Abstract
The C3(1) component of the rat prostate steroid binding protein has been used to target expression of the SV40 T/t-antigen to the mammary epithelium of mice resulting in pre-neoplastic lesions that progress to invasive and metastatic cancer with molecular features of human basal-type breast cancer. However, there are major differences in the histologic architecture of the stromal and epithelial elements between the mouse and human mammary glands. The rat mammary gland is more enriched with epithelial and stromal components than the mouse and more closely resembles the cellular composition of the human gland. Additionally, existing rat models of mammary cancer are typically estrogen receptor positive and hormone responsive, unlike most genetically engineered mouse mammary cancer models. In an attempt to develop a mammary cancer model that might more closely resemble the pathology of human breast cancer, we generated a novel C3(1)/SV40 T/t-antigen transgenic rat model that developed progressive mammary lesions leading to highly invasive adenocarcinomas. However, aggressive tumor development prevented the establishment of transgenic lines. Characterization of the tumors revealed that they were primarily estrogen receptor and progesterone receptor negative, and either her2/neu positive or negative, resembling human triple-negative or Her2 positive breast cancer. Tumors expressed the basal marker K14, as well as the luminal marker K18, and were negative for smooth muscle actin. The triple negative phenotype has not been previously reported in a rat mammary cancer model. Further development of a C3(1)SV40 T/t-antigen based model could establish valuable transgenic rat lines that develop basal-type mammary tumors.
Keywords: Breast cancer, C3(1)/SV40 T/t-antigen, C3(1)/Tag, Transgenic rat, Basal-type breast cancer, Triple-negative breast cancer
Introduction
Breast cancer is the second leading type of cancer and cause of cancer mortality of women in the United States (CDC 2009; Doane et al. 2006). There is a 12% cumulative lifetime risk for US women to develop breast cancer, with an overall age-adjusted mortality risk of 3.5% (Horner et al. 2009). While much progress has been made in the study of breast cancer at the molecular level, the complexity of the disease requires further understanding of the interactions of multiple pathways and processes within the complex organism. To address the complexity and heterogeneity of breast cancer in vivo, numerous genetically engineered mouse (GEM) models of mammary cancer have been developed to recapitulate some of the known genetic and biologic interactions occurring in human breast cancer (Van Dyke and Jacks 2002). These models provide tremendous insight into the molecular biology of breast cancer pathogenesis, including growth factors, oncogenes, tumor suppressor genes, and cell cycle regulators (Barkan et al. 2004; Kavanaugh et al. 2002).
GEM models have greatly advanced the study of human breast cancer in several ways (Ottewell et al. 2006). However, there are important biological and histological differences between the human and mouse that place limitations on the utility of certain GEM and chemically induced mouse models for the study of human breast cancer. First, the cellular composition of the mouse and human mammary glands differ significantly. While the mouse mammary gland is composed of scant stroma, glandular and ductular epithelial units embedded in large amounts of adipose tissue, the human gland contains significantly more epithelium and stroma. Additionally, the mouse models generally do not exhibit the same degree of inflammation and fibrosis as commonly observed in human breast cancer. Most importantly, while well over half of human breast cancers are estrogen receptor alpha (ERα) positive (Rakha et al. 2008), a vast majority of GEM- and chemically-induced mouse mammary tumors are ER-negative and hormone-independent (Cardiff 2001; Nandi et al. 1995). In contrast to the mouse, most mammary tumors in the rat (primarily through chemical carcinogenesis approaches) are hormone dependent, similar to their human counterparts (Smits et al. 2007). The terminal end bud (TEB) in the rat and the lobulo-alveoar unit (LA) in the mouse are the functional units in the mammary gland similar to the terminal ductal lobular unit (TDLU) in humans, and are the site of origin of the vast majority of mammary tumors in all three species (Cardiff and Wellings 1999; Russo et al. 1983, 1990).
More recently, global gene expression profiling has revealed that several distinct molecular sub-types of human breast cancer exist that may be classified as having a basal phenotype (including triple negative and most Her2+ tumors) or luminal features (ER+ or ER−) (Perou et al. 1999; Sorlie et al. 2001). While the classification and nosologic interpretation of breast tumors using these signatures remains controversial, basal type tumors lacking ER, PR and Her2/neu expression are particularly associated with a poor prognosis and limited treatment options exist to cure this sub-type of breast cancer (Doane et al. 2006; Perou et al. 1999; Skoog et al. 1987; Sorlie et al. 2001). Therefore, the development of animal models for basal-type breast cancer is particularly important to further study molecular pathways involved in this sub-type of breast cancer as well as for appropriate pre-clinical testing.
The C3(1)/SV40 T/t-antigen (C3(1)/Tag) transgenic mouse model of basal-type mammary cancer has been extensively studied and used for pre-clinical testing (Green et al. 2001; Kavanaugh and Green 2003; Kavanaugh et al. 2002; Wu et al. 2000). Mice carrying this transgene develop pre-invasive mammary intraepithelial neoplastic (MIN) lesions including atypical ductal hyperplasia (ADH) and ductal carcinoma in situ (DCIS) that are initially ER+, but lose ER expression as they progress to carcinoma (Green et al. 2000b; Maroulakou et al. 1994). Beginning around 8 weeks of age, the earliest lesions observable in female mice are early stage MIN, typically progressing to atypical high grade MIN and DCIS at around 12 weeks of age. These lesions further progress to invasive carcinoma beginning around 16 weeks of age. Animals generally require euthanasia due to the development of large mammary adenocarcinomas by 6 months of age, and metastasis to the lungs occurs in approximately 15% of animals in the FVB background, although metastatic lesions to the liver, adrenal gland, and heart have also been observed (Green et al. 2000b; Shibata et al. 1998).
