Abstract
Mutations in the MID1 gene are causally linked to X-linked Opitz BBB/G syndrome (OS), a congenital disorder that primarily affects the formation of diverse ventral midline structures. The MID1 protein has been shown to function as an E3 ligase targeting the catalytic subunit of protein phosphatase 2A (PP2A-C) for ubiquitin-mediated degradation. However, the molecular pathways downstream of the MID1/PP2A axis that are dysregulated in OS and that translate dysfunctional MID1 and elevated levels of PP2A-C into the OS phenotype are poorly understood. Here, we show that perturbations in MID1/PP2A affect mTORC1 signaling. Increased PP2A levels, resulting from proteasome inhibition or depletion of MID1, lead to disruption of the mTOR/Raptor complex and down-regulated mTORC1 signaling. Congruously, cells derived from OS patients that carry MID1 mutations exhibit decreased mTORC1 formation, S6K1 phosphorylation, cell size, and cap-dependent translation, all of which is rescued by expression of wild-type MID1 or an activated mTOR allele. Our findings define mTORC1 signaling as a downstream pathway regulated by the MID1/PP2A axis, suggesting that mTORC1 plays a key role in OS pathogenesis.
Keywords: ubiquitin ligase, proteasome-mediated degradation
Mutations in the MID1 gene are causally linked to X-linked Opitz BBB/G syndrome (OS), a congenital disorder that primarily affects the formation of ventral midline structures. The clinical manifestations of Opitz syndrome are characterized by a diverse spectrum of symptoms, including hypertelorism and hypospadias, cleft lip/palate, dysphagia, heart defects, and mental retardation (1).
The MID1 protein is a member of the RBCC/TRIM family, which contains a conserved module of the RING domain, followed by B-boxes and a coiled-coil domain (2). MID1 also contains fibronectin type III and B30.2/SPRY domains. The RING domain has been well characterized in ubiquitin-mediated protein degradation, whereas the other domains mediate protein-protein interactions (3–6). Opitz syndrome-derived mutations in MID1 have been identified throughout the protein and show several functional consequences such as compromised association with microtubules and/or transport along microtubules (4, 7–9). In addition to its microtubule-binding function, MID1 also functions as an E3 ligase that targets the microtubule-associated pool of the catalytic subunit of protein phosphatase 2A (PP2A-C) for ubiquitin-mediated degradation through an interaction with the protein α4 (3). Perturbation of the E3 ligase function of MID1 in OS cells leads to the accumulation of PP2A-C and the dramatic dephosphorylation of microtubule-associated proteins, which is postulated to contribute to OS pathogenesis (3).
Signaling from the mammalian target of rapamycin (mTOR) controls diverse cellular processes such as growth, autophagy, stress responses, cytoskeletal reorganization, cell motility, metabolism, and aging (10–12). mTORC1 consists of mTOR and the interacting proteins, Raptor, and mLST8. In particular, Raptor plays an essential scaffolding role toward mTORC1 substrates such as 4E-BP1 and S6K1 (13, 14). Studies in our laboratory have recently focused on the regulation of mTOR signaling by the ubiquitin proteasome system (UPS) (15). Here, we show that proteasome inhibition disrupts mTORC1 formation and signaling in a PP2A-dependent manner. Furthermore, we show that cells derived from Opitz syndrome patients harboring defects in the E3 ligase MID1 have profound defects in mTOR complex 1 (mTORC1) function, suggesting that the MID1/PP2A axis is an important regulator of mTORC1 signaling and that dysregulation of mTORC1 signaling plays a key role in the pathogenesis of OS.
Results
Proteasome Inhibition Disrupts mTORC1 Formation and Signaling.
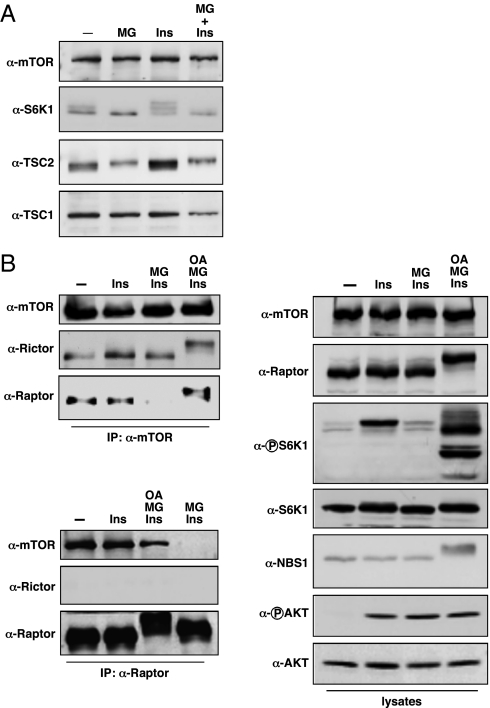
During the course of our studies on mTOR and its regulation by the ubiquitin proteasome system, we made the observation that proteasome inhibition decreased mTORC1 signaling (Fig. 1A). In serum-starved U2OS cells, MG132 treatment was able to block insulin-stimulated phosphorylation of the mTORC1 substrate S6K1, as judged by the stereotypic mobility shift. This observation was not completely unexpected, because negative regulators of mTORC1, such as TSC2 and REDD1, undergo ubiquitin-mediated degradation (15, 16). However, following MG132 treatment, TSC2 levels did not increase, suggesting that TSC2 was not responsible for decreased mTORC1 signaling. To formally address the possible contribution of TSC2 in this process, mTORC1 signaling was examined in TSC2−/− MEFs following MG132 treatment. In WT and TSC2−/− MEFs, MG132 treatment decreased S6K1 phosphorylation, demonstrating that this effect was TSC2 independent (Fig. S1A). Furthermore, this effect was also independent of REDD1, as these experiments were carried out under normoxic conditions, and TSC2 is required for REDD1-mediated mTORC1 down-regulation (17). In addition to our observations in U2OS cells and MEFs, we also observed down-regulation of mTORC1 signaling in both HCT-116 and MCF-7 cells following MG132 treatment of cells cultured in serum-replete media, suggesting that this phenomenon is independent of serum starvation/insulin restimulation and can be generalized to other cancer cells (Fig. S1B, lanes 1 and 2; compare to Fig. S3A, lanes 1 and 4). Therefore, proteasome inhibition reduces mTORC1 signaling through a mechanism that does not involve critical nodes in mTORC1 signaling previously ascribed to ubiquitin-proteasome regulation.
