Abstract
When a replicative DNA polymerase (Pol) is stalled by damaged DNA, a 'polymerase switch' recruits specialized translesion synthesis (TLS) DNA polymerase(s) to sites of damage. Mammalian cells have several TLS DNA polymerases, including the four Y-family enzymes (Polη, Polι, Polκ and REV1) that share multiple primary sequence motifs, but show preferential bypass of different DNA lesions. REV1 interacts with Polη, Polι, and Polκ and therefore appears to play a central role during TLS in vivo. Here we have investigated the molecular basis for interactions between REV1 and Polκ. We have identified novel REV1-interacting regions (RIRs) present in Polκ, Polι and Polη. Within the RIRs, the presence of two consecutive phenylalanines (FF) is essential for REV1-binding. PCNA-binding motifs found in many proteins also contain FF with some conserved residues N-terminal to FF, as frequently represented by Q-x-x-(I,L,M)-x-x-F-F (x, any residue). In contrast, our results show that the critical FF in RIR motifs are not flanked by specific conserved residues. Instead, the consensus sequence for REV1-binding is denoted by x-F-F-y-y-y-y (y, any residue but not proline). Our results identify structural requirements that are necessary for FF-flanking residues to confer interactions with REV1. A Polκ mutant lacking REV1-binding activity did not complement the genotoxin-sensitivity of Polk-null mouse embryonic fibroblast cells, thereby demonstrating that the REV1-interaction is essential for Polκ function in vivo.
Introduction
DNA in living organisms is continually exposed to a vast variety of genotoxic agents from endogenous and exogenous sources (Friedberg et al. 2005). Among such genotoxic agents, UV-irradiation has been most extensively studied for its biological consequences. UV irradiation induces the formation of dipyrimidine dimers, among which cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts {(6-4)PPs} are the predominant lesions. (6-4)PPs generate large distortions in the DNA double-helix structure and consequently, they are detected and repaired efficiently by the nucleotide excision repair pathway. In contrast, CPDs causing small distortions in the DNA structure are repaired more slowly. When the replication fork encounters a persisting DNA lesion, a high-fidelity replicative DNA polymerase (such as Polδ or Polε) with proof-reading 3’–5’ exonuclease activity often stalls at the damaged site. For continuation of replication, the stalled replicative DNA polymerase must be replaced with a DNA polymerase that is capable of carrying out translesion DNA synthesis (TLS). Mammalian cells have evolved multiple specialized TLS DNA polymerases including Polη, Polι, Polκ and REV1 (which comprise the Y-family of DNA polymerases) (Ohmori et al. 2001). Because TLS DNA polymerases are intrinsically error-prone due to the lack of proof-reading activity, their actions must be appropriately regulated to minimize mutations.
Of the Y-family DNA polymerases, Polη is able to bypass thymine-thymine (T-T) CPD efficiently and accurately in vitro by inserting two adenines opposite the dimer, yet is unable to bypass T-T (6-4)PP (Masutani et al. 1999a). XPV (Xeroderma Pigmentosum Variant) patients lacking active Polη are predisposed to sun-light induced skin cancer, suggesting that in the absence of Polη other DNA polymerase(s) perform mis-insertion opposite CPDs (Masutani et al. 1999b; Johnson et al. 1999). However, Polη may not cope with every kind of DNA lesion correctly; for instance, Polη was shown to insert A or G more frequently than C opposite benzo[a]pyrene (B[a]P)-adducted guanines (the major lesions induced by B[a]P) in vitro (Chiapperino et al. 2002; Rechkoblit et al. 2002). In contrast, Polκ preferentially inserts the correct C opposite B[a]P-adducted guanine in vitro (Rechkoblit et al. 2002; Suzuki et al. 2002; Zhang et al. 2000), yet cannot bypass CPDs or (6-4)PPs (Ohashi et al. 2000; Johnson et al. 2000 Gerlach et al. 2001). Consistent with results obtained from in vitro experiments, Polκ-deficient mouse embryonic stem (ES) cells are highly sensitive to B[a]P and generate more mutations upon B[a]P treatment than the wild-type cells (Ogi et al. 2002). Therefore, Polκ appears to be important for protecting cells against the lethal and mutagenic effects of B[a]P and in the absence of Polκ, alternative DNA polymerase(s) may conduct error-prone bypass of B[a]P-adducted DNA. Clearly cells must select appropriate TLS polymerase(s) for accurate lesion-specific bypass of DNA damage (Bi et al. 2005), thereby minimizing the aquisition of mutations following exposure to genotoxic agents. However the molecular basis for selection of appropriate TLS polymerases remains elusive.
In an attempt to identify factors involved in the recruitment of Polκ to sites of DNA damage, we carried out yeast two-hybrid (Y2H) screening using Polκ as bait. Unexpectedly, REV1 was identified as a Polκ-interacting partner. We confirmed the direct interaction between Polκ and a C-terminal domain (CTD) of REV1 using GST pull-down and co-immunoprecipitation assays (Ohashi et al. 2004). Furthermore, we and others found that the REV1-CTD interacts not only with Polκ but also with Polη and Polι and concluded that REV1 might play a central role in TLS events in vivo (Ohashi et al. 2004;Guo et al. 2003; Tissier et al. 2004).
In this report we have investigated the basis for interaction between REV1 and other Y-family TLS polymerases. We show that Polκ, Polι and Polη each have a REV1-interacting region (RIR) containing two consecutive phenylalanine (FF) residues that are critical for interaction with REV1-CTD. This is reminiscent of the PCNA-binding motif that also contains FF in many instances (Moldovan et al. 2007). However, we show that the novel FF motif required for recognition of the REV1-CTD is defined by alternative flanking sequences. Our identification of a novel motif for mediating interactions of Polκ, Polη, and Polι with REV1 may provide a molecular basis for coordination of the appropriate TLS polymerases for lesion-specific replication of damaged DNA.