Although C3(1)/Tag transgenic mice have been an extremely useful model of human breast cancer, developing a model of basal-type breast cancer in the rat would potentially offer particular advantages since the rat mammary gland is structurally more similar to that of the human than is the mouse. Secondly, a large amount of the pre-clinical toxicologic data has been collected from studies in rats, making a rat model particularly appropriate for the study of chemoprevention and chemotherapy. A majority of recently developed rat transgenic mammary cancer models employ overexpression of oncogenes such as Neu and Ras under the control of the MMTV promoter to mimic human breast cancer (Smits et al. 2007). However, our C3(1)/Tag rat model inactivates the p53 and Rb tumor suppressor pathways to generate mammary tumors that are triple-negative or Her2-positive. P53 and Rb functions are usually lost in human basal type breast cancers (Herschkowitz et al. 2008).
In this study, we report the development and characterization of C3(1)/Tag transgenic Sprague–Dawley rats that developed aggressive triple-negative or Her2-positive mammary tumors. Expression of this transgene, however, also led to the development of several additional types of epithelial cancers in the rat. These results demonstrate that it is feasible to generate a transgenic rat mammary cancer model with triple negative features, but modification of the transgene will likely be necessary to develop lines of rats that develop a more limited cancer phenotype that can be passed through germline transmission.
Materials and methods
Construction of the C(1)3-SV40 T/t-antigen transgene and creation of transgenic rats
Generation of the C3(1)/Tag transgene has previously been described (Maroulakou et al. 1994). The C3(1)/Tag transgene was excised from plasmid sequences and transgenic rats were generated by Chrysalis DNX Transgenic Sciences (Princeton, NJ) as described (Green et al. 2000a). Transgenic offspring were identified by Southern blot and slot blot techniques utilizing a 32P-labeled probe specific for SV40 sequences as previously described (Maroulakou et al. 1994). RNA was extracted from tissues using the guanidine thiocyanate/cesium chloride method (Chomczynski and Sacchi 2006). Twenty micrograms of total RNA was separated using a denaturing 1% agarose/formaldehyde gel, transferred to nitrocellulose membrane and SV40 transcripts were identified using the Sfi1-BamH1 fragment of the SV40 early region as 32P-labeled probe by as previously described (Maroulakou et al. 1994).
Necropsy, histopathology, and immunohistochemistry
Gross lesions and major organs were collected and fixed in 4% paraformaldehyde (PFA). Tissues were embedded in paraffin and 4um sections were stained with hematoxylin/eosin (H&E) or immunostained using the Vector ABC system (Vector Laboratories). After antigen retrieval by microwave in citrate buffer or distilled water and blocking with sheep or goat serum, mammary tumors were immunostained with cytokeratin (CK) 14 (1:200, The Binding Site, San Diego CA), alpha-smooth muscle actin (αSMA) (1:500, Sigma, St. Louis MO), estrogen receptor-alpha (ERα) (1:300, Santa Cruz Biotechnology, Santa Cruz CA), progesterone receptor (PR) (1:100, Dako, Carpenteria CA), SV40-Tag (1:25, Pharmingen, San Diego CA), Her2 (1:320, Cell Signaling Technology, Danvers MA), and p53 (1:500, Oncogene, Gibbstown NJ) overnight, and CK18 (1:1600, The Binding Site, San Diego CA), for 1 h at room temperature. Following secondary antibody application, diaminobenzidine (DAB) was used to visualize all immunohistochemical reactions.
Results
Identification of C3(1) −SV40 T/t-antigen transgenic rats
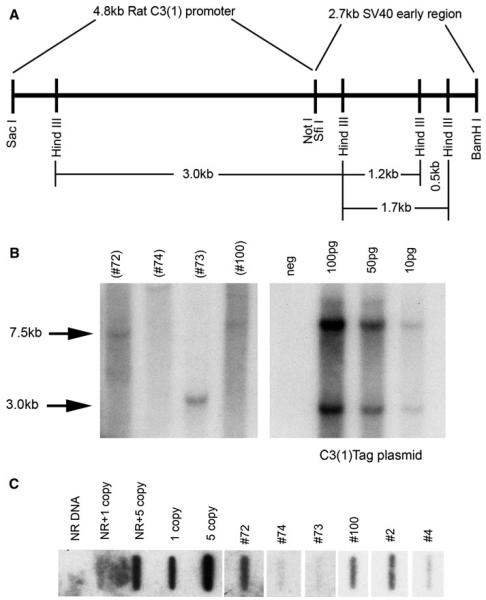
Six founder animals were produced as determined by Southern blot and slot blot analysis. Figure 1a illustrates the construction of the transgene, including the 4.8 kb rat C3(1) promoter and the 2.7 kb SV40 early region. Founder DNA was digested with SacI and BamHI to release the 7.5 kb C3(1)/Tag injection fragment (Fig. 1b). The control panel (right) depicts the cut plasmid containing the 7.5 kb transgene fragment and the 3 kb plasmid vector fragment. A total of four founder rats carrying the 7.5 kb full length transgene were identified and confirmed by Southern analyses (designated founders #2, #4, #72, and #100). A founder containing only a 3 kb transgenic fragment was observed (#73) in one animal that did not exhibit a phenotype. Transgene copy number was estimated by slot blot analysis (Fig. 1c) to between one and 5 copies in the four founder rats carrying the 7.5 kb transgene. The presence of the 7.5 kb transgene correlated with phenotypic abnormalities in the founder rats.
