23). Details of these analyses are available on request.
Intracellular ROS Measurements.
MEFs were harvested, washed with PBS containing 5 mM glucose, and incubated with DCF-DA (10 μM; Molecular Probes) for 15 min. Cells were then washed with PBS and analyzed by FACS. Modulators of ROS were tested at the following working concentrations: DFO 150 μM, Trolox 200 μM, BHA 100 μM, CoCl2 0.250 mM, H2O2 400 μM, and U0126 5 μM.
In Vivo Studies.
K-rasG12D;p53−/− MEFs (1 × 106) were injected s.c. into the flanks of nude mice. Mice harboring tumors measuring at least 1 cm in longest dimension were treated with either dH2O or LP (40 mg/kg twice daily, dissolved in dH2O). All mice were killed after 7 d.
Supplementary Material
Acknowledgments
We thank J. Lamb for help with the CMAP analysis, D. R. Mani for help with the statistical analysis of primary screening data, and C. Kim for critical reading of the manuscript. We are grateful to M. Zimmer and O. Iliopoulos for providing HIF-based reagents. This work was supported by National Institutes of Health (NIH) Grants 5K08CA111634-5 (to A.T.S) and 5-U01-CA84306. This work was also funded in part by the National Cancer Institute's Initiative for Chemical Genetics, NIH, under Contract N01-CO-12400. A.T.S is the Charles W. and Jennifer C. Johnson Koch Institute Clinical Investigator. M.M.W was a Merck Fellow of the Damon Runyon Cancer Research Foundation and a Genentech Postdoctoral Fellow. T.J. is a Howard Hughes Medical Institute and Daniel K. Ludwig Scholar.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105941108/-/DCSupplemental.
References
- 1.Mascaux C, et al. The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cengel KA, et al. Oncogenic K-Ras signals through epidermal growth factor receptor and wild-type H-Ras to promote radiation survival in pancreatic and colorectal carcinoma cells. Neoplasia. 2007;9:341–348. doi: 10.1593/neo.06823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winton T, et al. National Cancer Institute of Canada Clinical Trials Group National Cancer Institute of the United States Intergroup JBR.10 Trial Investigators Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massarelli E, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 6.Van Cutsem E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 7.Ji H, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 8.Rinehart J, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant KrasG12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohl NE, et al. Inhibition of farnesyltransferase induces regression of mammary and salivary carcinomas in ras transgenic mice. Nat Med. 1995;1:792–797. doi: 10.1038/nm0895-792. [DOI] [PubMed] [Google Scholar]
- 11.Adjei AA, et al. Phase II study of the farnesyl transferase inhibitor R115777 in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2003;21:1760–1766. doi: 10.1200/JCO.2003.09.075. [DOI] [PubMed] [Google Scholar]
- 12.Macdonald JS, et al. A phase II study of farnesyl transferase inhibitor R115777 in pancreatic cancer: A Southwest oncology group (SWOG 9924) study. Invest New Drugs. 2005;23:485–487. doi: 10.1007/s10637-005-2908-y. [DOI] [PubMed] [Google Scholar]
- 13.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 14.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Wu S, Liu J, Fang B. Identification of a small molecule with synthetic lethality for K-ras and protein kinase C iota. Cancer Res. 2008;68:7403–7408. doi: 10.1158/0008-5472.CAN-08-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Sakitama K, Ozawa Y, Aoto N, Tomita H, Ishikawa M. Effects of a new centrally acting muscle relaxant, NK433 (lanperisone hydrochloride) on spinal reflexes. Eur J Pharmacol. 1997;337:175–187. doi: 10.1016/s0014-2999(97)01289-2. [DOI] [PubMed] [Google Scholar]
- 21.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 22.Shaw AT, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 24.Hieronymus H, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kocsis P, et al. Tolperisone-type drugs inhibit spinal reflexes via blockade of voltage-gated sodium and calcium channels. J Pharmacol Exp Ther. 2005;315:1237–1246. doi: 10.1124/jpet.105.089805. [DOI] [PubMed] [Google Scholar]
- 28.Douer D, Tallman MS. Arsenic trioxide: New clinical experience with an old medication in hematologic malignancies. J Clin Oncol. 2005;23:2396–2410. doi: 10.1200/JCO.2005.10.217. [DOI] [PubMed] [Google Scholar]
- 29.Chou WC, et al. Role of NADPH oxidase in arsenic-induced reactive oxygen species formation and cytotoxicity in myeloid leukemia cells. Proc Natl Acad Sci USA. 2004;101:4578–4583. doi: 10.1073/pnas.0306687101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trachootham D, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skinner J, et al. Opposing effects of mutant ras oncoprotein on human fibroblast and epithelial cell proliferation: Implications for models of human tumorigenesis. Oncogene. 2004;23:5994–5999. doi: 10.1038/sj.onc.1207798. [DOI] [PubMed] [Google Scholar]
- 33.Koeller KM, et al. Chemical genetic modifier screens: Small molecule trichostatin suppressors as probes of intracellular histone and tubulin acetylation. Chem Biol. 2003;10:397–410. doi: 10.1016/s1074-5521(03)00093-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.