Results
FF Motif Is Essential for REV1-Interaction
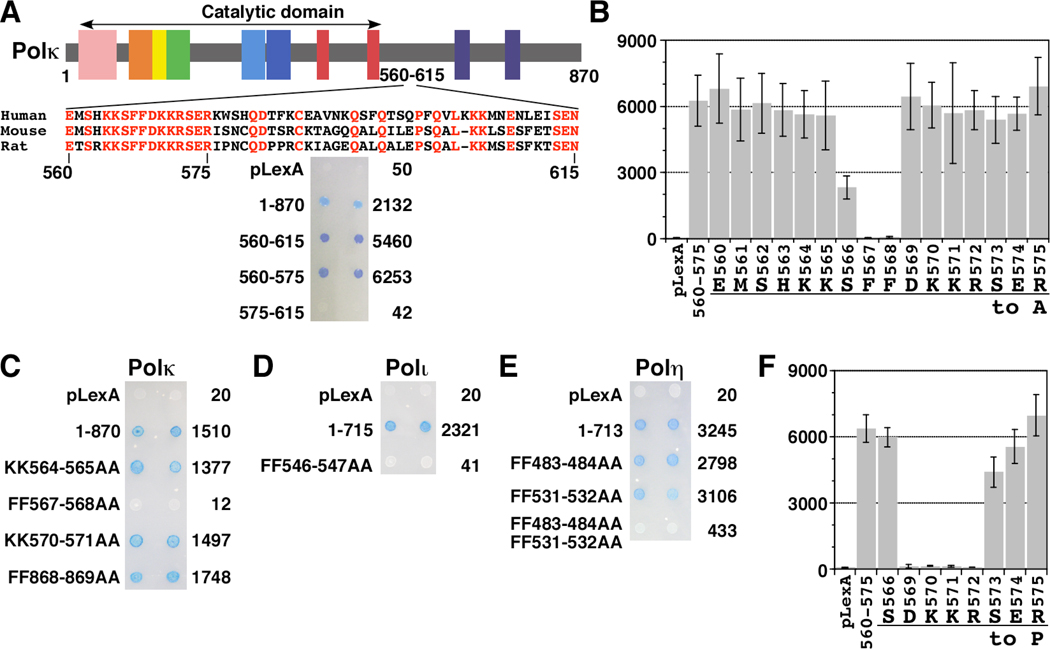
Using a Y2H assay with the pLexA (a vector with DNA binding domain, BD) and pB42AD (a vector with transcription activation domain, AD) plasmids (all of the plasmid constructs used for Y2H assay in this study carry an insert in the C-terminal region of the fusions), we previously showed that 56 amino acids (aa) spanning from 560 to 615 of the human Polκ protein were necessary and sufficient for mediating interaction with the REV1-CTD (Ohashi et al. 2004). Comparison of the amino acid sequences of human, mouse and rat Polκ homologues indicates that the residues 560–575 are highly conserved, whereas the 576–615 region is divergent (Fig. 1A). Further analysis using the same Y2H assay revealed that the highly conserved 560–575 region was sufficient for the REV1 interaction (Fig. 1A). We subsequently employed alanine scanning mutagenesis for each of the 16 amino acids (560–575) to determine which residues within the sequence are critical for the REV1-interaction. As shown in Fig. 1B, the interaction with REV1 was completely abolished when either one of the two phenylalanines (FF) at 567 and 568 was substituted to alanine. Substitution of serine 566 elicited more than 50% decrease of the interaction when the binding activity was measured by β-galactosidase assay, but substitutions of other residues in the 560–575 region did not affect the interaction. We then examined how the substitution of the FF residues at 567–568 to AA in the full-length Polκ (the total length is 870 aa) affects the REV1-interaction. Because Polκ has another FF sequence at 868–869 in the putative PCNA binding site at the extreme C-terminus, we changed the C-terminal FF to AA for comparison. The FF-to-AA substitution at 567–568 completely abolished the REV1 interaction, while the substitution at 868–869 did not affect the interaction (Fig. 1C). On the other hand, the KK-to-AA substitutions at either 564–565 or 570–571 did not affect the REV1-interaction (Fig. 1C). Thus, the FF residues at 567–568 in Polκ are essential for the interaction with REV1.
Figure 1.
Yeast two-hybrid assays showing interaction between REV1 and three Y-family DNA polymerases. Polκ, Polι, or Polη and REV1 were expressed in EGY48 as LexA DNA binding domain fusion protein and B42 transcription activation domain fusion protein, respectively. A, REV1-binding domain of Polκ. Truncation mutants of Polκ were examined for the interaction with REV1 (951–1251 aa) by semi-quantitative β-galactosidase assays (in Miller units). B, Alanine scanning mutagenesis of the Polκ 560–575 region. To perform alanine scanning mutagenesis all codons in the REV1-binding region of Polκ were individually substituted with “gct”. Alanine substitution mutants of Polκ were examined for interaction with REV1 (951–1251 aa) as described above. C, FF-to-AA mutation of Polκ abolishes the interaction with REV1. Wild type and mutants (KK564-565AA, FF575-576AA, KK570-571AA, and FF868-869AA) of Polκ were examined for interaction with REV1 (951–1251 aa) as described above. D, FF-to-AA mutation of Polι abolishes the interaction with REV1. Wild type and FF546-547AA mutant of Polι were examined for the interaction with REV1 (951–1251 aa) as described above. E, FF483-484AA, FF531-532AA double mutation of Polη reduces REV1-binding. Wild type and FF-to-AA double mutants of Polη were examined for interaction with REV1 (951–1251 aa) as described above. F, Proline scanning mutagenesis of the Polκ569-575 region. Proline scanning mutagenesis was performed using primers containing “cct” to substitute individual codons in the REV1-interacting region of Polκ. Mutants of Polκ were examined for the interaction with REV1 (951–1251 aa) as described above.
We then examined whether the two other TLS polymerases (Polι and Polη) have a similar motif in their REV1-interacting regions (RIRs). We previously reported that residues 449–589 of Polι and residues 509–557 of Polη are involved in REV1-binding, as determined by Y2H assays using the pGBKT7(BD)-pGADT7(AD) or pDBLeu(BD)-pPC(AD) vectors, respectively (Ohashi et al. 2004). In contrast, Tissier et al. (2004) showed, using the Y2H assay with the pGBKT7-pACT2 vectors, that amino acids 370–492 of Polη are most important for REV1-binding although the 492–595 region might be also involved in the interaction. We re-examined the REV1-binding of Polι and Polη by using the pLexA-pB42AD vectors. Human Polι has only one FF motif at 547–548 within the above RIR. As shown in Fig. 1D, when FF547-548 of Polι was substituted to AA, its interaction with REV1 was completely abolished. In the case of Polη, there is one FF motif in each of the 370–492 and 509–557 regions (FF483-484 and FF531-532). As shown in Fig. 1E, neither FF483-484AA nor FF531-532AA substitution caused any significant reduction in the REV1-interaction; however, the FF483-484AA FF531-532AA double mutant exhibited only 12% β-galactosidase activity of the wild type (Fig. 1E). This result implies that Polη has two major RIRs around FF483-484 or FF531-532, each of which can bind to REV1 independently.