Fig. 1.
Construction of the C3(1)/Tag transgene (a). Southern blot (b) identifying the 7.5 kb transgene in positive (#72, #100) founders, and a 3 kb partial transgene fragment (#73). Presence of the 7.5 kb transgene correlated with phenotype. Right panel shows the C3(1)/Tag plasmid control. The 2.9 kb fragment represents the plasmid backbone. Slot blot (b) confirming the transgene in positive founder rats. Southern and slot blots performed using 32P-labeled Sfi1-BamH1 SV40 early region as probe
Phenotypic abnormalities of founder rats and offspring
Transgenic founder rats demonstrated several different pathologies related to transgene expression at relatively early ages. Overall survival was directly related to the development of mammary carcinomas in the founders with an intact transgene. Of these four founder rats, survival ranged from 12 to 15 weeks for three of four founders (one male and two females), and one female founder survived until 29 weeks of age. Gross and histopathologic examination revealed the development of aggressive mammary carcinomas in one male and three female founders. Precancerous lesions in the mammary glands were characterized as atypical hyperplasia and ductal carcinoma in situ. Of the four founder rats with phenotype, only one transmitted the transgene to offspring. First generation offspring of this founder also developed similar mammary adenocarcinomas early in life. However, this line could not be propagated. Additionally, lesions in tissues other than the mammary glands were observed in the C3(1)/Tag founder rats and are listed in Table 1.
Table 1.
Non-mammary pathology in SV40 T-antigen founder rats
| Histopathologic lesion | Founder 1 72 Female |
Founder 2 002 Female |
Founder 3 004 Male |
Founder 4 100 Female |
|---|---|---|---|---|
| Age at death | 29 weeks | 14.8 weeks | 12.1 weeks | 15 weeks |
| Pituitary gland adenoma | X | X | X | |
| Thyroid gland follicular carcinoma | X | |||
| Parotid salivary gland carcinoma | X | |||
| Pancreatic islet cell carcinoma | X | X | X | |
| Parotid salivary gland atypical hyperplasia | X | |||
| Harderian gland atypical hyperplasia | X | |||
| Nasal submucosal gland atypical hyperplasia | X | |||
| Pulmonary alveolar hyperplasia | X | X |
Development of mammary adenocarcinomas in C3(1) SV40 T/t-antigen transgenic rats
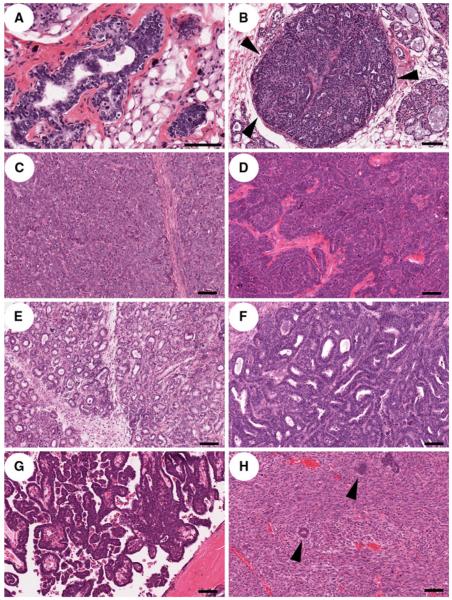
As previously reported for the C3(1)/Tag transgenic mouse model developed in our laboratory (Maroulakou et al. 1994), transgenic founder rats developed mammary carcinomas progressing from precancerous lesions (ADH, MIN [DCIS]) (Fig. 2a, b) to invasive carcinomas. While the solid phenotype was common in the rat tumors, there were numerous other histologic variants present within some tumors, including trabecular, tubuloacinar, papillary, and scirrhous variants. Solid mammary carcinomas (Fig. 2c) were characterized by lack of tubule formation, solid sheets and lobules of cells with scant faintly basophilic amorphous cytoplasm, high nuclear-cyto-plasmic ratio, and frequent mitoses that were often bizarre. There were often central foci to comedo-type zones of necrosis present. Areas of trabecular phenotype (Fig. 2d) were characterized by cords or projections of neoplastic cells surrounded by a fine fibrovascular stroma, and the tubuloacinar phenotype was characterized by clusters of numerous small round luminal structures (acini) (Fig. 2e) to rows of tubules lined by neoplastic mammary epithelial cells (Fig. 2f). The papillary phenotype (Fig. 2g) was characterized by fronds and arboriform projections into a luminal space, often containing proteinaceous fluid, and areas of scirrhous carcinoma (Fig. 2h) were composed of small clusters or nests of poorly-differentiated epithelial cells embedded within a dense desmoplastic stroma.
Fig. 2.
