Abstract
PTP1B is a protein tyrosine-phosphatase located on the cytosolic side of the endoplasmic reticulum that plays an important role in the regulation of the insulin receptor (IR). Replacement of the conserved Asp-181 by alanine is known to convert PTP1B into a substrate-trapping protein that binds to but cannot dephosphorylate its substrates. In this work, we have studied the effect of an additional mutation (Y46F) on the substrate-trapping efficiency of PTP1B-D181A. We observed that this mutation converts PTP1B-D181A into a highly efficient substrate-trapping mutant, resulting in much higher recovery of tyrosine-phosphorylated proteins coimmunoprecipitated with PTP1B. Bioluminescence resonance energy transfer (BRET) experiments were also performed to compare the dynamics of interaction of the IR with these mutants. Basal BRET, which mainly reflects the interaction of PTP1B with the IR precursor during its biosynthesis in the endoplasmic reticulum, was markedly increased with the PTP1B-D181A-Y46F mutant. In contrast, insulin-induced BRET was markedly reduced with PTP1B-D181A-Y46F. I125 insulin binding experiments indicated that PTP1B-D181-Y46F reduced the expression of IR at the plasma membrane. Reduced expression at the cell surface was associated with higher amounts of the uncleaved IR precursor in the cell. Moreover, we observed that substantial amounts of the uncleaved IR precursor reached the Tris-phosphorylated, fully activated form in an insulin independent fashion. These results support the notion that PTP1B plays a crucial role in the control of the activity of the IR precursor during its biosynthesis. In addition, this new substrate-trapping mutant may be a valuable tool for the identification of new PTP1B substrates.
Keywords: Endoplasmic Reticulum (ER), Hormone Receptors, Insulin, Receptor Tyrosine Kinase, Tyrosine Protein Phosphatase (Tyrosine Phosphatase), Bioluminescence Resonance Energy Transfer (BRET)
Introduction
Insulin exerts its biological effects through a plasma membrane receptor that possesses a tyrosine kinase activity. Binding of insulin to its receptor induces autophosphorylation of the insulin receptor (IR) on tyrosine residues (1). This stimulates the tyrosine kinase activity of the IR toward intracellular substrates involved in the transmission of the signal (1, 2). Phosphorylation of tyrosines 1158, 1162, and 1163, located in the kinase domain of the IR, is known to play a crucial role in the regulation of the activity of the receptor (3–5). Termination of the signal involves inactivation of the IR kinase by dephosphorylation of these tyrosines by protein tyrosine phosphatases (6). Several PTPases2, including PTP1B, PTPα, PTPϵ, leukocyte common antigen-related, and T cell protein tyrosine phosphatase, have been implicated in the regulation of the IR activity (7–10). Among them, PTP1B appears to play a major role in the control of insulin action. PTP1B is localized predominantly on the cytosolic side of the endoplasmic reticulum (ER) by means of a hydrophobic C-terminal targeting sequence (11). Involvement of PTP1B in insulin signaling was initially suggested by experiments in Xenopus oocytes (12) and in cultured cells (13, 14). Definitive evidence of the implication of PTP1B in insulin signaling was obtained with PTP1B knockout in mice, which resulted in a marked increase in insulin sensitivity associated with increased tyrosine phosphorylation of the IR and insulin receptor substrate-1 in liver and skeletal muscle (15, 16). Therefore, PTP1B clearly appears as a potential therapeutic target for the treatment of insulin resistance (17). However, although PTP1B has been shown to directly interact with the IR, the regulation of this interaction in living cells remains poorly studied. We previously developed a procedure (18) based on the BRET methodology that allows the study in real time, in living cells, of the interaction of the IR with a substrate-trapping version of PTP1B (PTP1B-D181A). We demonstrated that insulin induced a rapid and dose-dependent increase of the interaction between the IR and PTP1B. We also observed, in the absence of insulin, the presence of a substantial interaction between the IR and PTP1B (basal BRET signal). Cell fractionation experiments indicated that this basal BRET signal could be attributed to the interaction of PTP1B with the IR precursor during its biosynthesis in the ER (18). This result was thereafter confirmed in experiments using FRET instead of BRET (19).
