Abstract
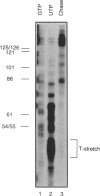
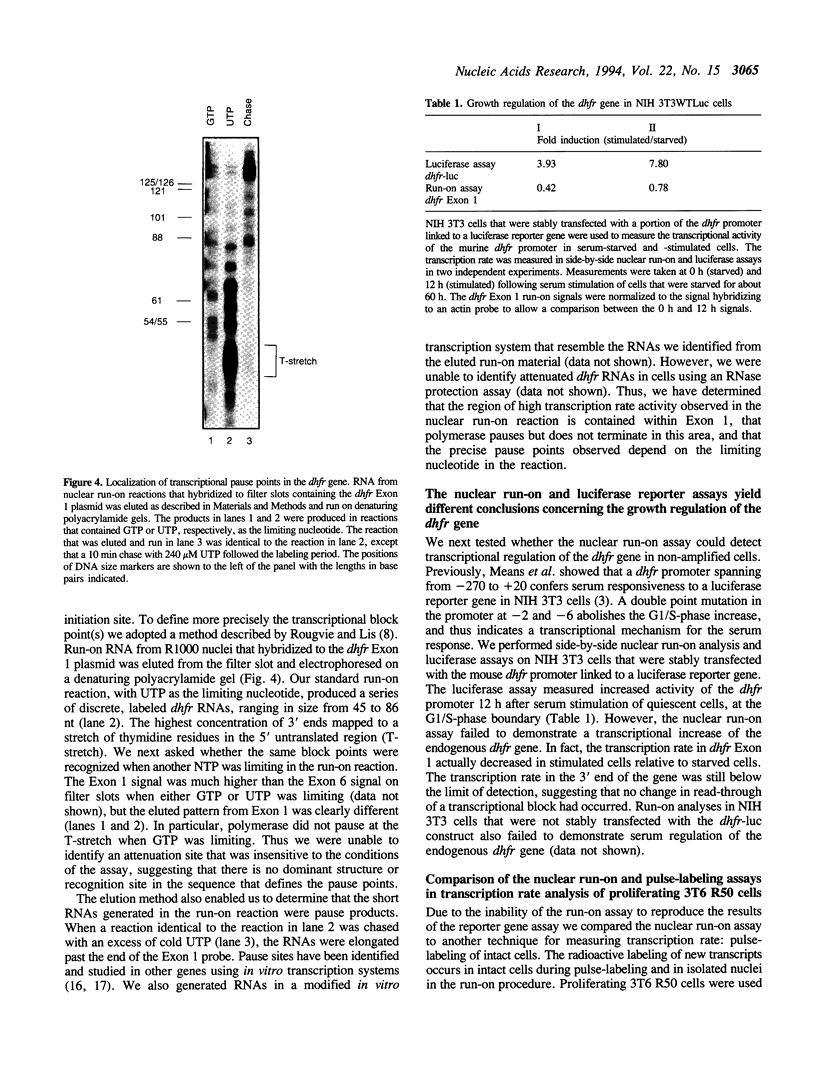
Using the nuclear run-on assay we found that in proliferating cells the transcription rate in the 5' end of the murine dihydrofolate reductase (dhfr) gene was approximately ten-fold higher than in the 3' end of the gene, suggesting transcriptional attenuation within the dhfr gene. However, when the transcription rate was measured by pulse-labeling, the rate was uniform throughout the gene, and the 5' dhfr signal was approximately ten-fold lower relative to a control gene signal than in the run-on assay. Previously, the activity of a dhfr promoter linked to a luciferase reporter gene was shown to increase about ten-fold at the G1/S-phase boundary following stimulation of serum-starved cells. To determine if the run-on procedure would detect growth regulation of the endogenous dhfr gene, serum-starved and -stimulated NIH 3T3 cells were analyzed. Using a dhfr 5' end probe no difference in transcription rate between these growth states was detected and the dhfr 3' end probe did not detect signal above background. In a cell line that was amplified at the dhfr locus, the transcription rate in the 5' end of the gene increased less than two-fold in stimulated cells, but the rate in the 3' end of the gene increased five- to seven-fold. Therefore, the dhfr gene is growth regulated at the level of transcription, but the nuclear run-on assay was only able to detect a difference in transcription rate in the 3' end of the gene in amplified cells. We suggest that isolation of nuclei may activate dhfr transcription complexes that normally are activated only at the G1/S-phase boundary.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Farnham P. J., Kollmar R. Characterization of the 5' end of the growth-regulated Syrian hamster CAD gene. Cell Growth Differ. 1990 Apr;1(4):179–189. [PubMed] [Google Scholar]
- Farnham P. J., Schimke R. T. Transcriptional regulation of mouse dihydrofolate reductase in the cell cycle. J Biol Chem. 1985 Jun 25;260(12):7675–7680. [PubMed] [Google Scholar]
- Feder J. N., Guidos C. J., Kusler B., Carswell C., Lewis D., Schimke R. T. A cell cycle analysis of growth-related genes expressed during T lymphocyte maturation. J Cell Biol. 1990 Dec;111(6 Pt 1):2693–2701. doi: 10.1083/jcb.111.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985 Jul 5;260(13):8163–8172. [PubMed] [Google Scholar]