Mammary intraepithelial neoplasia (MIN) and mammary carcinomas resulting from expression of T-antigen in the mammary epithelium. a Atypical ductal hyperplasia (ADH, low grade MIN lesion). Note multiple layers of disorganized and atypical epithelial cells with frequent mitotic figures (arrowheads). b Ductal carcinoma in situ (DCIS, high grade MIN lesion). Note filling and expansion of ductal lumens with poorly differentiated mammary epithelial cells without breach of the underlying basal lamina (arrowheads). c Solid mammary carcinoma characterized by sheets of neoplastic cells separated by fine fibrovascular stroma. d Trabecular carcinoma composed of rows of neoplastic cells without lumen formation. e Acinar adenocarcinoma composed of proliferation of small round luminal spaces without tubule formation. f Tubular adenocarcinoma characterized by formation of long branching luminal spaces. g Papillary adenocarcinoma characterized by arboriform branching of neoplastic cells overlying fronds of stroma projecting into a luminal space. h Scirrhous carcinoma characterized by clusters of neoplastic epithelial cells (arrowheads) embedded in a marked desmoplastic stroma. Bar = 100 μm
Expression of SV40 Tag and immunophenotype of mammary tumors
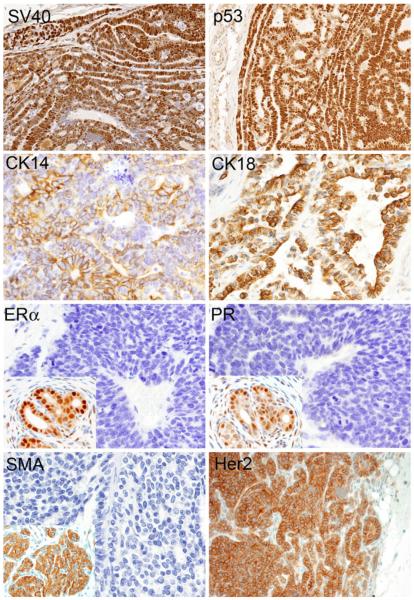
Mammary carcinomas from all animals expressed SV40 T/t-antigen by immunohistochemistry (Fig. 3). Additionally, all tumors were diffusely immunopositive for p53, CK14, and CK18. Tumors were also consistently negative for αSMA, ERα, PR, and were either Her2neu (C-erbB2) positive (Fig. 3) or negative (data not shown), consistent with the immunophenotype seen in triple-negative or Her2 positive human breast cancer. Two out of 12 animals examined by immunohistochemistry, however, exhibited very weak cytoplasmic ERα and PR immunostaining, which could be interpreted as negative by current criteria.
Fig. 3.
Immunohistochemical phenotype of C3(1)/Tag mammary tumors. Neoplastic cells show multifocal nuclear immunoreactivity to antibodies for SV40 and p53, and multifocal membrane immunoreactivity to antibodies for CK14 and CK18. Two our of four mammary tumors expressed Her2. Neoplastic cells are diffusely negative for antibodies to ERα, PR, and αSMA (Insets, positive controls)
Non-mammary pathology related to C3(1) SV40 T/t-antigen transgene expression
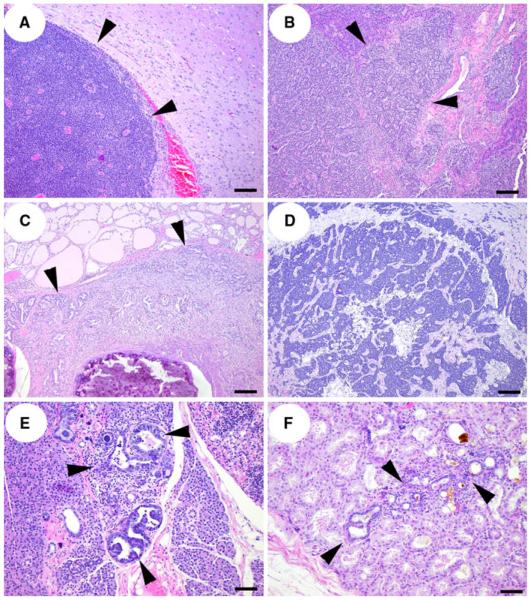
In addition to precancerous mammary lesions and mammary carcinomas, other pathology related to transgene expression included pituitary adeomas, islet cell carcinomas, thyroid follicular carcinoma, and parotid gland carcinoma (Table 1; Fig. 4). Additionally, several precancerous and hyperplastic lesions were observed, such as atypical hyperplasia in the salivary gland, Harderian gland, and nasal submucosal glands, and pulmonary alveolar hyperplasia. Expression of SV40 Tag protein was also detected by immunohistochemistry in proliferative lesions and tumors found in other organs (data not shown).
Fig. 4.