Several lines of evidence indicate that tyrosine phosphorylation of PTP1B may regulate its activity (20–22). Indeed, PTP1B was shown to be phosphorylated on tyrosines 66, 152, and/or 153 in an insulin-dependent manner, and replacement of these tyrosines by phenylalanine appears to affect its interaction with the IR (20). Tyr-46, located in the catalytic cleft of PTP1B, is highly conserved among protein tyrosine phosphatases. This tyrosine is believed to be important to define the specificity of the enzyme toward tyrosine-phosphorylated substrates (23). In this work, to further evaluate the role of tyrosine 46, we replaced this residue by a phenylalanine in the PTP1B-D181A trapping mutant and we studied, in living cells, the interaction of the IR with this double mutant (PTP1B-D181A-Y46F). We found that this additional mutation converts PTP1B-D181A into a highly efficient substrate-trapping mutant that precipitates a much larger number of tyrosine-phosphorylated proteins compared with the classical, widely used D181A single mutant. Moreover, we show that this new PTP1B double mutant traps the IR precursor in an intracellular compartment and markedly reduces the expression of the mature receptor at the cell surface.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals have been described previously (18, 24). Mouse monoclonal anti-GFP (clones 7.1 and 13.1) antibody was from Roche. The anti-IRβ antibody was from Santa Cruz Biotechnology, Inc. The antibody directed against the Tris-phosphorylated form of the activation loop of the IR (anti-3YP1158/62/63) was from BioSource.
Expression Vectors
Unless otherwise stated (supplemental Figs. S3 and S4), the IRB isoform of the IR (+ exon 11) was used throughout this study. cDNAs encoding IR, IR-Rluc, IR-YFP, and YFP-PTP1B-D181A have been described previously (18, 25). The YFP-PTP1B-D181A-Y46F mutant was generated by site-directed mutagenesis (QuikChange, Stratagene).
Cell Culture and Transfection
Cell culture was performed as described (26, 27). Briefly, HEK-293 cells were seeded at a density of 2 × 105 cells per 35-mm dish. One day later, cells were transfected with the appropriate plasmids as indicated in the figure legends.
For BRET experiments, 1 day after transfection, cells were transferred into 96-well microplates (CulturPlate-96, white, PerkinElmer Life Sciences) at a density of 3 × 104 cells per well. BRET measurements were carried out in these microplates on the following day.
BRET Measurements
BRET experiments were performed as described previously (18). Briefly, cells were preincubated for 15 min in PBS in the presence of 2.5 μm coelenterazine. Insulin was added, and light emission acquisition at 485 and 530 nm was started immediately. The dynamics of the interaction between the IR-Rluc and YFP-PTP1B mutants could be monitored for more than 30 min after insulin addition. The BRET signal was expressed in milliBRET units. The BRET unit has been defined previously as the ratio 530/485 nm, obtained when the two partners are present, corrected by the ratio 530/485 nm, obtained under the same experimental conditions when only the partner fused to Renilla luciferase is present in the assay (28–30). Each measurement corresponded to the signal emitted by the whole population of cells present in a well (i.e. approximately 40,000 cells).
Western Blotting Experiments
Precipitation of the IR on wheat germ lectin-agarose beads were performed as described previously (5). YFP-PTP1B mutants and IR-YFP were immunoprecipitated using anti-GFP antibody and detected by Western blotting as described previously (31).
I125 Insulin Binding
Mono-iodinated I125 insulin (32) was from PerkinElmer Life Sciences. HEK-293 cells transfected with the IR-Rluc and YFP-PTP1B-WT, YFP-PTP1B-D181A, or YFP-PTP1B-D181A-Y46F mutants were plated in 24-well culture dishes. A fraction of these cells was plated separately for determination of luciferase and fluorescence levels in parallel to insulin binding. For binding experiments, cells were washed three times with phosphate-buffered saline and incubated at 4 °C for 4 h in binding buffer (33) containing I125 insulin (0.1 μCi/well). Unbound ligand was removed by washing with ice-cold binding buffer. Cells were then lysed with 1 N NaOH, and the radioactivity was counted using a Berthold LB2111 counter.
Statistical Analysis
Results are expressed as mean ± S.E. Comparisons between experimental groups were made using the t test unless otherwise specified in the figure legends.