The above result also suggested that Polη has an extra weak RIR(s) because the FF483-484AA FF531-532AA double mutant showed some residual activity (12% of the wild type) when compared with the Polκ FF567-568AA and Polι FF547-548AA mutants that completely lack REV1-binding activity. Polη has two FF motifs in addition to FF483-484 and FF531-532; one at 17–18 near the dNTP binding site in the DNA polymerase catalytic domain and the other at 707–708 in the putative PCNA-binding sequence at the extreme C-terminus (Haracska et al. 2001). We examined whether the FF motif at 707–708 might be involved in REV1-binding. The FF707-708AA substitution alone or in combination with either one of FF483-484AA and FF531-532AA substitutions did not significantly decrease interaction with REV1-CTD. However, the FF483-484AA FF531-532AA FF707-708AA triple mutant showed 6% activity of the wild type (Supplementary Figure 1A). Thus, we conclude that the FF483-484 and FF531-532 motifs contribute much more significantly to the REV1-binding activity of Polη than the FF707-708 motif.
As shown in Fig. 1A, in the case of Polκ, the 16 amino acid region (560-EMSHKKSFFDKKRSER-575) was sufficient for REV1-binding. To examine whether the corresponding 16 amino acid regions of Polι (539-ASRGVLSFFSKKQMQD-554) and Polη (476-ATTSLESFFQKAAERQK-491 or 524-QSTGTEPFFKQKSLLL-539) had a similar REV1-binding activity, the DNA sequence of the respective region was cloned in the pLexA vector and examined for interaction with REV1. In our Y2H assays Polι residues 539–554 and Polη residues 524–539 each exhibited strong positive signals for the interaction with REV1, while the Polη 476–491 region showed a weaker REV1-interacting activity (Supplementary Figure 1B). Furthermore, when the FF motif in each of these 16 aa regions was substituted to AA, REV1-interaction activity was completely lost (Supplementary Figure 1B). Thus, in Polκ, Polι and Polη, sequences as short as 16 residues are sufficient for mediating interaction with REV1 (see below) and FF motifs are invariably crucial for REV1-binding. It should be noted that no conserved residue is found in the four RIR sequences, except for the central FF (see Table I). While some basic residues are present near the central FF motif, these do not seem to be essential for the REV1-binding (Fig. 1C).
Table I.
Peptides used in this study
| Name | Sequencea | REV1 bindingb / Kd | PCNA bindingc / Kd |
|---|---|---|---|
| κ560-575FF | + / 7.6 µM | ||
| η524-539FF | + / 13 µM | ||
| ι539-554FF | + / 69 µM | ||
| η476-491FF | + | ||
| κ560-575AA | − | ||
| κ562-575FF | + | ||
| κ564-575FF | + | ||
| κ558-571FF | − | ||
| κ856-870FF | − | ||
| η694-713FF | − | + / ~ 0.3 µM | |
| η694-713AA | − |
The N-terminal Cys residue was added for covalent linkage to sensor chip in SPR assay. FF or substituted AA is indicated in bold.
REV1-binding (+ or −) was examined by yeast two-hybrid or SPR assay. The Kd value was estimated from data by SPR assay.
PCNA binding was examined by SPR assay.
Direct Binding Assay by Surface Plasmon Resonance
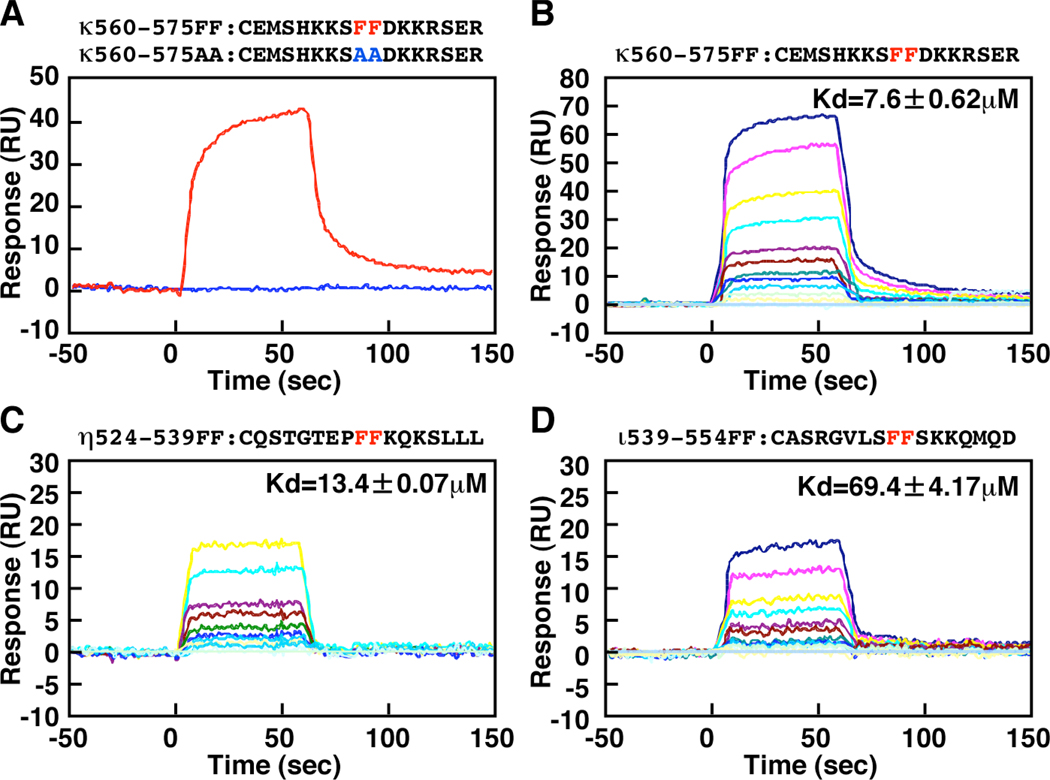
In order to show that peptides containing the above sequence directly bind to the REV1-CTD, we employed surface plasmon resonance (SPR) analysis in which we measured changes in mass on the surface of the sensor chips as a consequence of protein-protein interactions. Two 17 aa peptides corresponding to WT and mutant RIR motifs of Polκ (designated κ560-575FF, CEMSHKKSFFDKKRSER and κ560-575AA, CEMSHKKSAADKKRSER, which differed in the underlined residues) were coupled to the sensor chip via cysteine residues added to the N-termini. The resulting sensor chips were compared for binding with His-REV1(1130–1251). As shown in Fig. 2A, we detected binding of His-REV1(1130–1251) to the κ560-575FF peptide, but not to κ560-575AA. To control for the specificity of interaction with REV1, we used a REV1(1130–1218) derivative lacking a portion essential for the interaction with Polκ (Ohashi et al. 2004). Consistent with our previous results obtained by Y2H assays, REV1(1130–1218) failed to interact with κ560-575FF or κ560-575AA (data not shown). Therefore, we conclude that FF residues in the Polκ RIR are critically required to mediate specific interactions with REV1.
