Abstract
Phosphonates constitute a class of natural products that mimic the properties of the more common organophosphate ester metabolite, yet are not readily degraded owing to the direct linkage of the phosphorus atom to the carbon atom. Phosphonate hydrolases have evolved to allow bacteria to utilize environmental phosphonates as a source of carbon and phosphorus. The work reported in this paper examines one such enzyme, phosphonoacetate hydrolase. By using a bioinformatic approach we circumscribed the biological range of phosphonoacetate hydrolase to a select group of bacterial species from different classes of Proteobacteria. In addition, using gene context we identified a novel 2-aminoethylphosphonate degradation pathway in which phosphonoacetate hydrolase is a participant. The X-ray structure of phosphonoformate-bound phosphonoacetate hydrolase was determined to reveal that this enzyme is most closely related to nucleotide pyrophosphatase/diesterase, a promiscuous two-zinc ion metalloenzyme of the alkaline phosphatase enzyme superfamily. The X-ray structure and metal ion specificity tests showed that phosphonoacetate hydrolase is also a two-zinc ion metalloenzyme. By using site-directed mutagenesis and 32P-labeling strategies, the catalytic nucleophile was shown to be Thr64. A structure-guided, site-directed mutation based inquiry of the catalytic contributions of active site residues identified Lys126 and Lys128 as the most likely candidates for stabilization of the aci-carboxylate dianion leaving group. A catalytic mechanism is proposed which combines Lys12/Lys128 leaving group stabilization with zinc ion activation of the Thr64 nucleophile and the substrate phosphoryl group.
Keywords: Phosphonoacetate hydrolase, enzyme mechanism, alkaline phosphatase, phosphonate, divergent evolution, enzyme superfamily, phosphoryl transfer, nucleotide pyrophosphatase/diesterase, 2-aminoethylphosphonate, phosphonoacetaldehyde, phosphonoformate, nucleophilic catalysis, zinc ion
Environmental phosphonates are both synthetic and biogenic in origin (for recent reviews see references 1–4). Phosphonates differ from the more predominant organophosphates in that the phosphorus atom is bonded directly to a carbon atom. The unique properties of phosphonates derive from their C-P bond that, unlike the P-O-C linkage, is stable to acid hydrolysis, base hydrolysis, and to the action of enzymes that catalyze phosphoryl transfer from phosphate esters and anhydrides (5,6). Phosphonates are used commercially as detergent additives, agrochemicals, lubricant additives, antioxidants, adhesives, antiviral agents, antibiotics, and insecticides, and thus, they widely distributed in the environment (7–10).
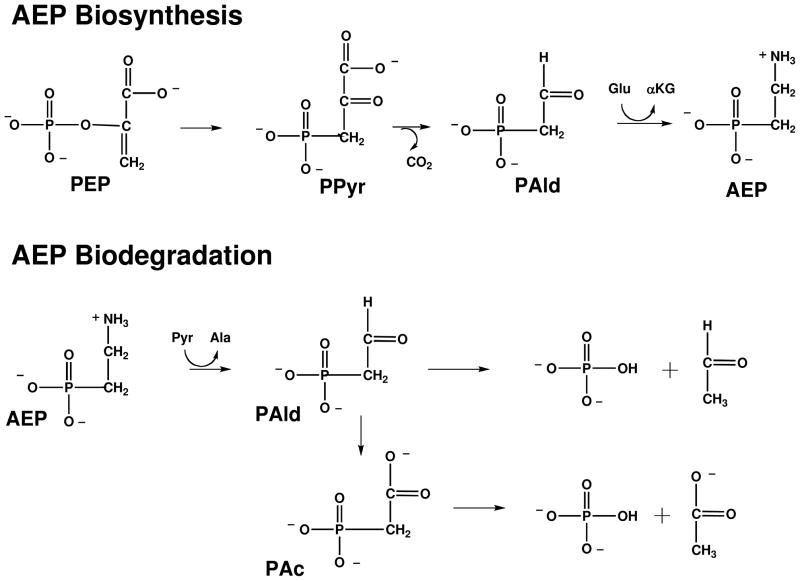
Biogenic phosphonates are produced by specialized microbes and invertebrates for niche adaptation, signaling, phosphorus storage or chemical warfare (7). 2-Aminoethylphosphonate (AEP)1, which is typically found conjugated to lipids or glycans, is the most ubiquitous of the phosphonate natural products (11). The P-C bond in AEP, as well as in many other biosynthetic phosphonates, is formed through the rearrangement of phosphoenol pyruvate (PEP) to phosphonopyruvate (PPyr) (Scheme 1) catalyzed by the enzyme PEP phosphomutase (12). This thermodynamically unfavorable reaction is driven forward by a coupling reaction. In the case of AEP biosynthesis this reaction is the decarboxylation of PPyr to form phosphoacetaldehyde (PAld), catalyzed by the enzyme phosphonopyruvate decarboxylase (13). The AEP is ultimately formed by AEP transaminase-catalyzed ammonia group transfer from L-glutamate to PAld (14).
Scheme 1.
The pathways associated with AEP biosynthesis and AEP biodegradation. Abbreviations used are PEP (phosphoenol pyruvate), PPyr (phosphonopyruvate), PAld (phosphonoacetaldehyde), Glu (L-glutamate), αKG (α-ketoglutarate), Pyr (pyruvate), Ala (L-alanine) and PAc (phosphonoacetate).
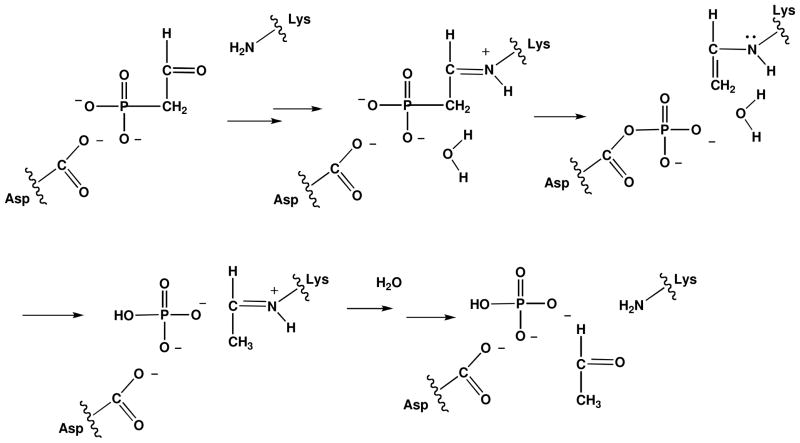
The nitrogen, carbon and phosphorus atoms of AEP are recycled in the environment by specialized bacteria, which employ the two-step chemical pathway shown in Scheme 1. PAld, the pathway intermediate, is formed from AEP by AEP transaminase-catalyzed ammonium group transfer to pyruvate (15). PAld undergoes hydrolytic cleavage at phosphorus thus liberating inorganic phosphate and acetaldehyde. The enzyme catalyst for this reaction, phosphonatase, combines covalent electrophilic catalysis (viz. Schiff base formation with the PAld carbonyl group), and nucleophilic catalysis (viz. formation of an aspartylphosphate intermediate) to mediate the transfer of the phosphoryl group to a water molecule (16–18) (Scheme 2). Phosphonatase is a member of the haloalkonic acid dehalogenase (HAD) enzyme superfamily, which is comprised primarily of phosphohydrolases that catalyze the hydrolysis of phosphate monoesters and anhydrides via an aspartylphosphate intermediate (19). By virtue of its ability to catalyze the hydrolytic cleavage of the PAld P-C bond, phosphonatase adds a novel chemical function to the HAD superfamily.
Scheme 2.
The mechanism for phosphonatase catalyzed hydrolysis of PAld (16–18).
The work that we report in this paper examines the chemical specialization of the P-C bond cleaving enzyme phosphonoacetate hydrolase (PAc hydrolase) (20–21), which is a member of the alkaline phosphatase enzyme superfamily (22). PAc hydrolase was first discovered in a bacterial isolate (Pseudomonas fluorescens 23F) from sludge produced by a laundry waste treatment plant located near Dunmurry, Northern Ireland (23–24). At the time it seemed that PAc hydrolase might have evolved to convert commercial PAc, introduced to the environment through waste disposal, to usable forms of carbon (acetate/glyoxylate cycle) and phosphorus (orthophosphate). However, later studies revealed the presence of the PAc hydrolase gene in bacterial isolates from remote and environmentally diverse geographical sites, which suggested that PAc might be synthesized in specialized bacteria (25–26). Functional analysis of the genes neighboring the P. fluorescens 23F PAc hydrolase gene did not, however, identify protein products that might produce PAc (27). Nonetheless, with the recent explosion of sequenced bacterial genomes we succeeded in locating orthologs of the P. fluorescens 23F PAc gene (by bioinformatic analysis) to gene clusters (in several species of Proteobacteria), which appear to support a novel 3-step chemical pathway for AEP degradation (Scheme 1). This pathway shares with the phosphonatase-centered pathway the transamination of AEP to form PAld (28), but deviates with the oxidation of the PAld to form phosphonoacetate (PAc) (Scheme 1). The hydrolytic cleavage of the P-C bond in PAc forms acetate and orthophosphate as the pathway end products rather than the acetaldehyde and orthophosphate produced by the hydrolytic cleavage of PAld.
In the text below, we examine the biological range and biochemical function of PAc hydrolase. Moreover, we report the results from an in depth structure-function analysis, which show that PAc hydrolase combines the catalytic strategies of the phosphohydrolases of the alkaline phosphatase superfamily, with a unique mechanism of substrate leaving group stabilization, to create a novel chemical function that supports a newly discovered AEP degradation pathway.
Materials and Methods
Materials
All biological buffers, enzymes, and chemicals used in the enzyme assays were purchased from Sigma. The [γ-32P]ATP was purchased from Amersham. All substrates and inhibitors were purchased from Sigma with the exception on phosphonopyruvate and phosphonoacetaldehyde, which were synthesized according to published procedure (vide infra).