RESULTS
PTP1B-D181A-Y46F Is a Highly Efficient Substrate-trapping Mutant
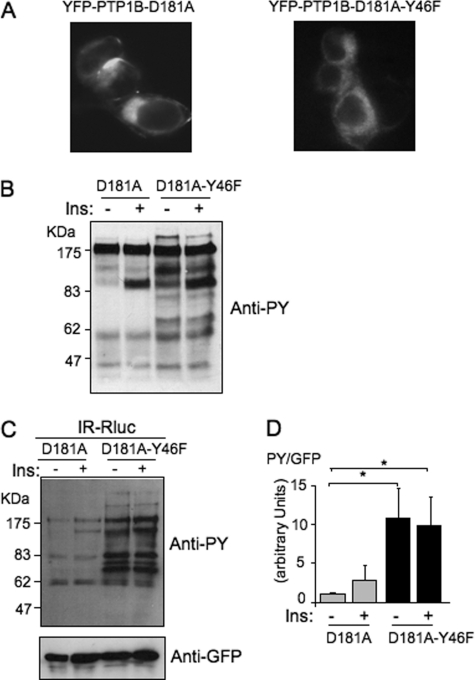
We previously observed that, when expressed in HEK 293 cells, YFP-PTP1B-D181A has an intracellular perinuclear distribution that corresponded to an endoplasmic reticulum localization (18). As shown in Fig. 1A, a similar distribution of the fluorescence was observed with YFP-PTP1B-D181A-Y46F, suggesting that the Y46F mutation did not affected the localization of the protein.
FIGURE 1.
The PTP1B-D181A-Y46F mutant is a much more efficient substrate-trapping mutant than PTP1B-D181A. A, localization of the YFP-tagged mutants of PTP1B was observed by fluorescent microscopy. B, HEK-293 cells transfected with the IR and either YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F were incubated for 5 min in the absence (−) or presence (+) of 100 nm insulin. 50 μg of proteins was loaded on a gel and submitted to Western blotting using an anti-phosphotyrosine antibody (4G10). C, HEK-293 cells transfected with IR-Rluc and either YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F were stimulated for 5 min with 100 nm insulin (Ins). Cells were extracted, and the YFP fluorescence was evaluated on an aliquot of the extract by measuring light emission at 530 nm after illumination at 485 nm. Equivalent amounts of fluorescent PTP1B were then immunoprecipitated using an anti-GFP antibody. Tyrosine phosphorylation of proteins coimmunoprecipitated with PTP1B was evaluated by immunoblotting using an anti-phosphotyrosine antibody (4G10). The amount of PTP1B loaded in each lane was then controlled by reprobing the membrane using an anti-GFP antibody. D, densitometric analysis of the total tyrosine phosphorylation level of immunoprecipitated proteins. The signal corresponding to the total level of tyrosine-phosphorylated proteins detected in each lane was corrected by the amount of PTP1B in the same lane as evaluated by using the anti-GFP antibody. Results are the mean ± S.E. of four independent experiments. *, p < 0.05 when compared with the unstimulated D181A control condition using analysis of variance followed by Dunnett's multiple comparison test.
It has been shown previously that when transfected in cells, substrate-trapping mutants of PTPases bind to tyrosine phosphorylated proteins and protect them from dephosphorylation by endogenous PTPases. To evaluate whether mutation of tyrosine 46 affected the efficiency of the PTP1B-D181 trapping mutant, HEK-293 cells transfected with either the YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F mutant were incubated for 5 min in the absence or presence of insulin. Cells were then extracted, proteins were separated by SDS-PAGE, and the tyrosine phosphorylation profile was evaluated by Western blotting using an anti-phosphotyrosine antibody. We observed that the level of tyrosine-phosphorylated proteins was much higher in PTP1B-D181A-Y46F- than in PTP1B-D181A-transfected cells, both in absence or presence of insulin (Fig. 1B). This result suggested that the double mutant was much more efficient than the classical D181A mutant in protecting tyrosine-phosphorylated residues from the activity of endogenous PTPases. Similar results were obtained with the untagged versions of these PTP1B substrate-trapping mutants (data not shown).
The ability of this new double mutant to coprecipitate tyrosine-phosphorylated proteins was then evaluated. HEK-293 cells cotransfected with IR-Rluc and either the YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F mutant were incubated for 5 min in absence or presence of insulin. Cells were extracted, and equivalent amounts of the PTP1B-D181A and D181A-Y46F mutants were immunoprecipitated using an anti-GFP antibody. We observed that the amount of coimmunoprecipitated tyrosine-phosphorylated proteins was much higher with YFP-PTP1B-D181A-Y46F than with YFP-PTP1B-D181A (Fig. 1C). This suggested that replacement of tyrosine 46 by a phenylalanine converts PTP1B-D181A into a much more efficient substrate-trapping mutant for coimmunoprecipitation of tyrosine-phosphorylated proteins (Fig. 1D).