Fig. 1.
MG132 treatment reduces mTORC1 formation and signaling. (A) Serum-starved U2OS cells were treated with MG132 (MG, 10 μM) for 3 h before insulin stimulation (Ins, 100 nM) as indicated. Cell lysates were immunoblotted with the indicated antibodies. (B) Serum-starved U2OS cells were treated with MG132 (MG, 10 μM) or okadaic acid (OA, 100 nM) as indicated before insulin stimulation (100 nM). α-mTOR immunoprecipitates, α-Raptor immunoprecipitates, and whole cell lysates were immunoblotted with the indicated antibodies.
Reduced S6K1 phosphorylation could be due to decreased mTORC1 kinase activity, increased phosphatase activity toward S6K1, or a combination of both mechanisms. We first tested whether formation of mTORC1 was disrupted in the presence of MG132, which would account for decreased mTORC1 activity toward S6K1. Indeed, treatment of U2OS cells with MG132 disrupted the interaction between mTOR and Raptor, whereas the interaction between mTOR and Rictor (mTORC2) was unchanged (Fig. 1B). Consistent with these findings, S6K1 showed reduced phosphorylation, whereas AKT phosphorylation was unaffected (Fig. 1B). We obtained similar results using another proteasome inhibitor, lactacystin (Fig. S1C). To confirm reduced mTORC1 kinase activity toward its substrates, we examined mTOR immunoprecipitates from MG132-treated cells for kinase activity in vitro and found decreased phosphorylation of the mTORC1 substrate 4E-BP1, consistent with reduced Raptor association (Fig. S1D).
Disruption of mTOR/Raptor Is Dependent on PP2A Activity.
Given the observation that MG132 caused a slight shift in TSC2 mobility indicative of phosphorylation changes and the previous involvement of PP2A in mTORC1 signaling (18–22), we tested whether PP2A was involved in disrupting mTORC1 formation. Treatment of cells with the PP2A inhibitor okadaic acid was able to restore mTOR/raptor complex formation and signaling, suggesting that PP2A regulates mTORC1 formation (Fig. 1B and Fig. S1B). We addressed the possibility that PP2A disrupted mTORC1 formation through a direct mechanism by examining whether mTOR/Raptor complex formation was dependent upon phosphorylation. Treatment of mTOR immunoprecipitates with λ-phosphatase in vitro disrupted the mTOR/Raptor interaction (Fig. S2), demonstrating that phosphorylation was important for mTORC1 formation.
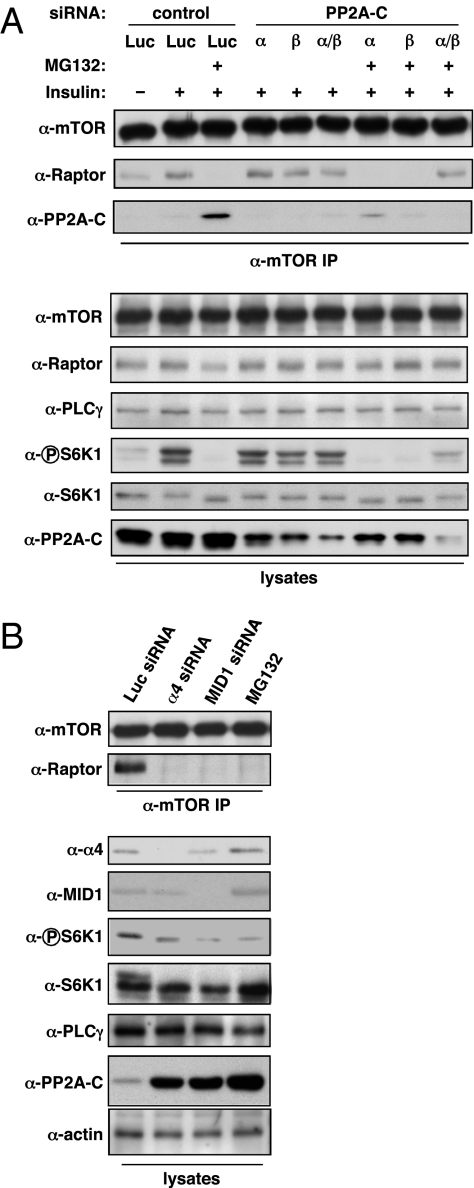
PP2A represents a family of holoenzyme complexes with different activities, subcellular localization, and substrate specificities (23). A given PP2A holoenzyme is made up of three distinct components: the aforementioned catalytic subunit (PP2A-C), a scaffolding subunit (PP2A-A), and a regulatory subunit (PP2A-B). Furthermore, additional complexity exists within each subunit class. Both the C and A subunit each have α- and β-isoforms, whereas the B subunit family encompasses B (PR55α, β, γ, δ), B′ (PR61α, β, γ, δ, ε), B′′ (PR72, PR130, PR48/70, G5PR), and B′′′ (PR93, PR110) subunits, allowing for a diverse repertoire of holoenzymes. To further assess the involvement of PP2A in the disruption of mTORC1, we used siRNA to knock down expression of the PP2A-C subunit, with the expectation that PP2A-C depletion would block the effect of MG132 (Fig. 2A and Fig. S3A). As observed previously, MG132 treatment disrupted complex formation between mTOR and Raptor and increased the amount of the PP2A-C subunit coimmunoprecipitating with mTOR. Depletion of the PP2A-Cα or -Cβ subunits alone or together in the absence of MG132 had little effect on mTORC1 formation or signaling. In contrast, depletion of both Cα and Cβ reduced the amount of PP2A-C associating with mTOR and was able to rescue both the coimmunoprecipitation of Raptor with mTOR and the phosphorylation of S6K1 in the presence of MG132. To further characterize the PP2A complex that regulates mTORC1, we used shRNA directed against specific A and B subunits to determine whether their depletion would block MG132-induced mTORC1 disruption (Fig. S3B). Codepletion of both PP2A-A subunit isoforms (α