Preparation of wild-type and mutant PAc hydrolase
The P. fluorescens 23F PAc hydrolase gene (29) was subcloned into pKK223-3 (Pharmacia) using a PCR based approach. Transformed Escherichia coli JM105 cells were grown in LB media at 37 °C for 16 h (OD600 = 1). Following the addition of IPTG (1 mM) the cells were incubated until the culture OD600 reached 1.8 (< 6 h) and then they were harvested by centrifugation (18000 g) (yield of 5 g/L). Forty grams of wet cells were suspended in 400 mL of ice-cold Buffer A (30 mM K+BICINE and 1 mM ZnCl2; pH 8.5) and then passed through a French press at 1000 psi. The lysate was clarified by centrifugation (18000 g for 30 min) at 4 °C prior to adding ammonium sulfate to 25% saturation. The resulting precipitate was removed by centrifugation before adding additional ammonium sulfate to 55%. The precipitated protein was harvested and then dissolved in 200 mL of Buffer A and dialyzed overnight at 4 °C against 2 L of Buffer A. The protein solution was chromatographed at 4 °C on a 5 × 45 cm DEAE-cellulose column pre-equilibrated with Buffer A. The column was eluted with 100 mL of Buffer A followed by a 2 L linear gradient of KCl (0 M - 0.5 M) in Buffer A. The PAc hydrolase containing fractions (eluting at 0.25 KCl) were identified using a spectrophotometric activity assay (vide infra) and/or SDS-PAGE analysis. The desired fractions were pooled, combined with ammonium sulfate (20% saturation) and then loaded onto a 2.5 × 30 cm Phenyl Sepharose column pre-equilibrated with Buffer A/ammonium sulfate (20% saturation). The column was eluted with a 2 L linear gradient 10 - 0% ammonium sulfate in Buffer A. The PAc hydrolase containing fractions (eluted at ca. 0% ammonium sulfate) were pooled and then concentrated using an Amicon ultrafiltration apparatus to a concentration of ~20 mg/mL. The concentrated enzyme was dialyzed against Buffer A overnight at 4 °C and then loaded onto a 2 cm × 20 cm Hydroxylapatite column pre-equilibrated with Buffer A. The column was washed with 10 mL of Buffer A, followed by 400 mL linear gradient 0 – 0.1 M phosphate in Buffer A. The desired fractions (eluted at ca. 0.02 M phosphate) were pooled and concentrated with an Amicon until the enzyme concentration reached ~30 mg/mL. The concentrated enzyme solution (1 mL) was then chromatographed at 4 °C on a 1.5 × 180 cm Sephracryl S-200 column using Buffer A as eluant. The desired fractions were combined, concentrated to ~20 mg/mL and passed through a 0.2 μm syringe filter before storing at 4 °C. The site-directed mutant genes were prepared from the wild-type gene by PCR using commercial primers and pfu polymerase (Stratagene). The purified DNA was ligated to the linearized pKK223-3 plasmid using T4 DNA ligase. The ligation product was used to transform competent E. coli JM105 cells. A positive clone isolated from the LB agar plate (100 μg/ml ampicillin) was verified by DNA sequencing. The mutant enzymes were prepared as described for wild-type PAc hydrolase.
PAc hydrolase MW determination
The N-terminal sequence was determined by automated peptide sequencing as TQLISVNSRSYRLS thus showing that the N-terminal Met was removed by posttranslational modification. The theoretical subunit mass of the recombinant hydrolase was calculated using the EXPASY Compute pI/MW program. The size of native hydrolase was determined by Sephacryl S-200 column chromatography using protein molecular weight standards to create a plot of log MW vs. elution volume.
Spectrophotometric activity assays
For all assays, control reactions in which the PAc hydrolase was omitted were carried out. PAc hydrolysis. These reactions were monitored at 25 or 30 °C by measuring the oxidation of β-NADH at 340 nm (ε = 6.22 mM−1cm−1) in 1 mL assay solutions initially containing PAc hydrolase, PAc, 10 u acetate kinase, 10 u pyruvate kinase, 10 u lactate dehydrogenase, 5 mM ZnCl2, 10 mM MgCl2, 1 mM PEP, 1 mM ATP, and 0.5 mM β-NADH in 50 mM K+ADA pH 7.0. PAld hydrolysis. This reaction was monitored at 25 °C by measuring the oxidation of β-NADH at 340 nm (ε = 6.22 mM−1cm−1) in a 1 mL assay solutions initially containing PAc hydrolase, PAld (30), 10 u alcohol dehydrogenase, 0.5 mM β-NADH, 5 mM ZnCl2, and 5 mM MgCl2 in 50 mM K+ADA (pH 7.0). Bis-p-nitrophenylphosphate, p-nitrophenylphosphate and p-nitrophenylsulfate hydrolysis. These reactions were monitored at 25 °C by measuring the formation of p-nitrophenolate at 410 nm (ε = 16200 M−1cm−1) in 1 mL assay solutions containing reactant and 1 mM ZnCl2 in 100 mM K+TRICINE (pH 8.0). ATP hydrolysis. This reaction was monitored at 25 °C by measuring the oxidation of NADH at 340 nm (ε = 6.22 mM−1cm−1) in 1 mL assay solutions initially containing PAc hydrolase, 1 mM ATP, 5 mM ZnCl2, 5 mM MgCl2,1 mM PEP, 10 u pyruvate kinase, 10 u lactate dehydrogenase, and 0.5 mM NADH in 50 mM K+ADA (pH 7.0). 2-Phosphoglycerate, 3-phosphoglycerol, and fosfomycin hydrolysis. The reaction solutions initially contained 40 μM PAc hydrolase, 2 mM reactant and 5 mM ZnCl2 in 50 mM K+ADA (25 °C, pH 7.0). The concentration of orthophosphate was measured by adding an aliquot of the solution to the Fiske-Subbarow assay solution (5 M H2SO4, 2.5 % V/W (NH4)2 Molybdate and 0.25 % V/W Fiske-Subbarow Reducer (Sigma)), then measuring the solution OD600 following 30 min of incubation. The orthophosphate concentration was calculated from a standard curve prepared with commercial KH2PO4.
Determination of PAc hydrolase metal ion activation
PAc hydrolase was exhaustively dialyzed at 4 °C against 30 mM K+BICINE (pH 8.0) prepared using deionized H2O (VWR Scientific Products). Reaction solutions initially containing 1 mM PAc, dialyzed PAc hydrolase and 3 mM divalent metal ion in 30 mM K+BICINE (pH 8.0) were incubated at 25 °C for a specified period and then assayed for acetate (using 10 u/ml acetate kinase, 10 u/ml pyruvate kinase, 10 u/ml lactate dehydrogenase, 5 mM ZnCl2, 10 mM MgCl2, 1 mM PEP, 1 mM ATP, and 0.5 mM β-NADH).
Steady-State Kinetic Constant Determinations
The Km and kcat values were determined from initial velocity data measured as a function of substrate concentration (0.5 – 10Km). The initial velocity data were fitted to equation 1. The kcat value was calculated by dividing the Vm value by the concentration of PAc hydrolase determined using the Bradford assay kit (Sigma).
| Eq. 1 |
where v is initial velocity, Vm is the maximum velocity, [S] is the substrate concentration, and Km is the Michaelis constant.
The inhibition constant (Ki) values were determined from the initial velocity data measured as a function of substrate concentration (0.5 – 10Km) and inhibitor concentration ([I] at 0, 1Ki, 2Ki). Data were fitted to equation 2 for competitive inhibition.
| Eq. 2 |
pH Rate Profile Analysis
The Vm and Vm/Km values were determined from initial velocity data measured as a function of substrate concentration (0.5 – 10Km) at solution pH values that ranged from 6.0 to 9.0. The reaction solutions were buffered using 50 mM ADA or 50 mM BICINE. The Vm/Km values were fitted to equation 3 to define the apparent ionization constants K1 and K2 and the Vm values were fitted to equation 4.
| Eq. 3 |
| Eq. 4 |
Where Y is the Vm/Km or Vm value, c is the pH-independent value of Y, [H] is the hydrogen ion concentration.
32P-Labeling of phosphonoacetate hydrolase with [γ32P]ATP
The purified PAc hydrolase was labeled using [γ-32P]ATP. The 400 μL reaction mixture initially contained 24 μM PAc hydrolase, 1 mM ZnCl2, 50 mM K+MES (pH 6.0) and 110 μM [γ32P]ATP. A 25 μL aliquot was removed at 5 s, 30 s, 1 min, 10 min, and 20 min and mixed with 25 μL of 1 M perchloric acid and stored on ice. For the control reaction, the perchloric acid was added to the enzyme followed by the addition of the [γ32P]ATP. The quenched samples were centrifuged at 10000 g for 10 min at 4 °C using a microcentrifuge. Each sample was washed with 50 μL 0.5 M K+MES (pH 6.0) twice without disturbing the protein pellet. The protein pellet was dissolved in 25 μL SDS loading buffer and chromatographed on a SDS-PAGE gel. The gel was washed four times with a 250 mL washing solution (1: 2 glacial acetic acid/ethanol), then placed on the filter paper and covered with a gel drying cellulose membrane. The dried gel was placed on the X-ray film (Kodak) with intensifier (Dupont) and stored at −80 °C for 14 h. The film was developed and the SDS-PAGE gel was then stained with Coomassie blue. The labeling reaction was also carried out with the T64A PAc hydrolase mutant and with the wild-type enzyme inhibited with 2 mM phosphonoformate.