Non-mammary lesions found in C3(1)/Tag founder rats. a pituitary adenoma (arrowheads) compressing adjacent neuropil. b Islet cell carcinoma infiltrating normal adjacent exocrine pancreatic parenchyma (arrowheads). c Thyroid follicular carcinoma (arrowheads) infiltrating adjacent tracheal submucosal tissues and surrounding tracheal rings. d Parotid salivary gland carcinoma. e Parotid salivary gland atypical hyperplasia (arrowheads) and f Harderian gland atypical hyperplasia (arrowheads). a, b, c, d: Bar = 100 μm, e, f: Bar = 50 μm
Discussion
The C3(1) 5′ region of the rat prostatic steroid binding protein has been previously used in our laboratory to direct the expression of SV40-T/t-antigens to the prostate and mammary glands of mice (Maroulakou et al. 1994). SV40-T/t-antigens induce cellular transformation in large part through its ability to bind to and inactivate p53 and Rb, two tumor suppressor genes critical for cell cycle regulation and genome stability. SV40-T/t-antigen binds p53, functionally inactivating it, as well as sequestering the tumor suppressor gene from degradation, thus increasing its expression in the nucleus Likewise, it also binds to unphosphorylated Rb, sequestering the active form from binding to E2F, thus abrogating the cell cycle checkpoint (Ali and DeCaprio 2001). A significant portion of human breast and many other human cancers have been shown to harbor mutations that effectively inactivate or mutate these tumor suppressor genes leading to abnormalities in cell cycle control, proliferation, and apoptosis (Ludlow 1993; Nielsen et al. 1997; Troester et al. 2006). In the present study, transgenic rats were produced that developed mammary carcinomas recapitulating important features of triple-negative or Her2-positive tumors.
Although the pathogenesis of breast cancer remains poorly understood, gene expression profiling has identified at least five basic molecular subtypes of human breast cancer: basal, Her2 positive, luminal A, luminal B, and normal breast subtypes (Dabbs et al. 2006; Perou et al. 1999, 2000; Sorlie et al. 2001,2003). The basal subtype in particular has been shown to be associated with hormone (ERα-, PR-) independence, negative Her2/neu (c-ErbB2) status, and a poor prognosis (Dabbs et al. 2006; Doane et al. 2006; Skoog et al. 1987). These tumors express markers present on normal basal cells (or myoepithelial cells) of the breast, including CK5/6, CK14, CK17, vimentin, and Her-1 (Dabbs et al. 2006; Gusterson et al. 2005; Livasy et al. 2006; Nielsen et al. 2004; Ribeiro-Silva et al. 2005). Although molecular categorization of the heterogeneous collection of breast cancers has been integrated into many articles, this categorization remains controversial. For example, some tumors that cluster within the Her2 type of breast cancers may lack Her2 protein expression, and ER and Her2 type breast cancers often fall into the luminal B subgroup (Weigelt and Reis-Filho 2009). Furthermore, the use of cytokeratins to characterize the origins of human breast cancer has suggested that basal breast cancers are positive for CK14, CK17, and CK5, and negative for the luminal markers CK8, CK18, and CK19. However, both luminal and basal markers may be expressed in some human breast cancers (Gusterson 2009; Gusterson et al. 2005). Normal rat mammary epithelium in this model consistently expressed CK18, and was negative for CK14, consistent with a previous report (Dundas et al. 1991). However, given that it is found in the human TDLU (Gusterson 2009; Gusterson et al. 2005), this could be reexamined in the rat.
The rat model recapitulated several important aspects of human breast cancer not generally observed in the C3(1)/Tag mouse model. In human breast cancer, as part of the multifactorial Nottingham prognostic index, histologic phenotype often correlates with malignant behavior and prognosis (Ellis et al. 1992; Elston and Ellis 2002). In the mouse model, tumors are often tubular adenocarcinomas, the most common human histologic subtype (Shibata et al. 1998). However, in the rat model, several histologic subtypes of mammary tumors were more readily observed ranging from tubular or tubuloacinar adenocarcinoma to scirrhous carcinoma, a tumor type in human breast cancer that has a poor prognosis (Kitagawa et al. 2004; Nozoe et al. 2007) with a prominent stromal component. These data indicate that the rat model may recapitulate more phenotypic heterogeneity as seen in human breast cancer.
Secondly and most importantly, tumors from the rat model recapitulate the immunophenotypes of triple negative (ER–, PR–, Her2–) or Her2-positive cancers observed in sub-sets of human breast cancers. Although several GEM models including the Brca+/−, p53+/−, Brcaco/co, MMTV-Cre/p53−/−, and some chemically induced tumor models display basal-type characteristics and certain molecular features similar to the human basal-type, triple-negative tumors (Bennett and Green 2008; Deeb et al. 2007; Herschkowitz et al. 2007; Xu et al. 1999), the C3(1)/Tag mouse mammary cancer model was identified as having the strongest molecular relationships to human basal-type, triple-negative breast cancer (Deeb et al. 2007; Herschkowitz et al. 2007). Although expression profiling could not be performed on the rat tumors, it seems likely that they share similar molecular signatures of the triple-negative tumors as documented for the mouse model.