and β) or depletion of the PP2A-Bα (PR55α) subunit rescued mTOR/Raptor complex formation and S6K1 phosphorylation. Although our use of the pan-reactive PP2A-B antibody complicates a precise assessment of the efficiency of the different PP2A-B isoform (α, β, γ, δ) knockdowns, our data nonetheless suggest that a holoenzyme containing the PP2A-Bα subunit participates in mTORC1 disruption. Finally, given that the mTOR N terminus contains HEAT repeats, a protein-protein interaction domain found in Huntingtin, elongation factor 3, PP2A-A subunit, and TOR, we hypothesized that the interaction we observed between mTOR and PP2A-C was mediated by this region. Based on a ClustalW alignment between the mTOR N terminus and PP2A-A (α- and β-isoform) sequences, we generated an mTOR fragment consisting of amino acids 600–1,450, which showed the highest homology to the HEAT repeats of the PP2A-A subunit. Expression of this fragment with PP2A-C showed that this region was sufficient to mediate interaction between mTOR and PP2A-C (Fig. S4). Taken together, our data suggest that a Bα (PR55α)-containing PP2A holoenzyme is recruited to mTORC1 via the mTOR HEAT repeats, where the catalytic activity of PP2A-C mediates mTORC1 association.
Fig. 2.
The MID1-PP2A axis mediates mTORC1 disruption. (A) U2OS cells were transfected with siRNAs targeting luciferase (Luc) or PP2A catalytic subunits (PP2A-Cα, –Cβ) as indicated. Transfected cells were serum starved for 18 h, incubated with MG132 as indicated, then restimulated with insulin (100 nM). α-mTOR immunoprecipitates and whole cell lysates were immunoblotted with the indicated antibodies. (B) U2OS cells were transfected with siRNAs targeting luciferase (Luc), α4, or MID1 or treated with MG132. α-mTOR immunoprecipitates and whole cell lysates were immunoblotted with the indicated antibodies.
MID1 E3 Ligase Regulates mTORC1 Signaling.
To further dissect the underlying molecular mechanism linking proteasome inhibition to the disruption of mTORC1 signaling via PP2A, we examined whether MID1, an E3 ubiquitin ligase that regulates PP2A stability (3), contributed to this effect. Depletion of either MID1 or its associated protein, α4, recapitulated the MG132 effect by disrupting mTOR/Raptor complex formation and decreasing S6K1 phosphorylation (Fig. 2B). In addition, α4 or MID1 knockdown also increased PP2A-C levels. These data suggest that alterations in PP2A stability affect mTORC1 formation and signaling.
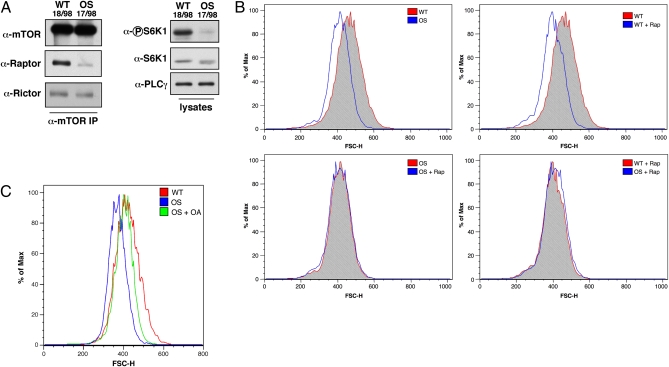
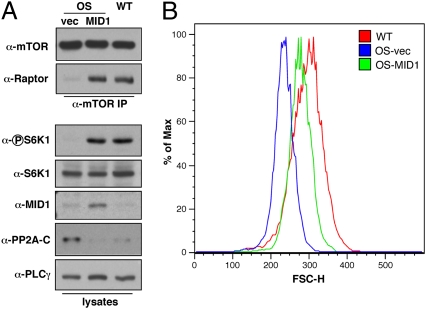
Given the link that we established between MID1 and mTORC1 signaling, we hypothesized that OS patients, which harbor deficiencies in MID1 function, may also have mTOR signaling defects. To test this, we examined mTORC1 formation in MID1-deficient OS and WT fibroblast cell lines derived from patients (3). Similar to our results in MID1 or α4-depleted cells, OS cells exhibited reduced amounts of mTOR/Raptor complex formation and reduced S6K1 phosphorylation (Fig. 3A and Fig. S5A). We next examined cell size, another mTORC1-dependent phenotype (24), in the OS and WT cell lines by measuring forward scatter of the G1 cell cycle population by FACS analysis (Fig. 3B and Fig. S5B). Consistent with reduced mTORC1 signaling, OS cells were smaller than the WT cells and treatment of WT cells with rapamycin reduced their size to that of the untreated OS cells. Interestingly, treatment of the OS cells with rapamycin did not cause any further change in cell size, which strongly suggests that the smaller size of OS cells is due to reduced mTORC1 signaling. To examine whether this size difference was mediated by PP2A, OS cells were treated with okadaic acid, which rescued OS cell size to WT levels (Fig. 3C). Finally, we determined whether these phenotypes could be rescued by WT MID1 reconstitution (Fig. 4 A and B). Expression of WT MID1 in OS cells increased the amount of Raptor that coimmunoprecipitated with mTOR similar to WT cells, restored S6K1 phosphorylation, and partially rescued the cell size defect.