PAc hydrolase crystallization and structure determination
Crystals of phosphonoacetate hydrolase in Buffer A were grown from a solution containing 10% poly(ethylene glycol) 8000, 100 mM Mg-acetate, and 5 mM phosphonoformate buffered at pH 7.0 with 100 mM MOPSO. Batch setups with macroseeding produced crystals in approximately two weeks with typical dimensions of 0.5 × 0.4 × 0.2 mm. The crystals belonged to the space group P21 with unit cell dimensions of a = 57.5 Å, b = 129.5 Å, and c = 133.4 Å, β = 96.9°. There were four molecules in the asymmetric unit corresponding to a crystalline solvent content of 56%. X-ray data from a single native crystal were collected to 2.8 Å resolution at 4 °C with a Bruker HiStar area detector system. The X-ray source was nickel-filtered CuKα radiation from a Rigaku RU200 X-ray generator operated at 50 kV and 90 mA and equipped with a 300 μm focal cup. The x-ray data were processed with the software package SAINT (Bruker AXS, Inc.) and internally scaled with XCALIBRE (31). The structure of phosphonoacetate hydrolase was solved by multiple isomorphous replacement with two heavy-atom derivatives: trimethyllead acetate and potassium hexachloroplatinate (II). The heavy atom derivative data sets, including Friedel pairs, were collected and processed to 2.8 Å resolution in a similar manner as described for the native X-ray data. The locations of the heavy atom binding positions were determined by inspection of difference Patterson maps calculated to 5.0 Å resolution and placed on a common origin by appropriate difference Fourier maps. Initial phases were subsequently calculated with the program SOLVE (32) and improved by solvent flattening with DM (33). The relationships between the four subunits in the asymmetric unit were determined with the program MUNCHKINS (34). Four-fold averaging with the program DM was subsequently utilized and produced a readily interpretable electron density map. After the model for phosphonoacetate hydrolase was constructed, crystals belonging to the monoclinic space group P21 were grown from 8% poly(ethylene) glycol 8000, 100 mM MgCl2, 250 mM tartrate, and 7 mM phosphonoformate buffered at pH 7.0 with 100 mM MOPSO. These crystals displayed unit cell dimensions of a = 57.8 Å, b = 129.3 Å, and c = 133.5 Å and diffracted to a nominal resolution of 2.1 Å. Again, a native X-ray data set was collected in a similar manner to that previously described for the orthorhombic crystal form. The structure was solved via molecular replacement with the software package, AMORE (35). The model was improved with alternating cycles of least squares refinement with the program TNT (36) and manual inspection with the modeling package, Turbo (37). Relevant X-ray data collection and least squares refinement statistics are presented in Table 1.
Table 1.
The X-ray data collection and refinement statistics for the structure determination of the PAc hydrolase bound with phosphonoformate and two Zn2+ ions.
| X-ray Data Collection Statistics | |||||
|---|---|---|---|---|---|
| Data set | Resolution (Å) | Independent Reflections | Completeness (%) | Ave I/Avg σ (I) | Rsyma,b |
| Native (I) | 30.0 - 2.8 | 44,619 | 88.0 | 10.7 | 5.2 (14.3) |
| Native (II) | 30.0 - 2.1 | 104,683 | 91.0 (81.0) | 13.7 | 5.3 (21.2) |
| (CH3)3PbCOOCH3 | 30.0 - 2.8 | 47,546 | 93.0 | 8.8 | 6.3 (15.8) |
| K2PtCl6 | 30.0 - 2.8 | 43,673 | 86.0 | 6.6 | 6.9 (21.3) |
| Refinement Statistics for Native Data Set | |||||
| Resolution limits (Å) | cR-factor (overall) %/# rflns | cR-factor (working) %/# rflns | cR-factor (free) %/# rflns | No. Protein Atoms | d No. Heteroatoms |
| 30.0 – 2.1 (2.1–2.2) | 19.0/127171 | 18.5/120751 | 23.7/6420 | 12222 | 537 |
| Ave B value (Å2) | Protein 33.3 | Zn2+ 22.5 | Ligand 44.4 | Solvent 34.5 | --------- |
| Weighted Root-Mean-Square Deviations from Ideality | |||||
| Bond Lengths (Å) | Bond Angles (Å) | Trigonal planes (Å) | General planes (Å) | Torsional angles (deg) | |
| 0.011 | 1.971 | 0.005 | 0.008 | 17.96 | |
Rsym = (Σ| I − Ī|/Σ I) × 100.
Statistics for the highest resolution bin
R-factor = (Σ|Fo − Fc|/Σ|Fo|) × 100 where Fo is the observed structure factor amplitude and Fc is the calculated structure factor amplitude.
These include 504 water molecules, three phosphonoformate molecules, one tartrate molecule and two zinc ions.
Results and Discussion
PAc hydrolase is a member of the alkaline phosphatase enzyme superfamily (22). This functionally diverse superfamily is comprised of phosphohydrolases, phosphotransferases, phosphomutases, and sulfatases (38–43). The family members share an α/β/α core domain and the use of metal ion catalysis and nucleophilic catalysis. PAc hydrolase is most closely related to family members nucleotide pyrophosphatase/phosphodiesterase (NPP) and alkaline phosphatase (AP). It is with these two members that PAc hydrolase will be compared in the text that follows.
X-ray Structure of the PAc Hydrolase-Phosphonoformate Complex
Quaternary and Tertiary Structure
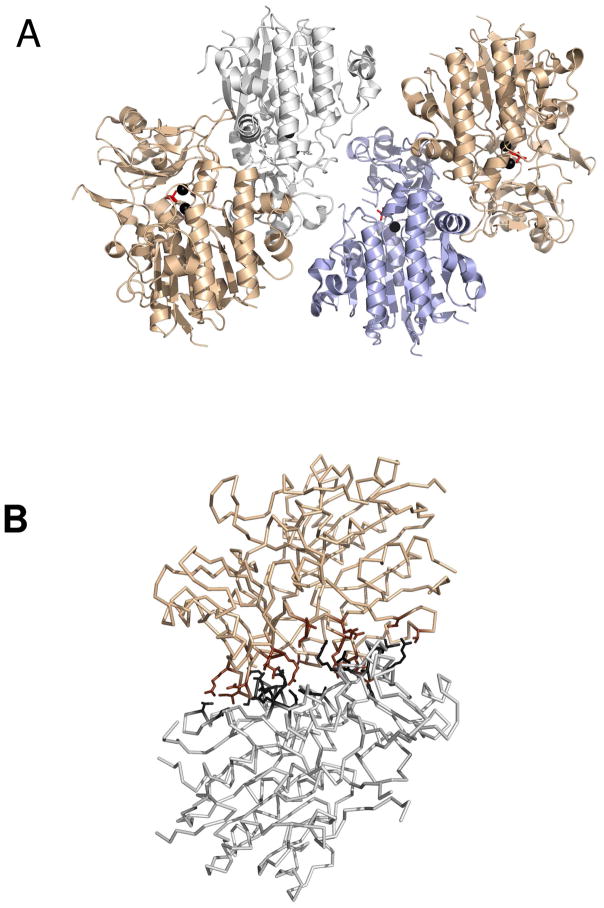
Recombinant P. fluorescens 23F PAc hydrolase was crystallized at neutral pH in the presence of Zn2+, Mg2+, phosphonoformate and tartrate. The crystals belonged to the space group P21 with unit cell dimensions of a = 57.5 Å, b = 129.5 Å, and c = 133.4 Å, β = 96.9°. The structure was solved, by multiple isomorphous replacement, at 2.1 Å resolution. The X-ray data collection and refinement statistics are listed in Table 1. Four subunits are observed in the asymmetric unit (Figure 1A). Two zinc ions are bound to each subunit. Three of the subunits are each bound with a phosphonoformate ligand and the fourth with a tartrate ligand. The theoretical monomer mass (−Met) is 44,239 Da and the mass of the native enzyme determined by gel filtration chromatography is ~93 kDa. Therefore, the predominant form of PAc hydrolase in solution is the homodimer. Inspection of the subunit-subunit contacts in the crystal structure suggests that residues Arg253, Asp249, Ile251, Thr248, Leu346, Pro346, Arg345, Ala209, Pro210, Glu28, Lys375, Glu30, Gln34, Asn33, Gln37, Ile36, Val53 and Thr48 form the dimerization surfaces (Figure 1B).
Figure 1.
(A). The four subunits observed in the asymmetric unit of the crystal of the PAc hydrolase(phosphonoformate complex). The zinc ions are shown as black spheres and the phosphonoformate ligand in red stick. (B). The PAc biological dimer with residues that comprise the subunit-subunit interface shown in black or brown colored stick.
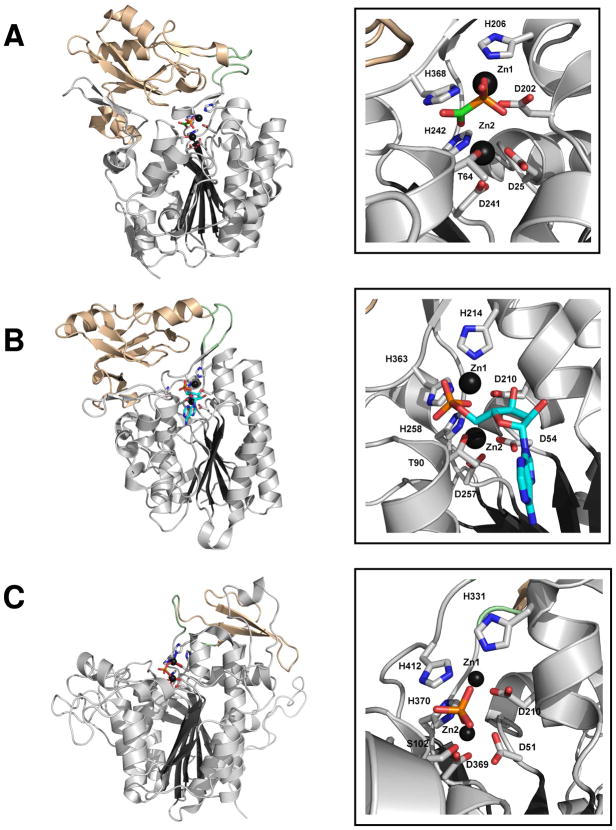
The PAc hydrolase monomer is comprised of a large α/β/α core domain (residues 1–245 and 367–388; 6-stranded mixed β-sheet) and a smaller α/β cap domain (residues 255–360; 4-stranded antiparallel β-sheet) (Figure 2A). The phosphonoformate ligand and two zinc ions are bound at the core domain (henceforth referred to as the catalytic domain) and are partially shielded from solvent by the cap domain. The locations of the respective active sites in the two superfamily members E. coli AP (PBD code 1ALK) (44) (Figure 2B) and Xanthomonas axonopodis NPP (PDB code 2GSU) (45) (Figure 2C) are conserved. However, an interesting divergence in structure is observed for AP, which unlike the PAc hydrolase and NPP does not possess a cap domain to cover the active site of the catalytic domain.