Figure 2.
Surface Plasmon Resonance Spectroscopy using His-REV1(1130–1251) protein and peptides. Binding of His-REV1(1130–1251) protein to immobilized peptides was measured using BIACORE 2000. Peptides were immobilized on the carboxylated dextran matrix of CM5 sensor chips. A, Binding to κ560-575FF or κ560-575AA. B, Binding to κ560-575FF. C, Binding to η524-539FF. D, Binding to ι539-554FF. In the experiments for A, the concentration of His-REV1(1130–1251) was 1.1 µM. In the experiments for B–D, the concentrations of His-REV1(1130–1251) were 1.6, 1.2, 0.8, 0.6, 0.4. 0.3, 0.2, 0.15, 0.1, and 0.05 µM. For experiments shown in B and D, two more concentrations (3.2 and 2.4 µM) of His-REV1(1130–1251) were used. For all binding assays shown in A–D, data are expressed as the signal obtained using each concentration of His-REV1(1130–1251) subtracted from the signal generated with the buffer containing no added protein.
Next, we compared the His-REV1(1130–1251)-binding activity of three different peptides corresponding to the RIR sequences of Polκ, Polι and Polη (see Table I). Based on the results shown in Fig. 2B, C and D, the dissociation constants (Kd) for REV1 binding of the Polκ-, Polι- or Polη-derived RIR peptides were estimated to be approximately 7.6, 13 or 69 µM, respectively. Therefore, the PolκRIR peptide has a higher affinity for REV1-CTD than the Polη and Polι RIR peptides. When a peptide carrying the FF-to-AA substitution in each case was used, we did not detect REV1-CTD binding (data not shown).
To further control for specificity of interactions between FF-containing peptides and REV1-CTD, we carried out similar experiments with a peptide designated η694-713FF (CKRPRPEGMQTLESFFKPLTH) which corresponds to the putative PCNA-binding sequence at the extreme C-terminus of Polη. While we could not detect binding of REV1-CTD to the η694-713FF peptide, PCNA was found to bind to the peptide with an estimated Kd value of approximately 0.4 µM (data not shown). PCNA failed to bind to a PIP box mutant peptide in which FF was substituted to AA (η694-713AA, CKRPRPEGMQTLESAAKPLTH) (data not shown), demonstrating that the interaction between PCNA and η694-713FF was specific.
Minimum Requirement for REV1-Interaction
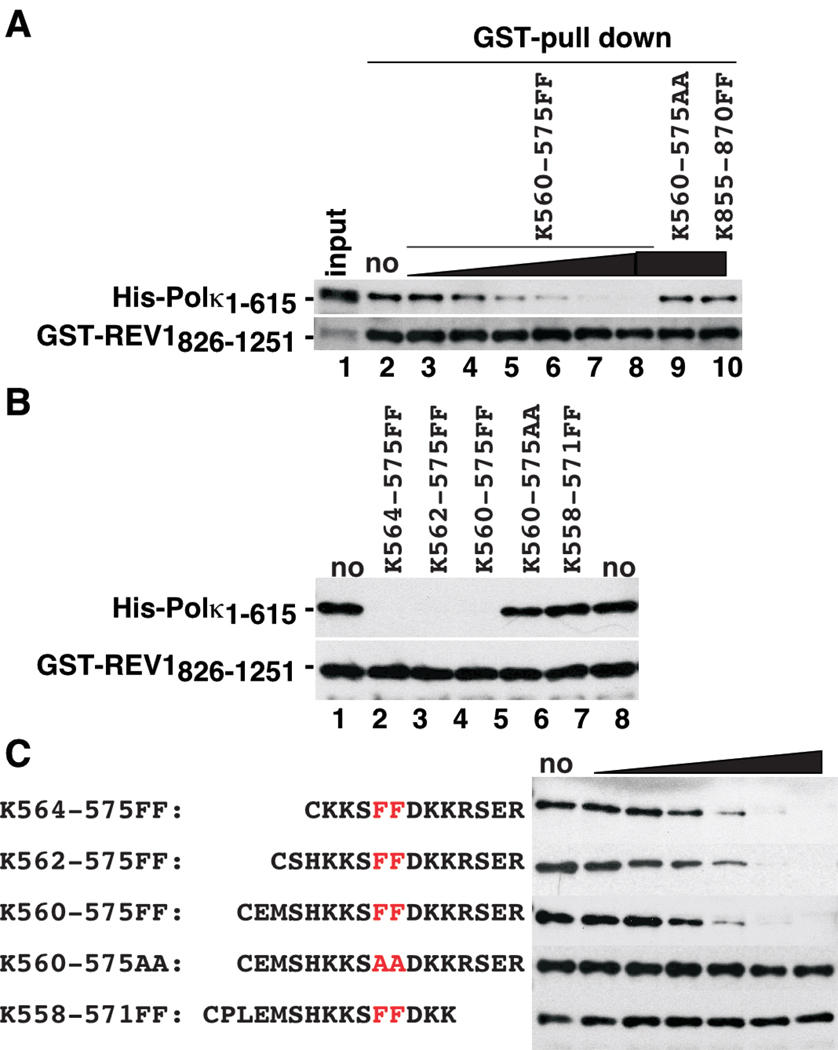
To independently confirm the interactions of Polκ-, Polη-, and Polι-derived sequences with REV1, we employed GST pull-down assays. We previously showed that His-Polκ(1–615) interacts strongly with GST-REV1(826–1251) in GST pull-down assays (Ohashi et al. 2004). Therefore, we determined whether various peptides could compete with His-Polκ(1–615) for binding to GST-REV1(826–1251). In this assay, we immobilized GST-REV1(826–1251) on Glutathione-Sepharose beads, incubated the beads with competitor peptide(s) (or appropriate controls) and then added His-Polκ(1–615) to the reaction mixture. As shown in Fig. 3A, κ560-575FF inhibited the binding of His-Polκ(1–615) to GST-REV1(826–1251) in a dose-dependent manner, while no inhibition was observed with κ560-575AA or κ856-870FF even at the highest concentration tested. This result further strengthens the notion that REV1 interacts with Polκ solely at a short sequence flanking FF567-568.