Fig. 3.
mTORC1 function is reduced in OS patient cells. (A) α-mTOR immunoprecipitates and whole cell lysates from WT (18/98) or OS (17/98) fibroblasts were immunoblotted with the indicated antibodies. (B) WT and OS cells were fixed and labeled with propidium iodide before FACS analysis. Rapamycin treatment (100 nM) for the indicated samples was for 18 h. For all samples, the G1 population was gated and the forward scatter height (FSC-H) was determined. Histogram plots were generated using FlowJo version 8.8.6. (C) As indicated, OS cells were treated with okadaic acid (OA,10 nM) for 18 h before fixation and propidium iodide labeling. For WT, OS, and OS cells treated with okadaic acid, the G1 population was gated and the FSC-H was determined. Histogram plots were generated using FlowJo.
Fig. 4.
Reconstitution of OS cells with MID1 restores mTOR signaling. (A) OS (17/98) cells were transfected with vector (vec) or WT MID1. α-mTOR immunoprecipitates and whole cell lysates from transfected OS cells or WT (18/98) cells were immunoblotted with the indicated antibodies. (B) WT and OS cells as in A were cotransfected with GFP. Cells were fixed, stained with propidium iodide, and FSC-H of the GFP+, G1 cells was determined by FACS. Histogram plots were generated using FlowJo version 8.8.6.
Activated mTOR Mutant Rescues mTORC1 Signaling Defects in OS Cells.
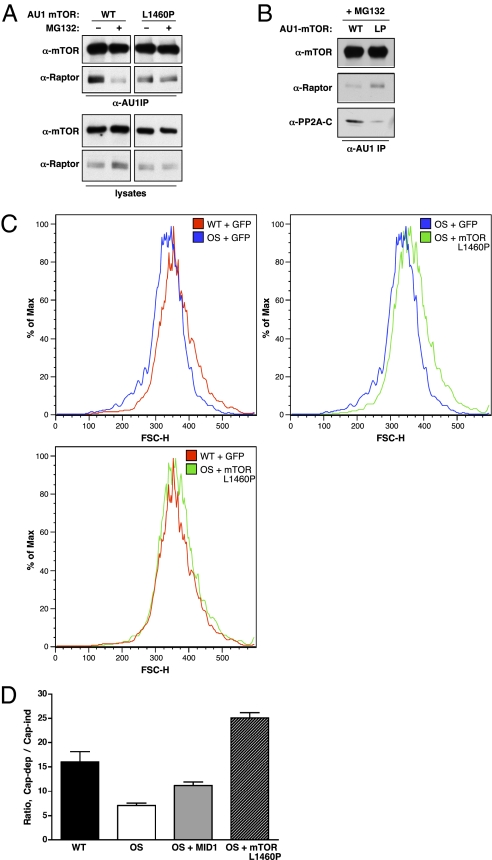
To determine whether increased mTORC1 signaling could reverse the phenotypes observed in OS cells, we took advantage of a constitutively active mTOR allele, L1460P (25). We first tested the association of the mTOR L1460P mutant with Raptor in the absence or presence of MG132. In contrast to disruption of the mTOR WT/Raptor complex, the mTOR L1460P/Raptor complex was unaffected in the presence of MG132 (Fig. 5A). Furthermore, the amount of PP2A-C coprecipitating with the mTOR L1460P mutant was reduced compared with WT (Fig. 5B), demonstrating that reduced recruitment of PP2A-C contributes to the lack of mTORC1 dissociation. We next examined whether expression of the mTOR L1460P mutant could rescue the cell size defect in OS cells. Ectopic expression of the mTOR L1460P mutant increased OS cell size to WT levels and restored mTORC1 signaling, as measured by S6K1 phosphorylation and cap-dependent protein translation (Fig. 5 C and D). Interestingly, although overexpression of an activated S6K1 allele (ΔN/C S6K1) (26) was also able to rescue OS cell size similar to mTOR L1460P, consistent with the role of S6K1 in regulating cell size (27), overexpression of Rheb in OS cells did not increase cell size or restore mTORC1 signaling (Fig. S6). These observations provide further support to our model that mTORC1 activation must occur at the level of, or downstream from, mTOR to overcome the PP2A-mediated disruption of mTORC1 signaling in OS cells.