Figure 2.
The backbone fold (left panel) and zinc ion-binding site (right panel) of (A.) PAc hydrolase bound with phosphonoformate, (B) NPP bound with AMP and (C) AP bound with orthophosphate. The catalytic domain is colored gray, the cap domain wheat and the linkers pale green. The oxygen atoms are colored red, the nitrogen atoms blue, the phosphorus atoms orange and the carbon atoms green or cyan. The zinc ions are shown as black spheres.
Active Site
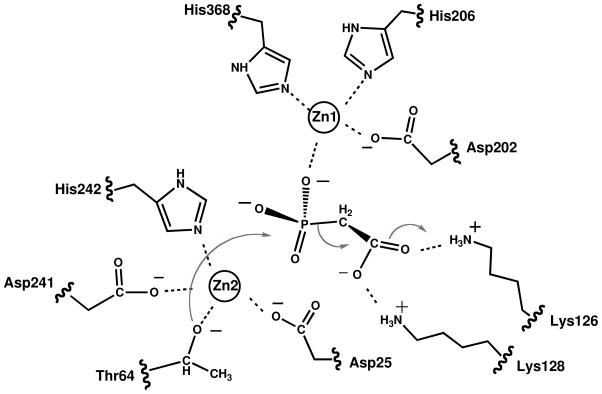
The PAc hydrolase active site contains two zinc ions, one of which (“Zn1”) is coordinated to His206 (2.2 Å), His368 (2.2 Å), Asp202 (1.9 Å) and to one oxygen atom (1.8 Å) of the phosphonoformate ligand phosphoryl group (Figure 2A). The second zinc ion (“Zn2”) is coordinated to His242 (1.8 Å), Asp25 (1.8 Å), Asp241 (2.0 Å) and to Thr64 (1.6 Å). X. axonopodis NPP (45) and E. coli AP (44) coordinate their zinc ions using the same constellation of His, Asp and Thr (Ser for AP) ligands (Figures 2B and 2C). The two zinc ions of AP (4.0 Å apart) are located closer together than are the two zinc ions of NPP (4.3 Å apart) and PAc hydrolase (4.6 Å apart). Consequently, the phosphate molecule bound to AP coordinates to both zinc ions whereas the phosphoryl group of the AMP molecule bound to NPP coordinates to only one zinc ion (Zn1), although the coordination to both zinc ions in the transition state has been proposed (45).
The substrate PAc was manually modeled in place of the phosphonoformate inhibitor in the PAc hydrolase active site. When the two phosphoryl groups are superimposed and the acetate group is overlayed with the phosphonoformate carboxylate group, the potential for steric clash with neighboring residues is evident. In the crystal structure, the phosphonoformate carboxylate group is positioned (at 2.7 Å) to engage in favorable electrostatic interaction with one of the two zinc ions (Zn2). We suspect that this interaction might direct the observed binding orientation of the carboxylate group. The model shown in Figure 3 retains the position of the phosphoryl group but directs the acetate group towards solvent. The orientation is consistent with the direction of the adenosine unit of the NPP-bound AMP ligand (45) and with the acetate group of the PAc ligand bound to AP (PDB code 1EW8) (46). The validity of this model was examined by carrying out the site-directed mutagenesis studies described in a later section. First however, we report the kinetic properties of the wild-type enzyme (vide infra).
Figure 3.
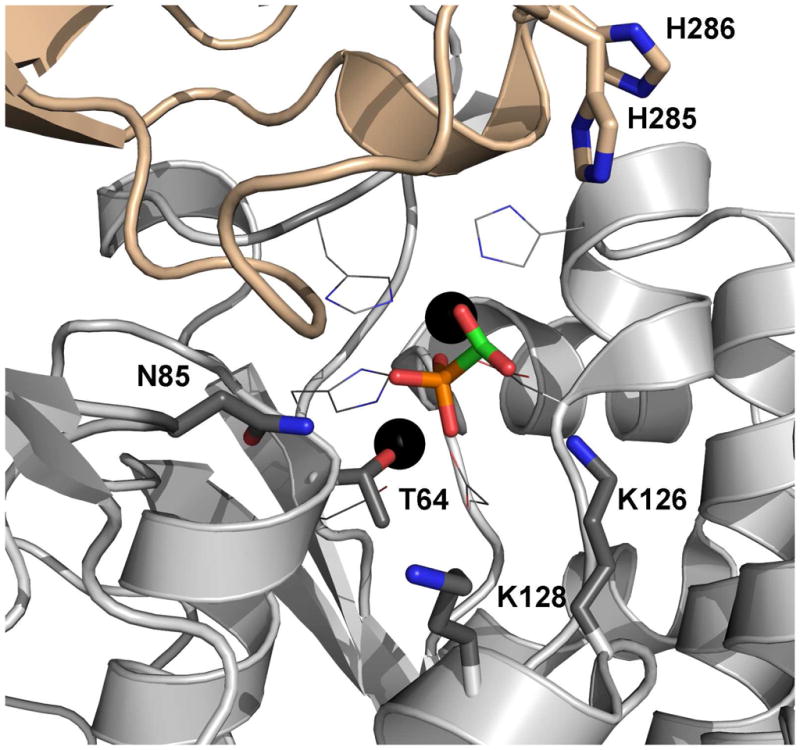
The model of PAc bound to the PAc hydolase active site. The oxygen atoms are colored red, the nitrogen atoms blue, the phosphorus atoms orange and the carbon atoms green. The zinc ions are shown as black spheres.
PAc Hydrolase pH and Metal Ion Dependence
pH Dependence
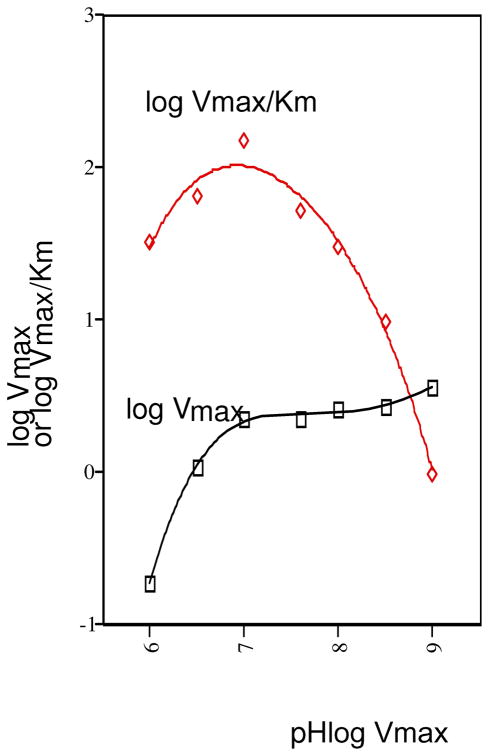
The Vm and Vm/Km pH rate profiles for PAc hydrolase-catalyzed PAc hydrolysis were measured in order to determine the optimal pH range for catalysis (Figure 4). The bell-shaped Vm/Km pH rate profile defines a narrow optimal pH range centered at pH 7. Activity assays were therefore carried out at pH 7 and only when required by the coupling assay employed, was a higher pH (pH 8) used. The pH rate profiles were fitted to equation 3 or 4 to define apparent pKa values (see figure legend) for the ionization of essential groups. Owing to what we view to be the complexities associated with the interpretation of pH rate profiles, we have not attempted to assign the apparent pKa values to PAc hydrolase catalytic groups.
Figure 4.
The Vm (●) and Vm/Km (○) pH rate profiles measured for PAc hydrolase catalyzed hydrolysis of PAc at 25 °C. The Vm/Km data were fitted to equation 3 to define an apparent pK1 and pK2 of 7 ± 2. The Vm pH rate data were fitted to equation 4 to define an apparent pKa = 6.4 ± 0.1.
Metal Ion Dependence
PAc hydrolase that had been exhaustively dialyzed against metal-free buffer was used to test the effect of a variety of divalent metal ions (Zn2+, Mg2+, Ca2+, Ni2+, Mn2+, Cr2+, Cu2+, Co2+) on the initial velocity of catalyzed hydrolysis of 1 mM PAc. At a concentration of 3 mM, Zn2+ and Co2+ produced a respective 10-fold and 2-fold increase in the initial velocity measured in the absence of added metal ion. The other metal ions tested did not enhance the initial velocity. An equal mixture of Zn2+ and Mg2+ (1.5 mM each) was no more effective than Zn2+ alone.2 PAc hydrolase kinetic experiments were therefore carried out using Zn2+ as the metal ion activator..
PAc Hydrolase Substrate Specificity
The substrate specificity constant (kcat/Km = 4 × 104 M−1 s−1 at pH 7 and 30 °C; Table 2) for PAc hydrolase-catalyzed PAc hydrolysis falls within the range of substrate specificity constants measured for other enzymes, which (like PAc hydrolase) function in secondary metabolism (viz. 1 × 103 to 1 × 105 M−1 s−1).
Table 2.
Steady-state kinetic constants for PAc hydrolase-catalyzed hydrolysis of PAc and various analogs at 25 °C and pH 7.0 (except where noted). See Materials and Methods for details.
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| phosphonoacetatea | 0.27 ± 0.1 | 0.060 ± 0.009 | 5 × 103 |
| phosphonoacetatea (30 °C) | 0.67 ± 0.1 | 0.015 ± 0.001 | 4 × 104 |
| p-nitrophenylphosphateb | 0.22 ± 0.1 | 14 ± 0.2 | 2 × 101 |
| 2-phosphoglyceratea,c | 4 × 10−5 | ------ | ------ |
| 3-phosphoglycerola,c | 4 × 10−5 | ------ | ------ |
| p-nitrophenylsulfateb | ~2 × 10−7 | ~ 10 | ~2 × 10−4 |
| ATPa | 2 ×10−4 | 1 | 2 × 10−1 |
| phosphonopyruvatea | NAd | ------ | ------ |
| phosphonoacetaldehydea | 0.027 ± 0.01 | 0.33 ± 0.01 | 8 × 101 |
| fosfomycina,c | NAd | ------ | ------ |
| bis-p-nitrophenylphosphateb,c | NAd | ------ | ------ |
The reaction buffer was 50 mM K+ADA (pH 7.0).