Figure 3.
Inhibition of the interaction between GST-REV1(826–1251) and His-Polκ(1–615) proteins by peptides. A, Inhibition of the interaction between the GST-REV1(826–1251) and His-Polκ(1–615) proteins (0.1 and 0.03 µM, respectively) by κ560-575FF (0.032, 0.16, 0.8, 4, 20, 100 µM), κ560-575AA (100 µM) or κ855-870FF (100 µM). A five-fold excess of bound fraction was applied to the gel, compared to the input fractions. Experiments were carried out as described under ‘Experimental Procedures’. B, Effect of shortening N-terminal sequences on binding of FF-peptides to REV1. Experiments were performed as described for A, except that κ560-575FF, κ562-575FF, κ564-575FF, κ560-575AA or κ558-571FF were added at a fixed concentration of 100 µM. C, Effect of shortening C-terminal sequences on binding of FF-peptides to REV1. Experiments were performed as described for A. The indicated peptides were added at various concentrations (0.032, 0.16, 0.8, 4, 20, 100 µM).
In peptide competition assays when we examined two shorter peptides truncated at the N-terminal of the FF motif in the Polκ RIR sequence (κ562-575FF, CSHKKSFFDKKRSER and κ564-575FF, CKKSFFDKKRSER), both inhibited the interaction between His-Polκ(1–615) and GST-REV1(826–1251) as efficiently as κ560-575FF peptide. Therefore, peptides containing as few as three amino acid residues N-terminal to the 567–568 FF are sufficient to compete for REV1-binding (Fig. 3B, C). However, in contrast, another peptide containing only three residues C-terminal to the FF motif (κ558-571FF, CPLEMSHKKSFFDKK) did not inhibit the binding of His-Polκ(1–615) to GST-REV1(826-1251). The failure of κ558-571FF to bind REV1-CTD was confirmed independently by SPR assay (data not shown), thus indicating that more than 3 residues C-terminal to the FF motif are necessary for REV1 interaction. The finding that several residues C-terminal to the FF motif in the Polκ RIR sequence are required for REV1 binding is rather unexpected because no conserved amino acids are found in the C-terminal to the FF motifs of the four RIRs derived from Polκ, Polι and Polη and also because alanine substitution of R572, S573, E574 or R575 in Polκ did not affect REV1-binding (Fig. 1B).
The apparent discrepancy between a requirement for several residues C-terminal to the critical FF and the lack of conservation in the RIR of different TLS polymerases might be explained if the Polκ-REV1 interaction depends on main-chain interactions, rather than on side-chain interactions of specific amino acid residues. To test this possibility, we performed proline scanning mutagenesis for each residue in the Polκ region 569–575. While proline is well known to disrupt helical structures, its presence also weakens the β sheet structure because of the lack of hydrogen for main-chain interactions and consequent failure to interact with oxygen in the opposing strand. The results shown in Fig. 1F clearly demonstrate that proline substitution of the D569, K570, K571 or R572 residue proximal to the critical FF motif abolished the interaction with REV1, while the substitution of the distal S573, E574 or R575 did not affect the interaction. While Ser precedes the FF motif in three of the four RIRs, the FF motif in the Polη sequence 524-QSTGTEPFFKQKSLLL-539 is preceded by Pro. As expected, the proline substitution of S566 did not affect the REV1-interaction (Fig. 1F). From these results, we conclude that a minimum of 4 residues at the C-terminal residues of the FF motif are required for binding to the C-terminal region of REV1, probably through interaction between main chains.
REV1-Interaction Is Essential for Polκ Function
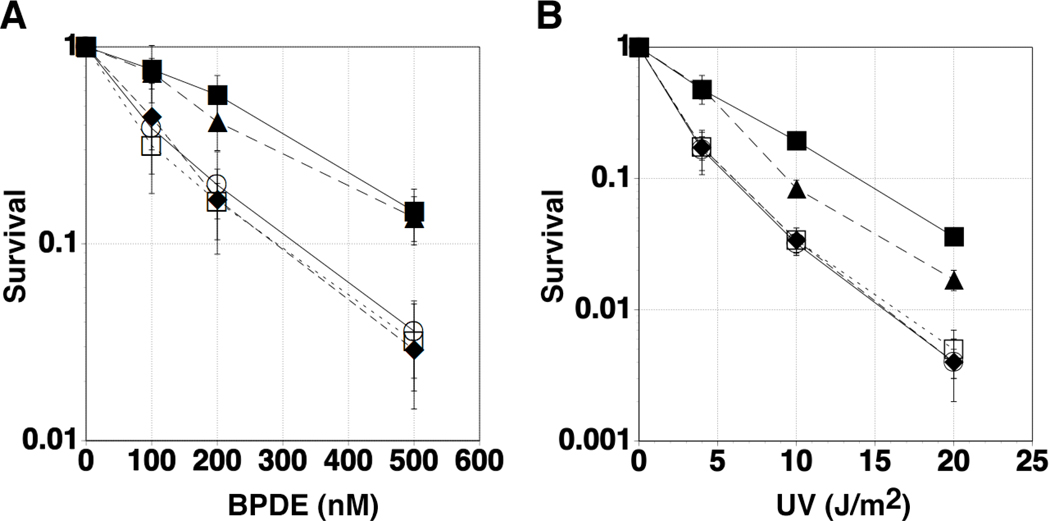
Finally, to address the biological relevance of the interaction between Polκ and REV1, we employed complementation assays using Polk−/− MEF (mouse embryonic fibroblast) cells. We have shown that as Polk−/− ES (embryonic stem) cells are sensitive to benzo[a]pyrene (Ogi et al. 2002), Polk−/− MEF cells are sensitive to BPDE, an activated form of benzo[a]pyrene (Bi et al. 2005). As expected, transient expression of GFP-Polκ(WT) in the Polk−/− MEF cells restored the BPDE-resistance of the cells, but expression of GFP-Polκ(FF567-568AA) or GFP (for control) failed to correct BPDE-sensitivity (Fig. 4A). Similarly, GFP-Polκ(WT), but not GFP-Polκ(FF567-568AA) or GFP, complemented UV-sensitivity of Polk−/− MEF cells (Fig. 4B), although it is still unclear why Polk−/− MEF cells are sensitive to UV-irradiation (Ogi & Lehmann 2006). We examined FLAG-Polκ(FF567-568AA) for co-immunoprecipitation with the endogenous REV1 protein in vivo and found that the mutant had a significantly reduced activity for REV1-binding compared with the wild-type (Supplementary Figure 2). We could not see any significant difference in PCNA binding between the mutant and wild-type Polκ in BPDE-treated cells (Supplementary Figure 3). It should be noted that both the mutant and wild-type Polκ bound to monoubiquitinated PCNA in BPDE-treated cells, indicating that at least one of the two ubiquitin-binding domains (designated UBZs) was involved in the binding. This result eliminates a trivial explanation that the reduced REV1-binding activity of the Polκ mutant protein is due to failure in protein folding. Thus, we conclude that the interaction with REV1 is essential for Polκ function in vivo.