Fig. 5.
An activated mTOR allele rescues mTORC1 signaling. (A) U2OS cells were transfected with AU1-tagged WT or L1460P mTOR. Following MG132 treatment as indicated, AU1 immunoprecipitates and whole cell lysates were immunoblotted for mTOR and Raptor. (B) U2OS cells were transfected with WT or L1460P mTOR. Following MG132 treatment, AU1 immunoprecipitates were blotted for mTOR, Raptor, and PP2A-C. (C) WT (18/98) and OS (17/98) cells were transfected with GFP and mTOR L1460P as indicated. Cells were fixed, stained with propidium iodide, and FSC-H of the GFP+, G1 cells was determined by FACS. Histogram plots were generated using FlowJo version 8.8.6. (D) WT (CCD) or OS (696) cells were transfected with a bicistronic dual luciferase translation (Cap-dep/IRES) reporter. OS cells were additionally cotransfected with WT MID1 or mTOR L1460P. At 48 h posttransfection, cells were assayed for Renilla luciferase (cap-dep) and firefly luciferase (cap-ind/IRES) activity. Data shown are the average ratio (from quadruplicate samples) of cap-dependent to cap-independent luciferase activity ± SEM.
Discussion
Degradation by the ubiquitin-proteasome system is emerging as an important regulatory theme in mTORC1 signaling. A growing number of E3 ligases that target upstream components of mTORC1 signaling (e.g., PTEN, AKT, TSC2, REDD1), downstream effectors of mTORC1 signaling (e.g., S6K1), as well as mTOR itself have been identified (15, 16, 28–32). Here, we demonstrate that the MID1 E3 ligase affects mTORC1 signaling at the level of mTOR/Raptor complex formation. MID1 loss of function increases the levels of PP2A-C, which, as part of a Bα-containing PP2A holoenzyme, disrupts mTOR/Raptor association and consequently inhibits mTORC1 signaling.
Importantly, our studies uncover a unique role for PP2A in regulating mTORC1 directly at the level of complex formation. Previous studies in yeast have placed PP2A downstream of TOR, where active TOR restricts PP2A activity by promoting the association of Tap42 (α4 ortholog) with the catalytic subunits Sit4 or Pph21/22, which displaces both the A and B subunits and results in an inactive phosphatase complex toward downstream TOR effectors such as NPR1 and GLN3 (33–35). Under conditions where TOR is inactivated (e.g., nutrient starvation or rapamycin treatment), the Tip41 protein binds Tap42, allowing for SIT4 dissociation and restoration of phosphatase activity (33, 36). Similarly, in mammalian cells, small molecule inhibition of PP2A activity blocks rapamycin-induced dephosphorylation of the mTORC1 substrates 4E-BP1 and S6K1 (18–22), suggesting that PP2A directly dephosphorylates mTORC1 substrates when mTORC1 activity is inhibited by rapamycin, or that mTORC1 constrains PP2A activity. However, with the latter scenario, it is unclear whether the analogous regulatory paradigm with the Tip41L/α4/PP2A axis in mammals is conserved from yeast (37–39). Our data are consistent with a regulatory loop involving the MID1/α4/PP2A degradation pathway, where PP2A acts on mTOR itself as opposed to acting on downstream mTORC1 substrates. Furthermore, an additional distinction of this regulatory loop from the previously described role of PP2A on downstream mTORC1 substrates is that the PP2A holoenzyme responsible for mTORC1 disruption involves the PP2A-Bα (PR55α) subunit rather than the PP2A-B′γ (PR61γ/PP2R5C) subunit implicated in S6K1 dephosphorylation (40).
We also found that the association between PP2A-C and mTOR is mediated via the mTOR HEAT repeats, a motif found in Huntingtin, elongation factor 3, A subunit of PP2A, and TOR (41, 42). Furthermore, binding of PP2A-C to the activated mTOR mutant L1460P is reduced compared with wild-type mTOR, accounting for the ability of this mutant to rescue mTORC1 signaling in OS cells. As the mTOR L1460P mutation lies in a region between the HEAT repeats and the FAT domain (25), we postulate that this mutation alters the conformation of the HEAT repeat region such that PP2A-C is unable to bind efficiently. Interestingly, a naturally occurring mTOR splice variant (mTOR-β) with oncogenic activity has been described, which contains an internal deletion of the HEAT repeats and the majority of the FAT domain (43). Based on our findings, we hypothesize that the enhanced cell proliferation and oncogenesis with this splice variant may result from a lack of PP2A-mediated regulation.
Our data also suggest that mTORC1 formation is regulated by phosphorylation, because PP2A-mediated mTORC1 disruption is blocked by the PP2A inhibitor okadaic acid, and isolated mTORC1 can be disrupted by phosphatase treatment in vitro. However, despite the plethora of mTOR and Raptor phosphorylation sites that have been identified to date, none appear to regulate complex formation (44–48), and our own attempts at identifying phosphorylation sites critical for mTORC1 formation have not yielded conclusive results. Whether complex formation is due to phosphorylation sites on mTOR or Raptor that are as yet unidentified, or if a combination of phosphorylation sites on either molecule are involved, remains to be determined. A further complication is the multimerization of mTORC1 (49), which could entail the involvement of phosphorylation sites on both mTOR and Raptor that are responsible for complex integrity. Nonetheless, the precise dissection of phosphorylation in regulating mTORC1 formation and signaling bears additional study.