The reaction buffer was 50 mM K+TRICINE (pH 8.0).
Tested at a concentration of 2 mM. For 2-phosphoglycerate and 3-phosphoglycerol the ratio of the initial velocity and the enzyme concentration is used to approximates the kcat value.
No activity (NA) was observed within the detection limit of ~ 1 × 10−7 s−1.
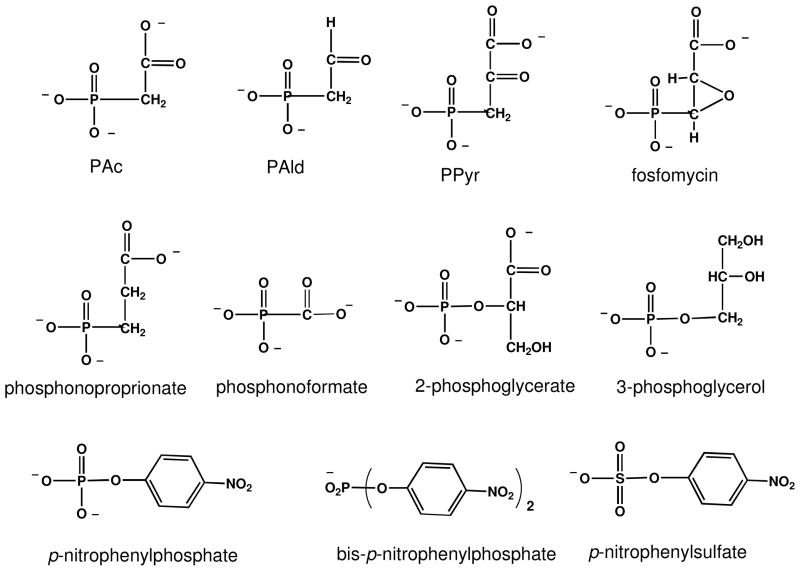
The structural determinants of PAc hydrolase substrate recognition were examined using structural analogs of PAc (structures are shown in Chart 1). The kcat and kcat/Km values measured for catalyzed PAld hydrolysis are 10-fold and 100-fold smaller, respectively than the corresponding values measured for PAc hydrolysis (Table 2). Thus, the charged oxygen atom of the PAc carboxylate group is an important, yet nonessential, structural feature for substrate recognition. Heterolytic cleavage of the P-C bond in PAld results in the formation of an enolate anion, in contrast to the aci-carboxylate dianion formed from PAc. P-C bond cleavage in PPyr would form a carboxy-enolate intermediate as would P-C bond cleavage in fosfomycin, if coupled with epoxide ring-opening. Neither fosfomycin nor PPyr are substrates for PAc hydrolase (detection limit for turnover is ~ 1 × 10−7 s−1). PPyr and phosphonoproprionate proved to be PAc hydrolase competitive inhibitors (vs PAc at pH 7 and 30 °C), both with a Ki = 1.2 mM (Table 3). Phosphonoformate was shown to be a considerably stronger competitive inhibitor (Ki = 0.19 mM at 30 °C and 0.049 mM at 25 °C) (Table 3). This tighter binding might however, be attributable to the small size of the phosphonoformate, which as pointed out earlier allows it to bind with its carboxylate group favorably interacting with a zinc ion (Zn2) (Figure 2A).
Chart 1.
The structures of the compounds tested as substrate or inhibitor.
Table 3.
Steady-state competitive inhibition constants measured for PAc analogs as inhibitors of PAc hydrolase catalyzed hydrolysis of PAc at pH 7.0 and 30 °C (except where noted). See Materials and Methods for details.
| Inhibitor | Ki (mM) | Type of Inhibition |
|---|---|---|
| phosphonoformate (25 oC) | 0.049 ± 0.008 | competitive |
| phosphonoformate | 0.19 ± 0.06 | competitive |
| phosphonoproprionate | 1.2 ± 0.8 | competitive |
| phosphonopyruvate | 1.2 ± 0.8 | competitive |
Next, PAc hydrolase was tested for hydrolytic activity towards substrates that are hydrolyzed by other members of the alkaline phosphatase superfamily, namely AP, NPP and arylsulfatase. PAc hydrolase catalyzed the hydrolysis of 2-phosphoglycerate and 3-phosphoglycerol, but at a very slow rate (kcat ~ 4 × 10−5 s−1).3 The turnover rate measured for these two substrates is four-orders of magnitude lower than that measured for the native substrate, PAc (Table 2). For comparison, the kcat/Km value reported E. coli AP catalyzed hydrolysis of ethylphosphate is 1 × 105 M−1 s−1 (47).
p-Nitrophenylphosphate constitutes an easier target for PAc hydrolase-catalyzed hydrolysis because the p-nitrophenolate anion is a good leaving group. Indeed, the kcat value measured for p-nitrophenylphosphate is similar to the kcat value measured for PAc (Table 2), however the Km value (17 mM) is quite large. For contrast, we note that the kcat/Km value reported for AP catalyzed p-nitrophenylphosphate hydrolysis (3 × 107 M−1 s−1) (47) is six orders of magnitude larger than the PAc hydrolase kcat/Km value.
ATP, which also possesses a good leaving group (ADP), proved to be a very slow, yet viable substrate for PAc hydrolase (1 × 10−1 M−1 s−1). This allowed us to use [γ-32P]ATP to test nucleophilic catalysis, which is reported in the following section.
Bis-p-nitrophenylphosphate was used as the substrate for testing PAc hydrolase phosphodiesterase activity. NPP and AP catalyze bis-p-nitrophenylphosphate hydrolysis with the kcat/Km values of 2 × 103 M−1 s−1 and 5 × 10−2 M−1 s−1, respectively (45). In contrast, no activity was detected for PAc hydrolase. Lastly, p-nitrophenylsulfate was tested as a substrate in order to access sulfatase activity. Only a very low level of activity was detected: kcat/Km = 2 × 10−4 M−1 s−1,3 which is on par with that reported for X. axonopodis NPP: kcat/Km = 2 × 10−5 M−1 s−1 (48).
PAc Hydrolase Catalytic Mechanism
Nucleophile
The alkaline phosphatase superfamily catalytic trait is nucleophilic catalysis, which for the phosphoryl transferases is mediated by a zinc ion (Zn2)-coordinated Ser (AP) (49) or Thr (NPP) (45, 50–51). The PAc hydrolase Thr64, shown positioned for in-line attack of the PAc phosphoryl group in Figure 3, is the likely nucleophile in PAc hydrolase catalysis. In order to confirm the role of Thr64, the following set of experiments were conducted. Firstly, the T64S, T64C and T64A mutant enzymes were prepared and tested for catalytic activity towards PAc hydrolysis. The CD spectra (not shown) and chromatographic behavior of the PAc hydrolase mutants were observed to be consistent with those of the wild-type enzyme, which indicates that the amino acid replacements had not disrupted the native fold. All three mutants were found to possess no detectable activity (detection limit 1 × 10−7 s−1), and thus the Thr64 was shown to be essential for PAc hydrolase catalysis (Table 4). Replacement of the Thr132 nucleophile in the Triticum aestivum NPP with Ala or Ser similarly removed all detectable activity (52).
Table 4.
Steady-state kinetic constants measured for wild-type and mutant PAc hydrolase catalyzed hydrolysis of PAc at pH 7.0 and 25 °C. See Materials and Methods for details.
| Substrate: phosphonoacetate | |||
|---|---|---|---|
| Enzyme | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
| Wild-type | 1.6 × 101 | 0.060 ± 0.001 | 5 × 103 |
| T64S | NA | ---- | ---- |
| T64C | NA | ---- | ---- |
| T64A | NA | ---- | ---- |
| N85A | NA | ---- | ---- |
| K126A | NA | ---- | ---- |
| K126L | NA | ---- | ---- |
| K126R | 8.4 × 10−2 | 0.060 ± 0.002 | 2 × 101 |
| K128A | NA | ---- | ---- |
| K128L | NA | ---- | ---- |
| K128R | 3.1 × 10−3 | 0.031 ± 0.008 | 2 |
| H285A | 7.2 × 10−1 | 0.047 ± 0.007 | 3 × 102 |
| H286A | 2.1 × 10−1 | 0.036 ± 0.004 | 1 × 102 |
| Substrate: phosphonoacetaldehyde | |||
|---|---|---|---|
| Enzyme | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
| Wild-type | 1.1 | 0.49 ± 0.05 | 4 × 101 |
| N85A | NA | ---- | ---- |
| K126A | NA | ---- | ---- |
| K126L | NA | ---- | ---- |
| K126R | NA | ---- | ---- |
| K128A | 1.3 × 10−1 | 9.2 ± 0.5 | 2 × 10−1 |
| K128L | 7.8 × 10−1 | 0.60 ± 0.1 | 2 × 101 |
| K128R | 1.1 × 10−2 | 0.48 ± 0.2 | 4 × 10−1 |
| Substrate: p-nitrophenylphosphate | |||
|---|---|---|---|
| Enzyme | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
| Wild-type | 1.3 × 101 | 17 ± 3 | 1 × 101 |
| N85A | 6.0 × 10−1 | 0.17 ± 0.2 | 6 × 101 |
| K126A | 1.1 × 10−1 | 11 ± 2 | 2 × 10−1 |
| K126L | 3 × 10−1 | 16 ± 2 | 3 × 10−1 |
| K126R | 1.4 | 6.0 ± 0.2 | 4 |
| K128A | 3.0 | 130 ± 30 | 4 × 10−1 |
| K128L | 1.0 | 130 ± 20 | 1 × 10−1 |
| K128R | 8 × 10−1 | 1.8 ± 0.1 | 7 |
Secondly, 32P-labeling of the enzyme via catalytic turnover of [γ-32P]ATP was tested with wild-type PAc hydrolase and the T64A mutant, in the presence and absence the inhibitor phosphonoformate (Figure 5). ATP is a slow PAc hydrolase substrate (Table 2). The reaction was carried out at pH 6 in order to retard the phosphoenzyme hydrolysis step, and thereby maximize the accumulation of the phosphorylated enzyme intermediate. The autoradiogram of the SDS-PAGE gel of the chromatographed [γ-32P]ATP-treated enzymes shows that wild-type PAc hydrolase was 32P-labeled and that the presence of saturating phosphonoformate precluded 32P-labeling. This result shows that the autophosphorylation occurs at the active site. The T64A PAc hydrolase mutant was not 32P-labeled. This observation shows that the T64 is required for the PAc hydrolase autophosphorylation. Earlier investigations of nucleophilic catalysis in mammalian NPPs showed, by kinetic analysis and by sequencing the 32P-labeled proteolytic peptide fragment, that the Thr64 counterpart (Thr204 in bovine liver NPP and Thr210 in human autotaxin, a NPP) undergoes transient phosphorylation during catalytic turnover of [γ-32P]ATP (50–51). By analogy, we assign Thr64 as the site of phosphorylation during PAc hydrolase catalyzed ATP turnover, and conclude that Thr64 performs the role of nucleophilic catalysis in PAc hydrolysis.