Figure 4.
Effect of FF567-568AA substitutions on biological activity of Polκ. Polk+/+ (filled square), Polk−/− (open circle), Polk−/− cells transiently expressing GFP (open square), GFP-Polκ(WT) (filled triangle), and GFP-Polκ(FF567-568AA) (filled diamond) were exposed to the indicated concentrations of BPDE (A) or were irradiated with the indicated doses of UVC (B). Five to eight days following genotoxin treatments, cells were fixed and stained as described under ‘Experimental Procedures’. Data points represent means of duplicate determinations and error bars are ± SDs.
Discussion
Here we have shown that very short sequences of Polκ, Polι and Polη containing an FF motif are sufficient for the interactions with REV1-CTD. The crucial role of FF in the RIRs is reminiscent of PCNA-interacting protein (PIP) box sequences, frequently represented by Q-x-x-(I, L, M)-x-x-F-F (for a recent review, see Moldovan et al. 2007). In PIP box sequences, conserved amino acids in the N-terminal side of FF are required for interacting with multiple residues of PCNA and adopting a 310 helical conformation that directs the side chain of the FF motif into a hydrophobic pocket of PCNA (Gulbis et al. 1996). Our alanine scanning experiments show that in contrast with PIP boxes, Polκ RIR residues N-terminal to the critical FF can be changed without any significant loss of REV1-binding activity. This finding is consistent with the lack of conservation of residues N-terminal to the FF in the four RIR sequences of Polκ, Polι and Polη. Furthermore, a peptide (κ564-575FF) containing only three residues (plus cysteine) N-terminal to the FF in the RIR of Polκ bound REV1-CTD efficiently. These results suggest that specific sequences of residues N-terminal to FF in the REV1-binding motif play little (if any) role in the interaction with REV1.
In PIP box sequences, residues C-terminal to the FF appear to be non-essential for PCNA-binding. However, in the RIR of Polκ, the presence of several residues in the C-terminal side of FF is essential for association with REV1; deletion of residues immediately C-terminal to the FF motif resulted in a complete loss of REV1-binding activity. Thus, although C-terminal RIR residues are required downstream of the critical FF, specific C-terminal sequences do not need to be conserved to confer REV1 interaction. Proline substitution of the four residues C-terminal to the FF within the Polκ RIR (D569, K570, K571 and R572) abrogated the REV1-interaction, whereas alanine substitution of the same four residues did not affect the interaction. Thus, the interaction with REV1-CTD requires the presence of several amino acid residues, other than proline, C-terminal to the critical FF motif. These results explain well why the C-terminal PIP box sequences of Polκ and Polη do not bind effectively to REV1-CTD, in spite of the fact that both contain FF. The PIP box of Polκ has only one residue C-terminal to the FF. While the PIP box of Polη has five residues, it contains proline at the second position following the FF. Therefore, the PIP box sequences of Polκ and Polη both lack the structural requirements for interactions with REV1.
The FF motif of RIR is likely to engage in a hydrophobic interaction with a portion of the REV1-CTD, similar to the FF sequence of PIP boxes which interact with PCNA (Gulbis et al. 1996). It is conceivable that the hydrophobic interaction is stabilized by the presence of several residues following the FF motif. However, the lack of any conserved residues C-terminal to FF implies that the putative stabilization does not depend critically on any specific side-chain interactions. The result that proline substitution immediately downstream of the FF motif disrupted the REV1-interaction supports our interpretation that the stabilization depends on main-chain interactions. Proline is an imino acid, rather than an amino acid, and therefore cannot act as a hydrogen donor in main-chain interactions for β sheet formation. Our SPR analysis showed that His-REV1(1130–1251) specifically interacts with κ560-575FF with a dissociation constant (Kd) value of 7.6 µM (Fig. 2B). In such a relatively weak protein-protein interaction, loss of a single hydrogen bond due to substitution by proline may result in significant weakening of the interaction. Furthermore, it should be noted that every other residue in a sequence, but not any two consecutive residues, acts as hydrogen donor in either parallel or anti-parallel β sheet interaction. Proline substitution in any one of the four consecutive residues following FF abrogated the REV1-interaction, suggesting that the Polκ RIR adopts a structure similar to mixed β sheets. Another possibility is that the presence of proline near the FF motif generates a curve in the orientation of the peptide chain, thereby reducing main-chain interactions by the downstream sequence.
Our results described here demonstrate that short FF-containing sequences of Polη, Polι, and Polκ are crucial for an interaction with the REV1-CTD. However, REV7, which is the accessory subunit of Polζ and also interacts with the REV1-CTD (Murakumo et al. 2001), lacks any FF motifs in the total 211 amino acid sequence. Because a minimal REV7 region of longer than 100 amino acids is necessary for its interaction with REV1-CTD (Murakumo et al. 2001 and our unpublished results), we believe that REV7 interacts with REV1-CTD differently from Polη, Polι or Polκ, and that both REV7 and REV1-CTD most probably recognize tertiary structure of the partner protein rather than short primary sequences.