The rat mammary tumors were also consistently negative for αSMA and multifocally positive for CK18, a luminal marker. Although these tumors as a group expressed markers of both luminal and basal phenotypes, it must be remembered that the basal immunophenotype and triple negative status are not synonymous. Approximately 20% of human triple-negative breast cancers express luminal markers. Triple negative status is based on clinical assays for ER, PR, and Her2 in human breast cancer, whereas the basal phenotype has been primarily based upon gene expression profiling (Anders and Carey 2008). While most triple negative tumors have a basal phenotype, and most basal breast cancers are hormone receptor and HER2 negative, a proportion of triple negative do not express basal markers, and alternatively, some basal breast cancers are hormone receptor positive (Bertucci et al. 2008; Rakha et al. 2008). Therefore, in human triple negative breast cancer, there is significant overlap between biological and clinical characteristics of sporadic triple-negative and basal-like cancers (Reis-Filho and Tutt 2008). Furthermore, basal-type carcinomas may express αSMA by immunohistochemistry, and this is more frequently associated with development of in situ carcinoma (Lerma et al. 2007). Additionally, Brcaco/co mouse model tumors are generally considered basal tumors, although some luminal marker expression can be observed (Herschkowitz et al. 2007), as we have observed in the rat tumors. Alternatively, the tumors may represent triple negative tumors that possess neither a distinct luminal nor basal immuno-phenotype. Importantly, early precancerous lesions of ADH and DCIS were negative for ERα and PR, in contrast to the mouse model, which often has hormone receptor positive precancerous lesions that gradually lose ERα immunoreactivity as they progress to carcinoma (Yoshidome et al. 1998). Histologic types of triple negative breast cancers are similar to those seen in basal breast cancer, most commonly infiltrating ductal carcinoma (Sasaki and Tsuda 2009; Thike et al. 2010). Most basal-type tumors are characterized by solid architecture, pushing margins, central to comedo-type necrosis, cellular pleomorphism with poorly differentiated nuclear features, high nuclear-cytoplasmic ratio and mitotic index, and scant stromal content, and central geographic or comedo-type necrosis (Banerjee et al. 2006; Dabbs et al. 2006; Rakha et al. 2008) The tumors in the C3(1)/Tag transgenic rat were characterized by a variable histologic phenotype, but contained large solid areas consistent with features of the basal subtype noted above.
Although expression of the SV40 T/t-antigen transgene using the C3(1) promoter consistently produced mammary precancerous lesions and carcinomas in rats, several other lesions related to transgene expression were also noted. These included atypical hyperplasias (parotid salivary gland, nasal submucosal glands, and Harderian gland) and tumors (pituitary adenomas, pancreatic islet cell carcinomas, thyroid follicular carcinoma and parotid salivary carcinoma) in very young animals. Lesions such as pituitary adenomas and islet cell tumors, otherwise considered age-related background pathology in the rat, occurred in animals at 3–4 months of age. This rapid onset of mammary and non-mammary neoplasia led to the early death and poor reproductive performance of the founder lines and their offspring.
The presence of these non-mammary lesions indicates that the 5′ flanking region of the C3(1) promoter is able to direct significant levels of expression of the SV40 early region to multiple epithelial tissues in the rat in a manner more promiscuous than when introduced into the mouse germline. It is important to note that prostatic precancerous lesions and neoplasia, common in the mouse model, were not present in the rat model at the time of their early death. It is possible that prostatic lesions may have developed had the rats survived longer. Alternatively, although the C3(1) gene is an endogenous rat gene expressed at extremely high levels in the rat prostate, a critical androgen response element contained within the first intron of the promoter was excised during the construction of the transgene, likely resulting in the loss of expression in the rat prostate (Maroulakou et al. 1994). It remains possible that additional hormone and/or prostate-specific regulatory elements not contained within the transgenic construct resulted in the lack of prostate expression. Therefore, the 5′ flanking region of the C3(1) promoter appears to contain regulatory elements that specify more general epithelial expression.
In summary, by utilizing the rat C3(1) promoter to drive the expression of SV40 T/t-antigens, we developed transgenic rats that recapitulated the multi-step progression of mammary carcinoma from pre-invasive through invasive carcinoma with features of triple negative breast and Her-positive basal breast cancer. Triple-negative breast cancer in particular remains a form of the disease for which few extremely effective therapies exist and for which improved pre-clinical models are very much needed. As the characterization of triple negative breast cancer is based on the immunophenotype of ER, PR, Her2 negative, and the basal phenotype has been based upon gene expression profiling, these tumor types may overlap but are not synonymous. And while it is controversial in terms of how these classifications may change clinical management of these types of breast cancer, it is hoped that through the use of models such as the C3(1)/Tag transgenic rat, further insights on potential therapeutic molecular targets may lead to more personalized medicine to combat this complex disease.
Unfortunately, the transgene used in this study resulted in very high levels of expression in the mammary glands leading to aggressive tumors developing in young animals that were subsequently unable to generate offspring to allow for the propagation of this model. However, it may be possible to alter the promoter region in future iterations to potentially reduce promiscuous transgene expression and increase mammary specificity. An additional possibility would be to directly target the mammary ductular epithelium with an SV40 Tag-expressing construct through the utilization of mammary duct infusion techniques developed by Gould et al. (1991) using retroviral vectors (Wang et al. 1991). This technique would circumvent many of the problems associated with pronuclear injection (Smits et al. 2007). Given the potential advantages of a rat model of triple-negative mammary cancer compared to existing mouse models, this would be an important step forward in studying the biology of triple-negative tumors.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute. We are grateful to Ron Lubet (NCI) and Clinton Grubbs (University of Alabama-Birmingham) for their assistance in providing control samples for immunohistochemistry, and Donna Bucher at PHL (NCI-Frederick) for technical assistance. We would like to thank Jerry Ward (Global Vet Pathology) for helpful discussions.