Although our studies primarily focused on the effect of MID1/PP2A on mTORC1 and its downstream effectors, it was a formal possibility that MID1/PP2A dysregulation of mTORC1 could influence additional pathways, particularly those subject to feedback inhibition by mTORC1, such as PI3K/AKT and Ras/ERK (50). Inhibition of mTORC1 signaling by rapamycin has been shown to lead to increased AKT activation due to the alleviation of a negative feedback loop involving S6K1 and IRS-1, although the magnitude of the feedback inhibition can vary between cell types (50, 51). We observed a varied effect of MG132 on AKT phosphorylation in cancer cell lines ranging from little to no effect in U2OS cells, to more pronounced up-regulation in HCT-116 and MCF-7 cells (Fig. S7). However, we did not observe AKT up-regulation in OS fibroblasts compared with WT counterparts, but rather a slight down-regulation of AKT phosphorylation, suggesting that alleviation of feedback inhibition does not occur in OS fibroblasts and/or is counteracted by the elevated levels of phosphatase activity due to MID1 loss of function. Therefore, the effect of mTORC1 dysregulation on upstream pathways may be an additional contributing factor to OS phenotypes.
Cells derived from Opitz syndrome patients show profound defects in mTORC1 signaling: reduced S6K1 phosphorylation, cell size, and cap-dependent translation, pointing to a critical role of the MID1/α4/PP2A axis in regulating mTORC1 function. Either restoration of MID1 function or mTORC1 activation that bypasses the PP2A-mediated regulation of mTOR/Raptor complex formation (i.e., mTOR mutation or downstream effectors such as S6K1) is sufficient to rescue the mTORC1 phenotypes in OS cells. Despite intensive study, the molecular pathways that link defective MID1 function to OS pathogenesis are incompletely understood. Our results presented here strongly suggest that the defect in mTORC1 signaling resulting from dysregulation of the MID1/PP2A axis is one of the avenues leading to OS pathogenesis. Of note, malformations occurring in OS patients have been attributed to defective migration patterns of neural crest cells needed to mediate ventral midline tissue formation in a very specific time window during development (1), and mTORC1 signaling not only is responsible for cytoskeleton dynamics and intracellular transport—both processes that significantly influence the migration potential of cells—but also has a direct regulatory function in migrating cells (52, 53). Dysfunctional mTORC1 signaling in OS patients with MID1 mutations is therefore likely to significantly contribute to the OS phenotype.
Methods
Antibodies and Reagents.
The α-mTOR antibody was generated by immunizing rabbits against a GST-mTOR fusion protein (amino acids 1,223–1,290). The α-AU1, α-HA (12CA5), and α-Myc (9E10) antibodies were purchased from Covance. The α-Raptor and α-Rictor antibodies were purchased from Bethyl. The α-PLCγ antibody has been previously described (15). The α-phospho-S6K1 (T389), α-phospho AKT (S473), α-AKT, and α-NBS1 antibodies were purchased from Cell Signaling Technology. The α-S6K1, α-TSC1, and α-TSC2 antibodies were purchased from Santa Cruz Biotechnology. The α-PP2A–A, –B, and –C antibodies were purchased from Millipore. The α-MID1 antibody was purchased from Abnova. The protein A agarose, anti-Flag agarose, insulin, and lactacystin were purchased from Sigma. MG132 was purchased from EMD Biosciences. Rapamycin was obtained from the National Cancer Institute, National Institutes of Health.
Cell Culture and Treatment.
The OS and control fibroblast cell lines have been described previously (3) and were maintained in DMEM (Irvine Scientific) supplemented with 10% FBS (HyClone). HEK 293, 293T, U2OS, and MCF-7 cells were maintained in DMEM supplemented with 10% FBS. TSC2+/+ and TSC2 −/− MEFs (generously provided by David Kwiatkowski, Harvard Medical School, Cambridge, MA) were maintained in DMEM supplemented with 10% FBS. HCT-116 cells were maintained in McCoy's 5A media supplemented with 10% FBS. For experiments involving serum starvation, cells were washed twice with PBS and then serum-starved for 16–18 h in DMEM supplemented with 0.1% FBS. For insulin stimulation, cells were incubated for 30 min with 100 nM insulin. For proteasome inhibitor treatment, cells were incubated with 10 μM MG132 or 10 μM lactacystin for 3 h before insulin stimulation. For okadaic acid treatment, cells were incubated with 100 nM okadaic acid for 1 h before incubation with proteasome inhibitor.
Immunoprecipitations, Immunoblotting, in Vitro Kinase Reactions, and Luciferase Assays.
Cells were lysed in lysis buffer [40 mM Hepes (pH 7.5), 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 0.3% CHAPS, 10 μg/mL aprotinin, 1 μg/mL pepstatin A, 10 μg/mL leupeptin, 2 mM phenylmethylsulfonyl fluoride, and 20 μM microcystin-LR]. Clarified extracts were immunoprecipitated with α-mTOR, α-Raptor, or α-AU1 antibodies followed by collection on protein A agarose or by immunoprecipitation with anti-Flag agarose. Immune complexes were washed three times with lysis buffer and resuspended in SDS/PAGE sample buffer. Samples were resolved by SDS/PAGE before immunoblotting as described (44). In vitro kinase reactions were carried out as described (54). For luciferase assays, cells were transfected with a bicistronic dual luciferase translation [cap-dependent/internal ribosome entry site (IRES)] reporter construct (provided by Ze'ev Ronai, Sanford-Burnham Medical Research Institute, La Jolla, CA). At 48 h posttransfection, cells were assayed for Renilla luciferase (cap-dependent) and firefly luciferase (cap-independent/IRES) activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
DNA Constructs, siRNA, and Transfections.