- Hisatake K., Hasegawa S., Takada R., Nakatani Y., Horikoshi M., Roeder R. G. The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993 Mar 11;362(6416):179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O., Gill G., Chen J. L., Dynlacht B. D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993 Jan 29;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Kokubo T., Gong D. W., Roeder R. G., Horikoshi M., Nakatani Y. The Drosophila 110-kDa transcription factor TFIID subunit directly interacts with the N-terminal region of the 230-kDa subunit. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):5896–5900. doi: 10.1073/pnas.90.13.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A., Meulia T., Brunvand M., Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992 Nov;6(11):2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- La Thangue N. B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994 Mar;19(3):108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- Linial M., Gunderson N., Groudine M. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science. 1985 Dec 6;230(4730):1126–1132. doi: 10.1126/science.2999973. [DOI] [PubMed] [Google Scholar]
- London L., Keene R. G., Landick R. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol Cell Biol. 1991 Sep;11(9):4599–4615. doi: 10.1128/mcb.11.9.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A. L., Slansky J. E., McMahon S. L., Knuth M. W., Farnham P. J. The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol Cell Biol. 1992 Mar;12(3):1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miltenberger R. J., Cortner J., Farnham P. J. An inhibitory Raf-1 mutant suppresses expression of a subset of v-raf-activated genes. J Biol Chem. 1993 Jul 25;268(21):15674–15680. [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990 Nov;10(11):6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Ruppert S., Wang E. H., Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993 Mar 11;362(6416):175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- Santiago C., Collins M., Johnson L. F. In vitro and in vivo analysis of the control of dihydrofolate reductase gene transcription in serum-stimulated mouse fibroblasts. J Cell Physiol. 1984 Jan;118(1):79–86. doi: 10.1002/jcp.1041180114. [DOI] [PubMed] [Google Scholar]
- Schmidt E. E., Merrill G. F. Transcriptional repression of the mouse dihydrofolate reductase gene during muscle cell commitment. J Biol Chem. 1989 Dec 15;264(35):21247–21256. [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Wiest D. K., Hawley D. K. In vitro analysis of a transcription termination site for RNA polymerase II. Mol Cell Biol. 1990 Nov;10(11):5782–5795. doi: 10.1128/mcb.10.11.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. S., Johnson L. F. Regulation of dihydrofolate reductase gene transcription in methotrexate-resistant mouse fibroblasts. J Cell Physiol. 1982 Feb;110(2):183–189. doi: 10.1002/jcp.1041100212. [DOI] [PubMed] [Google Scholar]
- Yoder S. S., Berget S. M. Posttranscriptional control of DHFR gene expression during adenovirus 2 infection. J Virol. 1985 Apr;54(1):72–77. doi: 10.1128/jvi.54.1.72-77.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]