Contributor Information
M. J. Hoenerhoff, Transgenic Oncogenesis and Genomics Section, Laboratory of Cancer Biology and Genetics, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Building 37, Room 4054, Bethesda, MD 20892, USA
M. A. Shibata, Department of Anatomy and Cell Biology, Division of Life Sciences, Osaka Medical College, Osaka, Japan
A. Bode, Carcinogenesis and Chemoprevention Program, Hormel Institute, University of Minnesota, Austin, MN 55912, USA
J. E. Green, Transgenic Oncogenesis and Genomics Section, Laboratory of Cancer Biology and Genetics, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Building 37, Room 4054, Bethesda, MD 20892, USA
References
- Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 2008;22:1233–1239. discussion 1239–1240, 1243. [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Reis-Filho JS, Ashley S, Steele D, Ashworth A, Lakhani SR, Smith IE. Basal-like breast carcinomas: clinical outcome and response to chemotherapy. J Clin Pathol. 2006;59:729–735. doi: 10.1136/jcp.2005.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan D, Montagna C, Ried T, Green JE. Mammary gland cancer. In: Holland EC, editor. Mouse models of human cancer. Hoboken; Wiley: 2004. pp. 103–131. [Google Scholar]
- Bennett CN, Green JE. Unlocking the power of cross-species genomic analyses: identification of evolutionarily conserved breast cancer networks and validation of pre-clinical models. Breast Cancer Res. 2008;10:213. doi: 10.1186/bcr2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. How basal are triple-negative breast cancers? Int J Cancer. 2008;123:236–240. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- Cardiff R. Validity of mouse mammary tumour models for human breast cancer: comparative pathology. Microsc Res Tech. 2001;52:224–230. doi: 10.1002/1097-0029(20010115)52:2<224::AID-JEMT1007>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Cardiff RD, Wellings SR. The comparative pathology of human and mouse mammary glands. J Mammary Gland Biol Neoplasia. 1999;4:105–122. doi: 10.1023/a:1018712905244. [DOI] [PubMed] [Google Scholar]
- CDC . In: US Cancer Statistics Working Group. United States Cancer Statistics: 1999-2005 Incidence and Mortality Web-based Report. D. N. C. I. Department of Health and Human Services, editor. Washington: 2009. [Google Scholar]
- Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- Dabbs DJ, Chivukula M, Carter G, Bhargava R. Basal phenotype of ductal carcinoma in situ: recognition and immunohistologic profile. Mod Pathol. 2006;19:1506–1511. doi: 10.1038/modpathol.3800678. [DOI] [PubMed] [Google Scholar]
- Deeb KK, Michalowska AM, Yoon CY, Krummey SM, Hoenerhoff MJ, Kavanaugh C, Li MC, Demayo FJ, Linnoila I, Deng CX, Lee EY, Medina D, Shih JH, Green JE. Identification of an integrated SV40 T/t-antigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res. 2007;67:8065–8080. doi: 10.1158/0008-5472.CAN-07-1515. [DOI] [PubMed] [Google Scholar]
- Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- Dundas SR, Ormerod MG, Gusterson BA, O’Hare MJ. Characterization of luminal and basal cells flow-sorted from the adult rat mammary parenchyma. J Cell Sci. 1991;100(Pt 3):459–471. doi: 10.1242/jcs.100.3.459. [DOI] [PubMed] [Google Scholar]
- Ellis IO, Galea M, Broughton N, Locker A, Blamey RW, Elston CW. Pathological prognostic factors in breast cancer. II. Histological type. Relationship with survival in a large study with long-term follow-up. Histopathology. 1992;20:479–489. doi: 10.1111/j.1365-2559.1992.tb01032.x. [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002;41:154–161. [PubMed] [Google Scholar]
- Green ES, Menz MD, LaVail MM, Flannery JG. Characterization of rhodopsin mis-sorting and constitutive activation in a transgenic rat model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000a;41:1546–1553. [PubMed] [Google Scholar]
- Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR, Wigginton J, Wiltrout R, Shibata E, Kaczmarczyk S, Wang W, Liu ZY, Calvo A, Couldrey C. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000b;19:1020–1027. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- Green JE, Shibata MA, Shibata E, Moon RC, Anver MR, Kelloff G, Lubet R. 2-difluoromethylornithine and dehydroepiandrosterone inhibit mammary tumor progression but not mammary or prostate tumor initiation in C3(1)/SV40 T/t-antigen transgenic mice. Cancer Res. 2001;61:7449–7455. [PubMed] [Google Scholar]
- Gusterson B. Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer. 2009;9:128–134. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- Gusterson BA, Ross DT, Heath VJ, Stein T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res. 2005;7:143–148. doi: 10.1186/bcr1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MJ, Ries LAG, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK. SEER cancer statistics review, 1975–2006. National Cancer Institute; Bethesda: 2009. [Google Scholar]
- Kavanaugh C, Green JE. The use of genetically altered mice for breast cancer prevention studies. J Nutr. 2003;133:2404S–2409S. doi: 10.1093/jn/133.7.2404S. [DOI] [PubMed] [Google Scholar]