What is the biological role of the REV1-binding of the Y-family DNA polymerases? As shown here, the FF567-568AA mutant of Polκ that cannot interact with REV1 failed to restore the BPDE and UV resistance of Polk−/− MEF cells. Because the GFP-fused Polκ proteins lacking RIR (for example, the derivative carrying only the 774–870 residues) still form nuclear foci (Ogi et al. 2005), formation of Polκ foci does not require the interaction with REV1. In fact, we found that the FF567-568AA mutant of Polκ formed nuclear foci in a genotoxin-inducible manner (Supplementary Figure 3C). Similarly, in the case of Polη, only the C-terminal 120 residues (594–713) are required for its focus formation in UV-irradiated cells (Kannouche et al. 2001), indicating that the two RIRs in Polη are dispensable for the focus formation. Furthermore, Vidal et al. (2004) showed that the mutation (YY426-427AA) in the PCNA-binding site (designated PIP1) of Polι abolished focus-forming ability in UV-irradiated cells, but another mutation (FF546-547AA), which was shown in this study to inactivate the REV1-interaction, did not affect the focus formation. These facts suggest that the REV1-interaction is not required for subcellular re-localization of the three polymerases in response to DNA damage. Instead, it appears that interactions of Y-family polymerases with REV1 are required for TLS at a stage subsequent to their PCNA-dependent recruitment to stalled replication forks.
A current model to explain how TLS polymerases are recruited to a site of DNA lesion suggests that PCNA, the sliding clamp which binds Polη, Polι and Polκ, is mono-ubiquitinated upon various DNA-damaging treatments by the Rad6–Rad18 complex (Kannouche et al. 2004; Watanabe et al. 2004). Indeed, Polη, Polι, Polκ and REV1 were all found to have either one or two ubiquitin-binding domains (Bienko et al. 2005). However, if all these enzymes are recruited by the same signal of mono-ubiquitinated PCNA (mUb-PCNA), it is unclear how the appropriate enzyme is selected for bypass of a given lesion. Recruitment and binding to mUb-PCNA of the 'wrong' enzyme for a given lesion could cause inefficient lesion bypass or even cessation of DNA synthesis. Therefore, it is likely that specific mechanisms exist to ensure that inappropriate recruitment of 'wrong' enzymes to DNA lesions is corrected if necessary. In such cases, we suggest that the inappropriately recruited enzyme is replaced with another TLS enzyme and that such exchange is facilitated via interaction with REV1 (Barkley et al. 2007). Furthermore, such a switching may occur between a TLS enzyme inserting a base opposite a DNA lesion and another enzyme (such as Polζ) mediating further chain elongation. This model seems to explain well why Polη, Polι and Polκ each have REV1-binding motif, independently of PCNA-binding motif and why REV7, the non-catalytic subunit of Polζ, also interacts with REV1.
Using the FF483-484AA FF531-532AA double mutant of Polη constructed in this study, Akagi et al. (J. Akagi, C. Masutani, K. Kataoka, T. Kan, E. O., T. Mori, H. O., and F. Hanaoka, submitted for publication) showed that the mutant Polη protein was defective for the interaction with REV1 in cultured human cells, while it exhibits normal enzyme activity when assayed in the crude lysate with a T-T CPD containing template DNA. In contrast with Polκ FF567-568AA (which did not complement the BPDE-sensitivity of Polk−/− cells), the REV1 interaction-deficient mutant of Polη corrected the UV-sensitivity of XPV cells as efficiently as wild-type Polη. Furthermore, upon UV-irradiation re-localization of endogenous REV1 required its interaction with Polη, while the re-localization of Polη occurred independently of REV1. These results suggest that Polη is recruited to sites of UV-induced DNA damage before REV1.
The highly elevated mutation frequency of XPV cells upon UV-irradiation was also corrected by either the wild-type or FF483-484AA FF531-532AA double mutant of Polη. Interestingly however, the enhanced frequency of spontaneous mutations in XPV cells was corrected by expressing wild-type Polη, but not the REV1 interaction-deficient Polη mutant. Therefore, REV1-Polη interactions are dispensable for bypass of UV-induced lesions but are required for TLS of spontaneously-arising DNA damage, such as oxidative damage. These findings are consistent with our model described above that the interaction with REV1 is necessary for replacing the inappropriately recruited enzyme with another TLS enzyme.
We suggest that Polη is sufficient to perform efficient bypass of UV-induced DNA lesions in the absence of REV1, but that bypass of other lesions such as those occurring spontaneously requires alternative TLS polymerases. REV1 likely plays a key role in facilitating the replacement of Polη with other TLS polymerases that are more appropriate for bypass of spontaneously-occurring damage. Similar to spontaneously-occurring lesions, BPDE adducts are not bypassed by Polη, thus explaining why REV1 is necessary for mediating replacement of Polη with Polκ (the appropriate TLS polymerase for bypass of BPDE lesions).
In summary, our findings reveal that short sequences containing FF are important for the weak interaction between REV1 and the other three Y-family DNA polymerases. Such weak interactions are likely to permit rapid and dynamic associations and dissociations among many factors involved in TLS events in vivo.
Experimental Procedures
Plasmids
For yeast two-hybrid assays, POLK-, POLI-, or POLH-cDNA and REV1-cDNA fragments were inserted into pLexA and pB42AD, respectively. For purification of His-tagged truncated Polκ proteins, a POLK-cDNA fragment encoding amino acids 1–615 was inserted into pQE9 (QIAGEN). A plasmid carrying GST-REV1(826–1251) was used for in vitro binding, as described (Ohashi et al. 2004). Bacterially-expressed His-REV1(1130–1251) was used for surface plasmon resonance assays. The cDNA encoding His-REV1(1130–1251) was amplified using a 5’-primer containing a 6×His tag sequence and a 3’-primer containing a XhoI site. A second PCR reaction was used to add a BamHI site at the 5’-end and the resulting DNA fragment was inserted into pGEX5X-1 to construct GST-His-REV1(1130–1251). For expression of Polκ in MEF cells, the WT or mutant POLK-cDNA sequence was inserted into pAC-CMV-GFP (14) or pEGFP (CLONTECH) to produce GFP-fusion proteins.