The AU1 mTOR WT construct has been described (44). The Flag-tagged mTOR 600–1,450 construct was generated by PCR amplification of the cDNA encompassing amino acids 600–1,450 with an N-terminal Flag epitope sequence followed by subcloning into the expression vector pcDNA3 (Invitrogen). The HA-tagged PP2A-C expression construct was generated by PCR amplification of the PP2A-C cDNA (Ultimate ORF collection; Invitrogen) with an N-terminal HA epitope sequence followed by subcloning into pcDNA3. The fidelity of both constructs was confirmed by sequencing. The AU1-mTOR L1460P construct was obtained from Fuyuhiko Tamanoi (University of California, Los Angeles, CA) via Addgene. The MID1 WT construct has been described (3). The pBB14-US9-GFP construct has been described (55). The PP2A-C (Cα, CCGAGUCCCAGGUCAAGAGUU; Cβ, UUGGUGUCAUGAUCGGAAUUU), PP2A-C mismatch (Cα mismatch, CCUAGUCGCAAGCCAUGAC; Cβ mismatch, UUGGUGUGAUGGUCGCAAU), and luciferase siRNAs were purchased from Dharmacon/Thermo Fisher. The predesigned MID1 (GGUCCUCUGCCAGUUUUGU, GCCACUCACCCGAAUAAGA, GGUUCUAAUUCCUGAAAUC) and α4 (GGCUAAAAUACAGAGAUAC, CGAAGUAGAAGUGGCGACU, GGUGUUCAAGGGCUUGGAC) siRNAs were purchased from Ambion. For the shRNA constructs, the following targeting sequences were subcloned into the pMKO.1-puro vector: PP2A–Aα, CCACCAAGCACATGCTACCCA; –Aβ, AATCCGTCAGCTCTCTCAGTC; PP2A–Bα, AACAGAGGTGATTACAGCAGC; –Bβ, AACACCTTCGTGTACAGCAGC, –Bγ, CATGGAGGCAAGACCCATAGA; –Bδ, AACATGGAGAGCAGGCCGGTG. Expression plasmids were transfected into cells using the calcium phosphate method or by Fugene 6 (Roche) according to the manufacturer's protocol. Cells were used in experiments 24–48 h posttransfection. For siRNA transfections, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer's protocol.
Determination of Cell Size.
To determine cell size, exponentially growing OS and control fibroblast cells were prepared and seeded under the same conditions and density. At 48 h postplating, cells were collected by trypsinization, fixed in 70% ethanol, stained with propidium iodide, and analyzed for size and cell cycle distribution by FACS (FACSort; BD Biosciences) with CellQuest software. Cell size was determined by forward scattering (FSC) of the G1-phase population using FlowJo version 8.8.6 software (Tree Star). Where indicated, the cells were harvested after treatment with either rapamycin (100 nM) or okadaic acid (10 nM) for 18 h. For MID1, AU1-mTOR-L1460P, and Rheb transfections, OS or control fibroblast cells were cotransfected with pBB14-US9-GFP at a density of 40% confluence. At 48 h posttransfection, cells were collected, and forward scatter of the GFP+, G1 phase population was analyzed by FACS.
Supplementary Material
Acknowledgments
We thank Drs. Wei Jiang, Robert Abraham, and Ze'ev Ronai for helpful discussions. This work was supported in part by National Institutes of Health Grants CA76193 and CA52995 (to G.G.C.) and American Cancer Society Grant RSG-11-069-01 (to G.G.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100131108/-/DCSupplemental.
References
- 1.Schweiger S, Schneider R. The MID1/PP2A complex: A key to the pathogenesis of Opitz BBB/G syndrome. Bioessays. 2003;25:356–366. doi: 10.1002/bies.10256. [DOI] [PubMed] [Google Scholar]
- 2.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger' E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 3.Trockenbacher A, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet. 2001;29:287–294. doi: 10.1038/ng762. [DOI] [PubMed] [Google Scholar]
- 4.Cainarca S, Messali S, Ballabio A, Meroni G. Functional characterization of the Opitz syndrome gene product (midin): Evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet. 1999;8:1387–1396. doi: 10.1093/hmg/8.8.1387. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 6.Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006;24:967–976. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Schweiger S, et al. The Opitz syndrome gene product, MID1, associates with microtubules. Proc Natl Acad Sci USA. 1999;96:2794–2799. doi: 10.1073/pnas.96.6.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Prickett TD, Elliott E, Meroni G, Brautigan DL. Phosphorylation and microtubule association of the Opitz syndrome protein mid-1 is regulated by protein phosphatase 2A via binding to the regulatory subunit alpha 4. Proc Natl Acad Sci USA. 2001;98:6650–6655. doi: 10.1073/pnas.111154698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aranda-Orgillés B, et al. The Opitz syndrome gene product MID1 assembles a microtubule-associated ribonucleoprotein complex. Hum Genet. 2008;123:163–176. doi: 10.1007/s00439-007-0456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang GG, Abraham RT. Targeting the mTOR signaling network in cancer. Trends Mol Med. 2007;13:433–442. doi: 10.1016/j.molmed.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapahi P, et al. With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nojima H, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 14.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 15.Katiyar S, et al. REDD1, an inhibitor of mTOR signalling, is regulated by the CUL4A-DDB1 ubiquitin ligase. EMBO Rep. 2009;10:866–872. doi: 10.1038/embor.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leicht M, Simm A, Bertsch G, Hoppe J. Okadaic acid induces cellular hypertrophy in AKR-2B fibroblasts: Involvement of the p70S6 kinase in the onset of protein and rRNA synthesis. Cell Growth Differ. 1996;7:1199–1209. [PubMed] [Google Scholar]