Hyperphosphorylation of PTP1B-D181A-Y46F on Tyrosine Residues by the Insulin Receptor
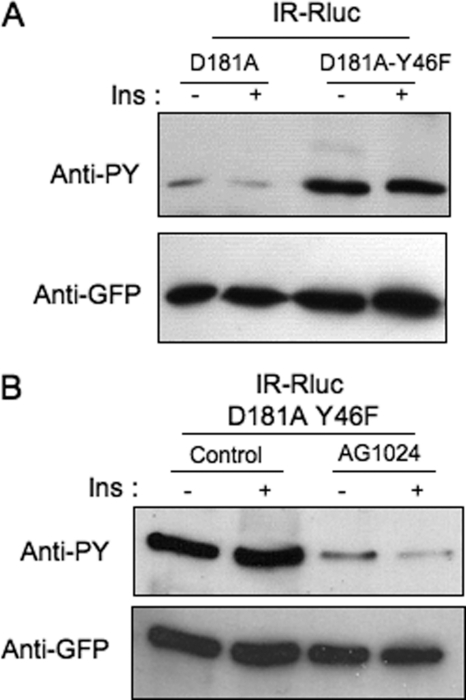
Previous studies have shown that PTP1B can be phosphorylated on tyrosine residues by the IR. As shown in Fig. 2A, immunoprecipitation with anti-GFP antibody showed that a modest tyrosine phosphorylation of PTP1B-D181A could be detected in HEK-293 cells cotransfected with IR-Rluc and YFP-PTP1B-D181A. In contrast, a much stronger tyrosine phosphorylation of PTP1B-D181A-Y46F mutant could be detected under similar conditions.
FIGURE 2.
Hyperphosphorylation of the PTP1B-D181A-Y46F mutant on tyrosine residues by the insulin receptor. A, HEK-293 cells transfected with IR-Rluc and either YFP-PTP1B-D181A or YFP-PTP1B-D181AY46F were incubated for 5 min in the absence (−) or presence (+) of 100 nm insulin (Ins). Proteins were extracted, and PTP1B was immunoprecipitated using an anti-GFP antibody. Tyrosine phosphorylation of PTP1B was evaluated by immunoblotting using an anti-phosphotyrosine antibody (4G10). The amount of PTP1B loaded in each lane was evaluated by reprobing the membrane using an anti-GFP antibody. Results are representative of five independent experiments. B, HEK-293 cells transfected with IR-Rluc and YFP-PTP1B-D181A-Y46F were preincubated for 1 h in the absence or presence of 100 μm AG1024 and incubated with 100 nm insulin for 5 min. Cells were then lysed, and PTP1B was immunoprecipitated and analyzed by Western blotting using an anti-phosphotyrosine antibody (4G10). The amount of PTP1B loaded in each lane was controlled using an anti-GFP antibody. Results are representative of three independent experiments.
To determine whether this hyperphosphorylation of the PTP1B-D181A-Y46 mutant was achieved by the insulin receptor kinase, HEK-293 cells cotransfected with IR-Rluc and YFP-PTP1B-D181A-Y46 were preincubated for 1 h with the tyrphostin AG1024, which selectively inhibits the tyrosine kinase activity of the IR (18). As shown in supplemental Fig. S1, autophosphorylation of the IR was indeed markedly inhibited by AG1024. Under these conditions, tyrosine phosphorylation of PTP1B-D181A-Y46F was also markedly inhibited (Fig. 2B), indicating that this phosphorylation was indeed achieved by the IR kinase.