Figure 5.
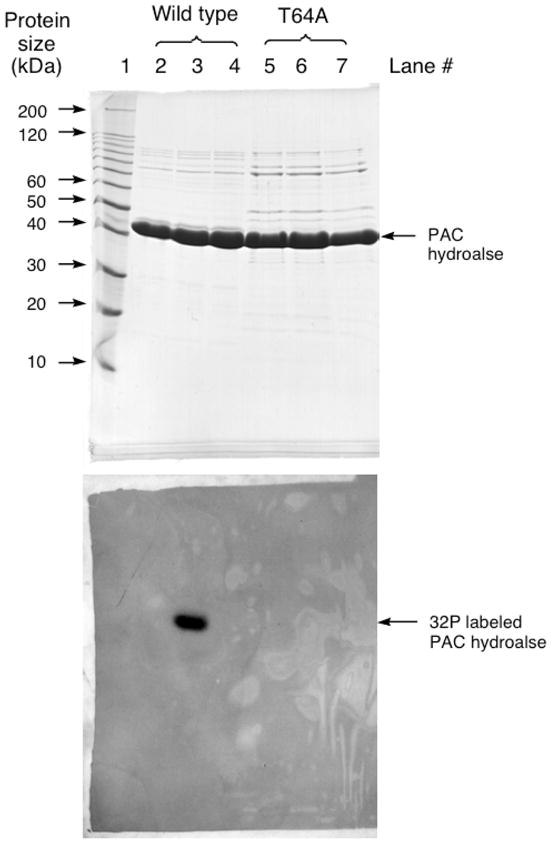
SDS-PAGE analysis of PAc hydrolase autophosphorylation with [γ 32P]ATP. The top panel is the Coomassie blue stained gel and the bottom panel is the audioradiograph of the gel. Lane 1: Protein molecular weight standards. Lane 2: The reaction of [γ 32P]ATP with acid-denatured wild-type PAc hydrolase. Lane 3: The reaction of [γ 32P]ATP with wild-type PAc hydrolase. Lane 4: The reaction of [γ 32P]ATP with wild-type PAc hydrolase in the presence of 2 mM phosphonoformate. Lane 5: The reaction of [γ 32P]ATP with acid-denatured T64A PAc hydrolase. Lane 6: The reaction of [γ 32P]ATP with T64A PAc hydrolase. Lane 7: The reaction of [γ 32P]ATP with T64A PAc hydrolase in the presence of 2 mM phosphonoformate.
Substrate Activation
The phosphoryl group of the phosphonoformate ligand is coordinated to one of the two PAc hydrolase active site zinc ions (Zn1). We retained this interaction in creating the model for PAc binding shown in Figure 3. The Asn85 side chain that is positioned across from the PAc phosphoryl group is close (6 Å), yet in this model it is not close enough for a hydrogen bond interaction. In order to reach the Asn85 side chain, the phosphoryl group would have to lose coordination to the zinc ion (Zn1). Other models were examined for the potential of an Asn85 hydrogen bond interaction with the PAc carboxylate group, while maintaining coordination of the phosphoryl group with the zinc ion and the correct positioning for in-line attack by the Thr64, but none was found to satisfy these requirements. An alignment of PAc hydrolase ortholog sequences (45% or above sequence identity) (SI Figure 1) indicates that Asn85 is stringently conserved. The counterpart to the PAc hydrolase Asn85 in X. axonopodis NPP is Asn111 (45) and in E. coli AP it is Arg186 (44). The side chains of both residues are engaged in a hydrogen bond interaction with the transferring phosphoryl group. The kcat/Km value for E. coli AP catalyzed hydrolysis of ethylphosphate is reduced 7000-fold in the R186S mutant, however for hydrolysis of p-nitrophenylphosphate the kcat/Km value of the R186S mutant is reduced only 170-fold (53). In the case of the Triticum aestivum NPP the impact of Ala replacement of Asn165 (counter part to the X. axonopodis NPP Asn111) on catalytic efficiency is substrate dependent: for nicotinamide adenine dinucleotide and nucleoside triphosphate substrates the activity is reduced ~10-fold, but for p-nitrophenylphosphate it is increased ~100-fold (52). The PAc hydrolase N85A mutant showed no significant catalytic activity towards PAc or PAld (Table 4). On the other hand, whereas the kcat measured for p-nitrophenolphosphate hydrolysis was reduced, inexplicably so was the Km (100-fold) for a net 10-fold increase in the kcat/Km value (Table 4). Although Asn85 contributes to catalysis, its role is not clear.
The side chains of stringently conserved PAc hydrolase residues Lys126 and Lys128 are located in the vicinity of the carboxylate group of the modeled PAc (Figure 3). The ammonium group of Lys126 is within hydrogen bonding distance of the carboxylate oxygen atom. The conformation of the side chain of Lys128 observed in the structure of the PAc hydrolase-phosphonoformate complex can be modeled to place the ammonium group within hydrogen bond interaction distance of the PAc carboxylate group. Each Lys was separately replaced with Arg, Leu and Ala and the kinetic properties of the site directed mutants were measured (Table 4). All three Lys126 mutants are inactive as catalysts of PAc or PAld hydrolysis, but each retained significant activity towards p-nitrophenylphosphate hydrolysis. The PAc hydrolase K128A and K128L mutants do not hydrolyze PAc whereas the kcat/Km value for K128R mutant is 1000-fold lower than that of the wild-type enzyme. The three mutants do, however retain activity towards catalysis of PAld hydrolysis. The kcat values for p-nitrophenylphosphate hydrolysis are similar to those measured for wild-type PAc hydrolase (Table 4). Overall, the PAc hydrolysis reaction is most sensitive to Lys126 and Lys128 amino acid replacement, which is consistent with the need for two positively charge residues to stabilize of the aci-carboxylate dianion displaced by the Thr64.
The alkaline phosphatase enzyme superfamily member cofactor independent phosphoglycerate mutase utilizes a large, dynamic cap domain to bind the substrate glycerate unit as it closes over the catalytic domain (41). The observation that PAc hydrolase and NPP both possess a cap domain (AP does not) (Figure 2) prompted us to probe the impact of the replacement of the PAc hydrolase cap domain residues that are directed towards the catalytic site, on catalytic efficiency. His285 and His286 were targeted because they maintain hydrogen bond forming potential, are stringently conserved among PAc hydrolase orthologs (SI Figure 1) and are positioned over the substrate (Figure 3). If the cap domain was to clamp down further on the catalytic domain one or both of the His residues might be positioned to favorably interact with the PAc carboxylate. However, Ala replacement of either His reduced the kcat by only an order of magnitude (Table 4). This small reduction does not support a direct role for His85 or His86 in catalysis. We therefore conclude that the snapshot of the cap and catalytic domains observed in the crystal structure represent the catalytically active form of the enzyme.
Proposed Model for Catalysis
The model that we propose for the PAc hydrolase catalytic mechanism is a variation on the mechanism of NPP catalysis proposed by Herschlag and coworkers (45), which in turn is based on the NPP crystal structure (45) and on the large volume of work that has been carried out to define the catalytic mechanism of the E. coli AP (47, 49, 53–56). The key difference between the chemical reactions catalyzed by PAc hydrolase and NPP/AP is the first step, which involves P-C bond cleavage rather than P-OR bond cleavage. It is on this step that we will focus our discussion.
The cleavage of the P-OR bond catalyzed by AP and NPP occurs by attack the catalytic Ser or Thr, respectively. The active form of the nucleophile is the alkoxide anion, which is stabilized and oriented by coordination to one of the two zinc ion cofactors (Zn2) (Figure 2B and 2C). The phosphorus is activated for nucleophilic attack by interaction of its oxygen atoms with one or both of the zinc ions and with the properly positioned Arg (AP) or Asn (NPP) side chain. The stabilization of the alkoxide leaving group is necessary for the P-OR bond cleavage in phosphate ester metabolites, and this role has been assigned to one of the two zinc ions (Zn2), which must form a coordination bond with the departing oxygen atom. The zinc ion cannot engage in such an interaction with the PAc methylene group. Therefore, an alternate strategy to leaving group stabilization in PAc hydrolase catalysis must exist. The leaving group is an aci-carboxylate dianion. Aci-carboxylate dianion intermediates are known to be formed and stabilized in the active sites of members of the enolase enzyme superfamily (57) and in the active sites of members of the isocitrate lyase/PEP mutase superfamily (58). Enzymes from both these families use electropositive groups (metal ion cofactor and/or hydrogen bond donating amino acid residues) to delocalize charge via interaction with the carboxylate group of aci-carboxulate dianion. The best candidates for this role in PAc hydrolase are the respective ammonium groups of Lys126 and Lys128. Thus, the proposed model for the PAc hydrolase substrate complex (Scheme 3) incorporates the coordination of the substrate phosphoryl group to one zinc ion (Zn1), the coordination of the Thr64 nucleophile to the other zinc ion (Zn2), and the hydrogen bond interactions of the PAc carboxylate group with the Lys126 and Lys128 side chains.
Scheme 3.
A proposed model for the catalytic mechanism used by PAc hydrolase in the first partial reaction.