Yeast two-hybrid assay
Polκ, Polι, or Polη and REV1 were expressed in the yeast strain EGY48 as LexA DNA binding domain fusion proteins and a B42 transcription activation domain fusion protein respectively, using CLONTECH yeast two-hybrid system. Approximately 105 cells were spotted onto SD/−Ura/−His/−Trp/−Leu/+X-gal plates to select positive clones. Semi-quantitative β-galactosidase assays were performed according to the manufacturer’s protocol.
Surface plasmon resonance assay
Surface plasmon resonance assay was performed with BIACORE 2000. Synthetic peptides were immobilized on the carboxylated dextran matrix of CM5 sensor chips through the sulfhydryl group of the N-terminal Cys by thiol activation chemistry, according to the manufacturer’s protocol. GST-His-REV1(1130–1251) was purified through a Glutathione-Sepharose column and then cleaved between the GST and His tags using Factor Xa protease (Amersham Biosciences). Subsequently, His-REV1(1130–1251) was purified through Ni-NTA column. Finally, to eliminate uncleaved GST-His-REV1(1130–1251), His-REV1(1130–1251) was passed through Glutathione-Sepharose column. Varying concentrations of His-hREV1(1130–1251) protein in running buffer (10mM HEPES, pH7.5, 150mM NaCl, 3mM EDTA, 0.005% surfactant P20) were injected over the sensor chip surface at a flow rate of 20 µl/min at 25°C. The contact and dissociation times were 60 and 120 sec, respectively. Binding constants were determined from the titration curves using BIAevaluation 3.0 software. The peptide-protein interaction signal minus the control cell signal was analyzed. The Kd value was calculated according to equilibrium value analysis by averages of measurement signals at respective concentrations.
In vitro GST-pull down assay
His-tagged Polκ(1–615) protein was purified as described previously (Ohashi et al. 2004). GST-REV1(826–1251) protein was expressed in E. coli cells after induction with 0.5 mM IPTG. After incubation for 2 hr at 30°C, the E. coli cells were harvested and lysed in bacterial lysis buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM PMSF, 5 µg/ml leupeptin) by sonication. The lysates were centrifuged at 15,000g for 10 min and the GST-REV1(826–1251) protein was immobilized on Glutathione-Sepharose beads (Amersham Biosciences). 20 µl of Glutathione-Sepharose beads containing 0.1 nmol of the GST-REV1(826–1251) protein were incubated with one of the peptides at 4°C for 2 hr and then with 0.03 nmol of the purified His-Polκ(1–615) protein at 4°C for 4 hr in 1 ml of binding buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.1% Tween-20, 0.75 mg/ml bovine serum albumin (BSA), 1 mM PMSF, 5 µg/ml leupeptin). The beads were washed 5 times with 1 ml of wash buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.1% Tween-20, 1 mM PMSF, 5 µg/ml leupeptin), and bound proteins were eluted by boiling in 1× sample buffer and analyzed by SDS-PAGE followed by immunoblotting assay with anti-His antibody (QIAGEN) or anti-GST antibody (MBL).
Immunoblotting assay
After SDS-PAGE, separated proteins were transferred to an Immobilon P membrane (Millipore) and probed with anti-His, or anti-GST antibody. The target proteins were visualized using the ECL western blotting analysis system (Amersham Biosciences).
Cell culture and reagents
Mouse embryonic fibroblast (MEF) cells were grown in D-MEM medium supplemented with 10% fetal bovine serum. For transient expression, cells were grown on 10-cm culture dishes and transfected with 12 µg of plasmid DNA by using Lipofectamine 2000 (Invitrogen) or TransFectin lipid reagent (Bio-Rad) according to the manufacturer’s protocol. Cells were trypsinized 24 hr after transfection and plated onto 6-well plates. After 12 hr, cells were exposed to BPDE or UV light. Colonies were counted after incubation for 5 to 8 days.
Supplementary Material
Acknowledgement
We thank Drs. Roger Woodgate and Fumio Hanaoka for providing us with Polι and Polη cDNA, respectively and Dr. Yuji Masuda for anti-REV1 antibody. This study was supported by grants-in-aids (17013041, 16370077 to H.O. and 17770003 to E.O.) from the Japanese Ministry of Education, Culture, Sports, Science and Technology and by grant ES09558 from the NIH (to C. V.).
The abbreviations used are
- AD
transcription activation domain
- B[a]P
benzo[a]pyrene
- BD
DNA binding domain
- BPDE
benzo[a]pyrene dihydroxy epoxide
- CPD
cyclobutane pyrimidine dimer
- ES
embryonic stem
- Kd
dissociation constant
- MEF
mouse embryonic fibroblast
- PIP
PCNA-interacting protein
- Pol
DNA polymerase
- REV1-CTD
REV1 C-terminal domain
- RIR
REV1-interacting region
- SPR
surface plasmon resonance
- TLS
translesion DNA synthesis
- XPV
Xeroderma Pigmentosum Variant
- Y2H assay
yeast two-hybrid assay
References
- Barkley LR, Ohmori H, Vaziri C. Integrating S-phase checkpoint signaling with trans-lesion synthesis of bulky DNA adducts. Cell Biochem. Biophys. 2007;47:393–408. doi: 10.1007/s12013-007-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase κ is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Chiapperino D, Kroth H, Kramarczuk IH, Sayre JM, Masutani C, Hanaoka F, Jerina DM, Cheh AM. Preferential misincorporation of purine nucleotides by human DNA polymerase η opposite benzo[a]pyrene 7,8-diol 9,10-epoxide deoxyguanosine adducts. J. Biol. Chem. 2002;277:11765–11771. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. Washington D.C.: ASM Press; 2005. [Google Scholar]
- Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol κ, a DNA polymerase encoded by the human DINB1 gene. J. Biol. Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase θ. Proc. Natl. Acad. Sci. U S A. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999a;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999b;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Moldovan G-L, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- Ogi T, Kannouche P, Lehmann AR. Localisation of human Y-family DNA polymerase κ: relationship to PCNA foci. J. Cell Sci. 2005;118:129–136. doi: 10.1242/jcs.01603. [DOI] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR. The Y-family DNA polymerase κ (pol κ) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polκ protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl. Acad. Sci. U S A. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. Translesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Ohashi E, Kolbanovskiy A, Geacintov NE, Grollman AP, Ohmori H, Shibutani S. Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-N2-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol η and REVl protein. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J. Biol. Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.