Replacement of Tyrosine 46 by Phenylalanine Markedly Increases the Basal BRET Signal
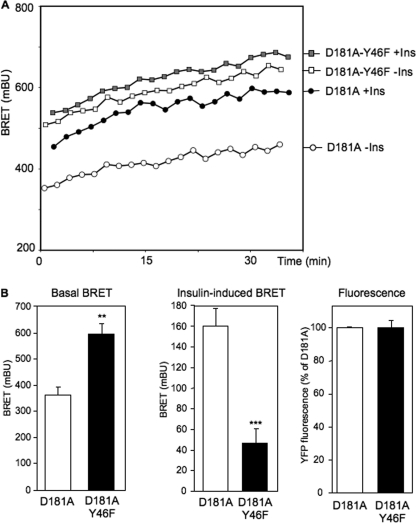
To evaluate the effect of the Y46F mutation on the dynamics of interaction between the IR and PTP1B, we compared basal and insulin-stimulated BRET signals in cells expressing IR-Rluc and either YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F. As shown in our previous study, in cells expressing YFP-PTP1B-D181A, a robust basal BRET signal was observed (Fig. 3A). In agreement with our previous work, insulin (100 nm) markedly stimulated this signal (about 180–200 milliBRET units above basal at 20 min). Interestingly, in cells expressing the YFP-PTP1B-D181A-Y46F mutant, the basal BRET signal was much higher. Moreover, insulin had only a modest effect on this signal (Fig. 3A). Fig. 3B shows the mean ± S.E. of the basal and the insulin-induced BRET signal at 20 min in five independent experiments. We observed that the basal BRET signal was much higher with YFP-D181A-Y46F than with YFP-PTP1B-D181A, despite similar levels of expression of the two tyrosine phosphatases (determined by measuring YFP fluorescence). In contrast, the insulin-induced BRET signal (increase in BRET signal above basal at 20 min) was markedly lower with the Y46F mutant.
FIGURE 3.
Interaction between the IR and PTP1B mutants in intact living cells. A, HEK-293 cells were transfected with IR-Rluc and either YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F expression vectors. BRET measurements were performed in real time after addition of either 100 nm insulin (Ins) or vehicle. Results are representative of five independent experiments. B, basal BRET, insulin-induced (increased above basal at 20 min) BRET, and YFP fluorescence levels in cells expressing YFP-PTP1B-D181A or YFP-PTP1B-D181A-Y46F. Results are the mean ± S.E. of five independent experiments. **, p < 0.01; ***, p < 0.001 when compared with D181A mutant under similar conditions.
Effect of the Tyrphostin AG1024 on BRET between IR and PTP1B-D181A-Y46F
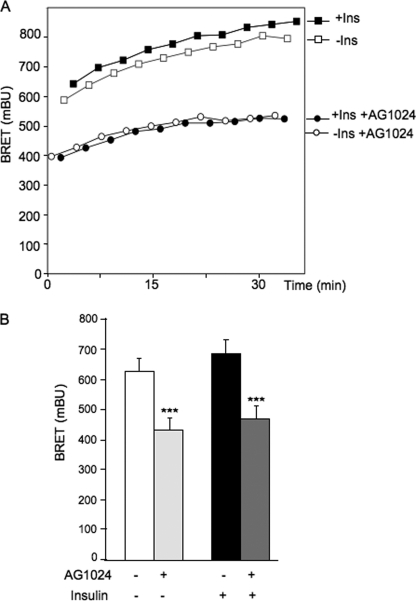
To determine whether this high basal BRET signal was dependent on the tyrosine kinase activity of the IR, we investigated the effect of the tyrphostin AG1024 on BRET between IR and PTP1B-D181-Y46F. Cells cotransfected with IR-Rluc and YFP-PTP1B-D181A-Y46F were preincubated for 1 h in the absence or presence of AG1024. BRET experiments were then performed as described previously. We observed that the basal and the insulin-stimulated BRET signals were markedly decreased by AG1024 (Fig. 4). This indicates that the high-constitutive BRET signal observed with this mutant is at least in part dependent on the kinase activity of the IR.
FIGURE 4.
Effect of tyrphostin AG1024 on BRET between IR and PTP1B-D181A-Y46F. A, HEK-293 cells were transfected with IR-Rluc and YFP-PTP1B-D181A-Y46F. 48 h after transfection, cells were preincubated for 1 h in the absence (−) or presence (+) of 100 μm AG1024. BRET measurements were performed in real time, after addition of either 100 nm insulin (Ins) or vehicle. Results are representative of five independent experiments. B, graphic representation of BRET signals measured 20 min after addition of 100 nm insulin. Results are the mean ± S.E. of five independent experiments. ***, p < 0.001 when compared with cells not treated with AG1024.