It is interesting to contrast the catalytic strategy used by phosphonatase to catalyze P-C bond cleavage in PAld (Scheme 2) with the catalytic strategy depicted in Scheme 3 for the PAc hydrolase. Because PAc hydrolase also catalyzes PAld hydrolysis, albeit less efficiently than PAc hydrolysis (Table 2), in effect PAld hydrolase activity has evolved within the catalytic scaffold of the alkaline phosphatase enzyme superfamily in addition to evolving within the catalytic scaffold of the HAD enzyme superfamily. The C=O of PAld can in principle engage one of the PAc hydrolase active site Lys side chains in hydrogen bond formation for stabilization of the enolate anion formed with P-C bond cleavage. The PAld hydrolase activity of PAc hydrolase is considerably less than that of phosphonatase (18) (kcat/Km = 1 × 105 M−1 s−1 vs kcat/Km = 8 × 101 M−1 s−1) and although interesting from an evolution-mechanism perspective, this activity is not expected to be of biological significance.
PAc Hydrolase Biological Range and Function
To gain insight into the metabolic function of PAc hydrolase we first carried out a BLAST search of the bacterial genomes deposited in NCBI, using the P. fluorescens 23F PAc hydrolase amino acid sequence as query, to identify bacteria that produce PAc hydrolase. In this manner we were able to trace the biological range of PAc hydrolase within the bacterial species represented in the NCBI genome bank. The sequence identity boundary separating orthologs from paralogs was well defined (> 45% vs < 30%). Interestingly, PAc hydrolase was found only in Proteobacteria. Within the beta subdivision, Bordetella, Burkholderia, Cupriavidus, Ralstonia, Alicycliphilus, Polaromonas, and Verminephrobacter species possess orthologs that share 50–85% sequence identity with the P. fluorescens 23F enzyme. Within the alpha subdivision Mesorhizobium, Oligotropha, Rhodobacterales, Rhodopseudomonas, Roseovarius, Thalassiobium, Agrobacterium, and Sinorhizobium species possess orthologs that share 55–46% sequence identity with the P. fluorescens 23F enzyme. In the delta subdivision Sorangium, Pseudomonadaceae and Azotobacter are the only representatives found to contain a PAc hydrolase ortholog (47% sequence identity).
Next, we searched among the annotated genomes for PAc hydrolase gene neighbors that might encode proteins that function in the synthesis or breakdown of PAc. We discovered gene clusters in the genomes of Sinorhizobium medicae and meliloti that encode AEP transaminase, PAc hydrolase and an aldehyde dehydrogenase. In numerous Burkholderia species the genes annotated AEP transporter, PAc hydrolase and PAld dehydrogenase are juxtaposed. Together these enzymes might form the components of a novel AEP transport and degradation pathway, as is illustrated in Scheme 1. A BLAST of the genomes of the PAc hydrolase containing bacteria using AEP transaminase and the putative PAld dehydrogenase sequences as queries confirmed that all three of the pathway genes are present. The spotty distribution of the PAc hydrolase gene among the various subdivisions of Proteobacteria is suggestive of its distribution by lateral gene transfer (59). This is also indicated by the co-location of a putative transposon as was observed for a few of the inspected genomes. If clustered together, all three of the pathway genes might efficiently be transferred by a single mobile genetic element.
A brief check was made to determine if the phosphonatase gene is present in the genomes of the bacteria that harbor the PAc hydrolase gene. With only a few exceptions (Burkholderia graminus being one) it is not. A BLAST search of the NCBI genome bank using the phosphonatase sequence as query identifies phosphonatase in a wide range of Gram-negative and Gram-positive bacteria. This suggests that the AEP degradation pathway that proceeds via P-C cleavage in PAld appears to be more wide spread than that which proceeds via P-C bond cleavage in PAc (Scheme 1).
Conclusion
Of the known members of the alkaline phosphatase enzyme superfamily, the PAc hydrolase tertiary structure and active site structure most closely resembles those of the NPP. However, the lack of significant sequence identity between the PAc hydrolase and NPP (~15%) compared to relatively high sequence identity between PAc hydrolase orthologs (> 45%) suggests early divergence within this lineage of the alkaline phosphate enzyme superfamily. Nevertheless, two basic elements of catalysis, namely activation of the substrate phosphoryl group by zinc ion coordination and nucleophilic attack by a zinc ion activated Thr are conserved (Scheme 3). Whereas the active site Asn that binds the substrate phosphoryl group in NPP is also present in PAc hydrolase, it does not appear to participate in PAc binding. In NPP, a zinc ion stabilizes the oxygen anion leaving group whereas in PAc hydrolase, the carbanion leaving group is stabilized by delocalization to the carboxylate oxygen atoms and hence to the charged Lys residues (Scheme 3).
Supplementary Material
Footnotes
This work was supported by N.I.H. grant GM 28688 (D. D-M), NIH grant DK47814 (HMH) and BBSRC (UK) grant 81/P11488 (J.P.Q.) and intramural funding from the IRP, NIMH (B.M.M.).
Abbreviations used are: PAc, phosphonoacetate; PAc hydrolase, phosphonoacetate hydrolase; ATP, adenosine 5′-triphosphate; ADP, adenosine 5′-diphosphate; AMP, adenosine 5′-monophosphate; PEP, phosphoenol pyruvate; NADH, dihydronicotinamide adenine dinucleotide; HEPES, N-[2-hydroxyethyl]piperazine-N′-[2 ethanesulfonic acid]; ADA, N-(2-acetamido) iminodiacetic acid; BICINE, N,N-Bis(2-hydroxyethyl)glycine; MOPSO, 3-(N-morpholino)-2-hydroxypropanesulfonic acid; PPyr, phosphonopyruvate; AP, alkaline phosphatase; NPP, nucleotide pyrophosphatase/diesterase; PAld, phosphonoacetaldehyde; AEP, 2-aminoethylphosphonate; HAD, haloalkonic acid dehalogenase; HPLC, high performance liquid chromatography.
AP binds a magnesium ion cofactor at its active site. The absence of a bound magnesium ion in the PAc hydrolase X-ray structure and the absence of a requirement for magnesium ion for catalysis are evidence that the third metal cofactor used in AP catalysis is not employed in PAc hydrolase catalysis.
The PAc hydrolase was judged to be pure based on SDS-PAGE analysis, however we did not take additional steps to rule out the possibility that the low activities observed with these substrates originate in a contaminating, high activity hydrolase.
Supporting Information Available
The supporting information contains Figure SI1, the multiple alignment of phosphonoacetate hydrolase amino acid sequences. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.White AK, Metcalf WW. Microbial Metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 2.Metcalf WW, van der Donk WA. Biosynthesis of phosphonic and phosphinic acid natural products. Ann Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair SK, Van der Donk WA. Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos and phosphinothricin. Arch Biochem Biophys. 2011;505:13–21. doi: 10.1016/j.abb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinn JP, Kulakova AN, Cooley NA, McGrath JW. New ways to break an old bond: the bacterial carbon-phosphorus hydrolases and their roles in biogeochemical phosphorus cycling. Environ Microbiol. 2007;9:2392–2400. doi: 10.1111/j.1462-2920.2007.01397.x. [DOI] [PubMed] [Google Scholar]
- 5.Kittredge MS, Robert E, Simonsen DG. The occurrence of free 2-aminoethylphosphonic acid in the sea anemone, Anthopleura elegantissima. Biochemistry. 1962;1:624–628. doi: 10.1021/bi00910a013. [DOI] [PubMed] [Google Scholar]
- 6.Horiguchi M, Kandatsu M. Isolation of 2-aminoethane phosphonic acid from rumen protozoa. Nature. 1959;184:901–902. doi: 10.1038/184901b0. [DOI] [PubMed] [Google Scholar]
- 7.Hilderbrand RL. In: The role of phosphonates in living systems. Hilderbrand RL, editor. C.R.C. Press, Inc; Boca Raton, Florida: 1983. [Google Scholar]
- 8.Orsini F, Sello G, Sisti M. Aminophosphonic acids and derivatives. Synthesis and biological applications. Curr Med Chem. 2010;17:264–289. doi: 10.2174/092986710790149729. [DOI] [PubMed] [Google Scholar]
- 9.De Clercq E, Holy A. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat Rev Drug Discov. 2005;4:928–940. doi: 10.1038/nrd1877. [DOI] [PubMed] [Google Scholar]
- 10.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. Fosfomycin for the treatment of multi-drug resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis. 2010;10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy KE, Thompson GA. Phosphonolipids: Localization in surface membranes of Tetrahymena. Science. 1970;168:989–991. doi: 10.1126/science.168.3934.989. [DOI] [PubMed] [Google Scholar]
- 12.Bowman E, McQueney M, Barry R, Dunaway-Mariano D. Catalysis and thermodynamics of the rearrangement of phosphoenol pyruvate to phosphonopyruvate. Entry into the phosphonate class of naturally occurring organophosphonates. J Amer Chem Soc. 1988;110:5575–5577. [Google Scholar]
- 13.Zhang G, Dai J, Lu Z, Dunaway-Mariano D. The phosphonopyruvate decarboxylase from Bacteroides fragilis. J Biol Chem. 2003;278:41302–41308. doi: 10.1074/jbc.M305976200. [DOI] [PubMed] [Google Scholar]
- 14.Kim J. PhD Thesis. University of Maryland; 1994. Investigations of the 2-aminoethylphosphonate biosynthetic enzymes in Tetrahymena pyriformis; pp. 96–106. [Google Scholar]