- Kavanaugh CJ, Desai KV, Calvo A, Brown PH, Couldrey C, Lubet R, Green JE. Pre-clinical applications of transgenic mouse mammary cancer models. Transgenic Res. 2002;11:617–633. doi: 10.1023/a:1021159705363. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Sakuma H, Ishida N, Hirano T, Ishihara A, Takeda K. Contrast-enhanced high-resolution MRI of invasive breast cancer: correlation with histopathologic subtypes. AJR Am J Roentgenol. 2004;183:1805–1809. doi: 10.2214/ajr.183.6.01831805. [DOI] [PubMed] [Google Scholar]
- Lerma E, Peiro G, Ramon T, Fernandez S, Martinez D, Pons C, Munoz F, Sabate JM, Alonso C, Ojeda B, Prat J, Barnadas A. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas) Mod Pathol. 2007;20:1200–1207. doi: 10.1038/modpathol.3800961. [DOI] [PubMed] [Google Scholar]
- Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- Ludlow JW. Interactions between SV40 large-tumor antigen and the growth suppressor proteins pRB and p53. Faseb J. 1993;7:866–871. doi: 10.1096/fasebj.7.10.8344486. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci USA. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Guzman RC, Yang J. Hormones and mammary carcinogenesis in mice, rats, and humans: a unifying hypothesis. Proc Natl Acad Sci USA. 1995;92:3650–3657. doi: 10.1073/pnas.92.9.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen NH, Emdin SO, Cajander J, Landberg G. Deregulation of cyclin E and D1 in breast cancer is associated with inactivation of the retinoblastoma protein. Oncogene. 1997;14:295–304. doi: 10.1038/sj.onc.1200833. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- Nozoe T, Oyama T, Mori E, Uramoto H, Takenoyama M, Hanagiri T, Sugio K, Yasumoto K. Clinicopathologic significance of an immunohistochemical expression of p27 in scirrhous carcinoma of the breast. Breast Cancer. 2007;14:277–280. doi: 10.2325/jbcs.14.277. [DOI] [PubMed] [Google Scholar]
- Ottewell PD, Coleman RE, Holen I. From genetic abnormality to metastases: murine models of breast cancer and their use in the development of anticancer therapies. Breast Cancer Res Treat. 2006;96:101–113. doi: 10.1007/s10549-005-9067-x. [DOI] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro-Silva A, Ramalho LN, Garcia SB, Brandao DF, Chahud F, Zucoloto S. p63 correlates with both BRCA1 and cytokeratin 5 in invasive breast carcinomas: further evidence for the pathogenesis of the basal phenotype of breast cancer. Histopathology. 2005;47:458–466. doi: 10.1111/j.1365-2559.2005.02249.x. [DOI] [PubMed] [Google Scholar]
- Russo J, Tay LK, Ciocca DR, Russo IH. Molecular and cellular basis of the mammary gland susceptibility to carcinogenesis. Environ Health Perspect. 1983;49:185–199. doi: 10.1289/ehp.8349185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J, Russo IH, Rogers AE, van Zwieten MJ, Gusterson B. Pathology of tumours in laboratory animals. Tumours of the rat. Tumours of the mammary gland. IARC Sci Publ. 1990:47–78. [PubMed] [Google Scholar]
- Sasaki Y, Tsuda H. Clinicopathological characteristics of triple-negative breast cancers. Breast Cancer. 2009;16:254–259. doi: 10.1007/s12282-009-0153-5. [DOI] [PubMed] [Google Scholar]
- Shibata MA, Jorcyk CL, Liu ML, Yoshidome K, Gold LG, Green JE. The C3(1)/SV40 T antigen transgenic mouse model of prostate and mammary cancer. Toxicol Pathol. 1998;26:177–182. doi: 10.1177/019262339802600121. [DOI] [PubMed] [Google Scholar]
- Skoog L, Humla S, Axelsson M, Frost M, Norman A, Nordenskjold B, Wallgren A. Estrogen receptor levels and survival of breast cancer patients. A study on patients participating in randomized trials of adjuvant therapy. Acta Oncol. 1987;26:95–100. doi: 10.3109/02841868709091747. [DOI] [PubMed] [Google Scholar]
- Smits BM, Cotroneo MS, Haag JD, Gould MN. Genetically engineered rat models for breast cancer. Breast Dis. 2007;28:53–61. doi: 10.3233/bd-2007-28106. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning P Eystein, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123–133. doi: 10.1038/modpathol.2009.145. [DOI] [PubMed] [Google Scholar]
- Troester MA, Herschkowitz JI, Oh DS, He X, Hoadley KA, Barbier CS, Perou CM. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer. 2006;6:276. doi: 10.1186/1471-2407-6-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Wang B, Kennan WS, Yasukawa-Barnes J, Lindstrom MJ, Gould MN. Frequent induction of mammarycarcinomas following neu oncogene transfer into in situ mammary epithelial cells of susceptible and resistant rat strains. Cancer Res. 1991;51:5649–5654. [PubMed] [Google Scholar]
- Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- Wu K, Kim HT, Rodriquez JL, Munoz-Medellin D, Mohsin SK, Hilsenbeck SG, Lamph WW, Gottardis MM, Shirley MA, Kuhn JG, Green JE, Brown PH. 9-cis-Retinoic acid suppresses mammary tumorigenesis in C3(1)-simian virus 40 T antigen-transgenic mice. Clin Cancer Res. 2000;6:3696–3704. [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Yoshidome K, Shibata MA, Maroulakou IG, Liu ML, Jorcyk CL, Gold LG, Welch VN, Green JE. Genetic alterations in the development of mammary and prostate cancer in the C3(1)/Tag transgenic mouse model. Int J Oncol. 1998;12:449–453. [PubMed] [Google Scholar]