- 19.Lin TA, Lawrence JC., Jr Control of PHAS-I phosphorylation in 3T3-L1 adipocytes: Effects of inhibiting protein phosphatases and the p70S6K signalling pathway. Diabetologia. 1997;40(Suppl 2):S18–S24. doi: 10.1007/s001250051391. [DOI] [PubMed] [Google Scholar]
- 20.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 21.Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci USA. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause U, Bertrand L, Hue L. Control of p70 ribosomal protein S6 kinase and acetyl-CoA carboxylase by AMP-activated protein kinase and protein phosphatases in isolated hepatocytes. Eur J Biochem. 2002;269:3751–3759. doi: 10.1046/j.1432-1033.2002.03074.x. [DOI] [PubMed] [Google Scholar]
- 23.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urano J, et al. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci USA. 2007;104:3514–3519. doi: 10.1073/pnas.0608510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheatham L, Monfar M, Chou MM, Blenis J. Structural and functional analysis of pp70S6k. Proc Natl Acad Sci USA. 1995;92:11696–11700. doi: 10.1073/pnas.92.25.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suizu F, et al. The E3 ligase TTC3 facilitates ubiquitination and degradation of phosphorylated Akt. Dev Cell. 2009;17:800–810. doi: 10.1016/j.devcel.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Chong-Kopera H, et al. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem. 2006;281:8313–8316. doi: 10.1074/jbc.C500451200. [DOI] [PubMed] [Google Scholar]
- 31.Panasyuk G, Nemazanyy I, Filonenko V, Gout I. Ribosomal protein S6 kinase 1 interacts with and is ubiquitinated by ubiquitin ligase ROC1. Biochem Biophys Res Commun. 2008;369:339–343. doi: 10.1016/j.bbrc.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Mao JH, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 37.Smetana JH, Zanchin NI. Interaction analysis of the heterotrimer formed by the phosphatase 2A catalytic subunit, alpha4 and the mammalian ortholog of yeast Tip41 (TIPRL) FEBS J. 2007;274:5891–5904. doi: 10.1111/j.1742-4658.2007.06112.x. [DOI] [PubMed] [Google Scholar]
- 38.McConnell JL, Gomez RJ, McCorvey LR, Law BK, Wadzinski BE. Identification of a PP2A-interacting protein that functions as a negative regulator of phosphatase activity in the ATM/ATR signaling pathway. Oncogene. 2007;26:6021–6030. doi: 10.1038/sj.onc.1210406. [DOI] [PubMed] [Google Scholar]
- 39.Yoo SJ, et al. The alpha4-containing form of protein phosphatase 2A in liver and hepatic cells. J Cell Biochem. 2008;105:290–300. doi: 10.1002/jcb.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn K, et al. PP2A regulatory subunit PP2A-B′ counteracts S6K phosphorylation. Cell Metab. 2010;11:438–444. doi: 10.1016/j.cmet.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci USA. 2010;107:2467–2472. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: Insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panasyuk G, et al. mTORbeta splicing isoform promotes cell proliferation and tumorigenesis. J Biol Chem. 2009;284:30807–30814. doi: 10.1074/jbc.M109.056085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem. 2009;284:14693–14697. doi: 10.1074/jbc.C109.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster KG, et al. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J Biol Chem. 2010;285:80–94. doi: 10.1074/jbc.M109.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Acosta-Jaquez HA, et al. Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol Cell Biol. 2009;29:4308–4324. doi: 10.1128/MCB.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gwinn DM, Asara JM, Shaw RJ. Raptor is phosphorylated by cdc2 during mitosis. PLoS ONE. 2010;5:e9197. doi: 10.1371/journal.pone.0009197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carracedo A, Baselga J, Pandolfi PP. Deconstructing feedback-signaling networks to improve anticancer therapy with mTORC1 inhibitors. Cell Cycle. 2008;7:3805–3809. doi: 10.4161/cc.7.24.7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kladney RD, et al. Tuberous sclerosis complex 1: An epithelial tumor suppressor essential to prevent spontaneous prostate cancer in aged mice. Cancer Res. 2010;70:8937–8947. doi: 10.1158/0008-5472.CAN-10-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, et al. Rapamycin inhibits cytoskeleton reorganization and cell motility by suppressing RhoA expression and activity. J Biol Chem. 2010;285:38362–38373. doi: 10.1074/jbc.M110.141168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulhati P, et al. mTORC1 and mTORC2 regulate EMT, motility and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-10-4058. 10.1158/0008-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiang GG, Abraham RT. Determination of the catalytic activities of mTOR and other members of the phosphoinositide-3-kinase-related kinase family. Methods Mol Biol. 2004;281:125–141. doi: 10.1385/1-59259-811-0:125. [DOI] [PubMed] [Google Scholar]
- 55.Kalejta RF, Brideau AD, Banfield BW, Beavis AJ. An integral membrane green fluorescent protein marker, Us9-GFP, is quantitatively retained in cells during propidium iodide-based cell cycle analysis by flow cytometry. Exp Cell Res. 1999;248:322–328. doi: 10.1006/excr.1999.4427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.