- 15.Kim AD, Baker AS, Metcalf WW, Wanner BL, Martin BM, Dunaway-Mariano D. The 2-aminoethylphosphonate-specific Transaminase of the 2-aminoethylphosphonate degradation pathway. J Bacteriology. 2002;184:4134–4140. doi: 10.1128/JB.184.15.4134-4140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen DB, Hepburn TW, Moos M, Mariano PS, Dunaway-Mariano D. Investigation of the Bacillus cereus phosphonoacetate hydrolse. Evidence for a Schiff Base Mechanism and sequence analysis of an active site peptide containing the catalytic lysine residue. Biochemistry. 1988;27:2229–2236. doi: 10.1021/bi00406a063. [DOI] [PubMed] [Google Scholar]
- 17.Lee S-L, Hepburn TW, Schwartz WH, Ammon HL, Mariano PS, Dunaway-Mariano Stereochemical probe for the mechanism of P-C bond cleavage catalyzed by the Bacillus cereus phosphonoacetaldehyde hydrolase. J Amer Chem Soc. 1992;114:7346–7354. [Google Scholar]
- 18.Baker AS, Ciocci MJ, Metcalf WW, Kim JB, Babbitt PC, Wanner BL, Martin BM, Dunaway-Mariano D. Insights into the mechanism of catalysis by the P-C bond cleaving enzyme phosphonoacetaldehyde hydrolase derived from gene sequence-analysis and mutagenesis. Biochemistry. 1998;37:9305–9315. doi: 10.1021/bi972677d. [DOI] [PubMed] [Google Scholar]
- 19.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 20.McMullan G, Quinn JP. In vitro characterization of a phosphate starvation-independent carbon-phosphorus bond cleavage activity in Pseudomonas fluorescens 23F. J Bacteriol. 1994;176:320–324. doi: 10.1128/jb.176.2.320-324.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrath JW, Wisdom GB, McMullan G, Larkin MJ, Quinn JP. The purification and properties of phosphonoacetate hydrolase, a novel carbon-phosphorus bond-cleavage enzyme from Pseudomonas fluorescens 23F. Eur J Biochem. 1995;234:225–30. doi: 10.1111/j.1432-1033.1995.225_c.x. [DOI] [PubMed] [Google Scholar]
- 22.Galperin MY, Bairoch A, Koonin EV. A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases. Protein Science. 1998;7:1829–1835. doi: 10.1002/pro.5560070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCullan G, Harrington F, Quinn JP. Metabolism of phosphonoacetate as the sole carbon and phosphorus source by an environmental bacterial isolate. Appl Environ Microbiol. 1992;58:1364–1366. doi: 10.1128/aem.58.4.1364-1366.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullan G, Quinn JP. Detection of a novel carbon-phosphorus bond cleavage activity in cell-free extracts of an environmental Pseudomonas fluorescens isolate. Biochem Biophys Res Commun. 1992;184:1022–1027. doi: 10.1016/0006-291x(92)90693-f. [DOI] [PubMed] [Google Scholar]
- 25.Panas P, Ternan NG, Dooley JS, McMullan G. Detection of phosphonoacetate degradation and phnA genes in soil bacteria from distinct geographical origins suggest its possible biogenic origin. Environ Microbiol. 2006;8:939–945. doi: 10.1111/j.1462-2920.2005.00974.x. [DOI] [PubMed] [Google Scholar]
- 26.Thomas S, Burdett H, Temperton B, Wick R, Snelling D, McGrath JW, Quinn JP, Munn C, Gilbert JA. Evidence for phosphonate usage in the coral holobiont. ISME J. 2010;4:459–461. doi: 10.1038/ismej.2009.129. [DOI] [PubMed] [Google Scholar]
- 27.Kulakova AN, Kulakov LA, Akulenko NV, Ksenzenko VN, Hamilton JT, Quinn JP. Structural and functional analysis of the phosphonoacetate hydrolase (phnA) gene region in Pseudomonas fluorescens 23F. J Bacteriol. 2001;183:3268–3275. doi: 10.1128/JB.183.11.3268-3275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim AD, Baker AS, Metcalf WW, Wanner BL, Martin BM, Dunaway-Mariano D. The 2-aminoethylphosphonate-specific transaminase of the 2-aminoethylphosphonate degradation pathway. J Bacteriology. 2002;184:4134–4140. doi: 10.1128/JB.184.15.4134-4140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulakova AN, Kulakov LA, Quinn JP. Cloning of the phosphonoacetate hydrolase gene from Pseudomonas fluorescens 23F encoding a new type of carbon-phosphorus bond cleaving enzyme and its expression in Escherichia coli and Pseudomonas putida. Gene. 1997;195:49–53. doi: 10.1016/s0378-1119(97)00151-0. [DOI] [PubMed] [Google Scholar]
- 30.Isbell AF, Englert LF, Rosenberg H. Phosohonacetaldehyde. J Org Chem. 1969;34:755–756. [Google Scholar]
- 31.Wesenberg G, Rayment I. unpublished results. [Google Scholar]
- 32.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr. 1998;D54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 34.Rypniewski WR, Breiter DR, Benning MM, Wesenberg G, Oh BH, Markley JL, Rayment I, Holden HM. Crystallization and structure determination to 2.5-A resolution of the oxidized [2Fe-2S] ferredoxin isolated from Anabaena 7120. Biochemistry. 1991;30:4126–4131. doi: 10.1021/bi00231a003. [DOI] [PubMed] [Google Scholar]
- 35.Navaza J. Amore - an Automated Package for Molecular Replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 36.Tronrud DE, Ten Eyck LF, Matthews BW. Acta Crystallogr Sect A. 1987;43:489–501. [Google Scholar]
- 37.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. Silicon Graphics; 1991. [Google Scholar]
- 38.Millan JL. Alkaline Phosphatases. Structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signalling. 2006;2:335–341. doi: 10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galperin MY, Jedrzejas MJ. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins. 2001;45:318–324. doi: 10.1002/prot.1152. [DOI] [PubMed] [Google Scholar]
- 40.Jedrzejas MJ, Setlow P. Comparison of the binuclear metalloenzymes diphosphoglycerate-independent phosphoglycerate mutase and alkaline phosphatase:their mechanism of catalysis via a phosphoserine intermediate. Chem Rev. 2001;101:607–618. doi: 10.1021/cr000253a. [DOI] [PubMed] [Google Scholar]
- 41.Rigden DJ, Lamani E, Mello LV, Littlejohn JE, Jedrzejas MJ. Insights into the catalytic mechanism of cofactor-independent phosphoglycerate mutase from X-ray crystallography, simulated dynamics and molecular modeling. J Mol Biol. 2003;328:909–920. doi: 10.1016/s0022-2836(03)00350-4. [DOI] [PubMed] [Google Scholar]
- 42.Gosh D. Human sulfatases: a structural perspective. Cell Mol Life Sci. 2007;64:2013–2022. doi: 10.1007/s00018-007-7175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bojarova P, Williams SJ. Sulfotransferases, sulfatases and formyglycine-generating enzymes: a sulfation fascination. Curr Opin Chem Biol. 2008;12:573–581. doi: 10.1016/j.cbpa.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 44.Kim EE, Wyckoff HW. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 45.Zalatan JG, Fenn TD, Brunger AT, Herschlag D. Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: implications for mechanism and evolution. Biochemistry. 2006;45:9788–9803. doi: 10.1021/bi060847t. [DOI] [PubMed] [Google Scholar]
- 46.Holtz KM, Stec B, Myers JK, Antonelli SM, Widlanski TS, Kantrowitz ER. Alternate modes of binding in two crystal structures of alkaline phosphatase-inhibitor complexes. Protein Sci. 2000;9:907–915. doi: 10.1110/ps.9.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brien PJ, Hershlag D. Alkaline phosphatase revisited: Hydrolysis of alkyl phosphates. Biochemistry. 2002;41:3207–3225. doi: 10.1021/bi012166y. [DOI] [PubMed] [Google Scholar]
- 48.Lassila J, Herschlag D. Promiscuous sulfatase activity and thio-effects in a phosphodiesterase of the alkaline phosphatase superfamily. Biochemistry. 2008;47:12853–12859. doi: 10.1021/bi801488c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coleman JE. Structure and mechanism of alkaline phosphatase. Annu Rev Biophys. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 50.Oda Y, Kuo MD, Huang SS, Huang JS. The major acidic fibroblast growth factor (aFGF)-stimulated phosphoprotein from bovine liver plasma membranes has a aFGF-stimulated kinase, autoadenylation, and alkaline nucleotide phosphodiesterase activities. J Biol Chem. 1993;268:27318–27326. [PubMed] [Google Scholar]
- 51.Clair T, Lee HY, Lioatta LA, Stracke ML. Autotaxin is an exoenzyme possessing 5′-nucleotide phosphodiesterase/ATP pyrophosphatase and ATPase activities. J Biol Chem. 1997;272:996–1001. doi: 10.1074/jbc.272.2.996. [DOI] [PubMed] [Google Scholar]
- 52.Joye IJ, Belien T, Brijis K, Soetaert W, Delcour JA. Mutational analysis of wheat (Triticum aestivum L.) nucleotide pyrophosphatse/phosphodiesterase shows the roles of six amino acids in the catalytic mechanism. Appl Microbiol Biotechnol. 2010 doi: 10.1007/s00253-010-2962-z. Published on line Dec 29 2010. [DOI] [PubMed] [Google Scholar]
- 53.O’Brien PJ, Lassila JK, Fenn TD, Zaltan JG, Herschlag D. Arginine coordination in enzymatic phosphoryl transfer: evaluation of the effect of Arg166 mutations in Escherichhia coli alkaline phosphatase. Biochemistry. 2008;47:7663–7672. doi: 10.1021/bi800545n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stec B, Holtz KM, Kantrowitz ER. A revised mechanism for the alkaline phosphatase reaction involving three metal ions. J Mol Biol. 2000;299:1303–1311. doi: 10.1006/jmbi.2000.3799. [DOI] [PubMed] [Google Scholar]
- 55.Zaltan J, Fenn TD, Herschlag D. Comparative enzymology in the alkaline phosphatase superfamily to determine the catalytic role of an active site metal ion. J Mol Biol. 2008;384:1174–1189. doi: 10.1016/j.jmb.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Kantrowitz ER. Trapping the tetrahedral intermediate in the alkaline phosphatase reaction by substitution of the active site serine with threonine. Protein Sci. 2006;15:2395–2401. doi: 10.1110/ps.062351506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 58.Liu S, Lu Z, Han Y, Melamud E, Dunaway-Mariano D, Herzberg O. Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution. Biochemistry. 2005;44:2949–2962. doi: 10.1021/bi0479712. [DOI] [PubMed] [Google Scholar]
- 59.Huang J, Su Z, Xu Y. The evolution of microbrial phosphate degradative pathways. J Mol Evol. 2005;61:682–690. doi: 10.1007/s00239-004-0349-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.