YFP-PTP1B-D181A-Y46F Reduces Insulin Binding at the Cell Surface and Impairs Maturation of the Insulin Receptor Precursor
Our previous work (18, 34) had indicated that an important part of the basal BRET signal could be attributed to an interaction between PTP1B and the insulin receptor precursor during its biosynthesis. In this study, mutation of PTP1B-D181A on Tyr-46 resulted in a further increase in the basal BRET signal, associated with a marked decrease in insulin effect above basal (insulin-induced BRET, Fig. 3B). This suggested that the double mutant may have trapped the IR precursor and reduced the expression of the mature receptor at the cell surface. In agreement with this notion, decreasing the amount of PTP-1B-D181A-Y46F transfected in cells (from 600 ng to 150 ng of cDNA/well) partially restored insulin-induced BRET signal (supplemental Fig. S2).
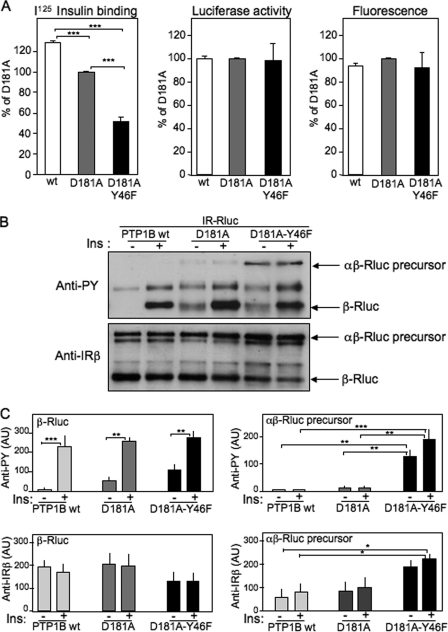
To determine whether the expression of the Y46F mutant indeed impaired the expression of the mature receptor at the plasma membrane, cell surface I125 insulin binding experiments were performed (Fig. 5A). In cells expressing the PTPB-D181A mutant, cell surface insulin binding was decreased by about 25% when compared with cells expressing PTP1B-WT, suggesting that the interaction of the IR precursor with PTP1B-D181A resulted in a significant reduction of the expression of the mature receptor at the cell surface. In cells expressing PTP1B-D181A-Y46F, insulin binding was markedly reduced compared with cells expressing WT or PTP1B-D181A, despite a similar level of total IR expression, as evaluated by measuring luciferase activity (Fig. 5A). This result strongly suggested that in PTP1B-D181A-Y46F-transfected cells, substantial amounts of insulin receptors remained trapped in an intracellular compartment.
FIGURE 5.
Effect of the PTP1B-D181A-Y46F mutant on cell surface expression of the insulin receptor. A, cell surface I125 insulin binding was measured 48 h after transfection of HEK-293 cells with IR-Rluc and YFP-PTP1B-WT, YFP-PTP1B-D181A, or YFP-PTP1B-D181A-Y46F. Luciferase activity and YFP fluorescence levels were measured in parallel for evaluation of total IR-Rluc and YFP-PTP1B expression on the same cells. Results are the mean ± S.E. of four to nine independent experiments. B, 48 h after transfection, cells were incubated in absence (−) or presence (+) of 100 nm insulin (Ins) for 5 min. After cell extraction, the IR was partially purified on wheat germ lectin-agarose beads and submitted to Western blotting. The tyrosine phosphorylation level of the receptor was evaluated using an anti-phosphotyrosine antibody (4G10). The amounts of uncleaved IR precursor (αβ-Rluc) and mature cleaved receptor (β-Rluc) were evaluated using an anti-receptor β-subunit antibody. C, densitometric analysis of the autoradiograms. Results are the mean ± S.E. of five independent experiments. AU, arbitrary units. *, p < 0.05; **, p < 0.01; ***, p < 0.001 using analysis of variance followed by Tukeys's multiple comparison test.
Wheat germ lectin purification of the insulin receptor from HEK-293 cells cotransfected with IR-Rluc and either the wild-type or mutant versions of PTP1B was then performed (Fig. 5, B and C). Blotting with an anti-phosphotyrosine antibody revealed a band of about 260 kDa in PTP1B-D181A-Y46F-transfected cells. This band was hardly detectable in PTP1B-D181-transfected cells and was absent in WT-PTP1B-transfected cells. Reprobing the membrane with an anti-IRβ antibody revealed that this band indeed corresponded to the uncleaved αβ-Rluc precursor. Together with insulin binding experiments, these results indicated that the double mutant version of PTP1B trapped the IR precursor and reduced the expression of the mature IR at the cell surface.
The Immature Insulin Receptor Precursor Can Reach the Fully Activated Tris-phosphorylated Form
The insulin receptor possesses a number of tyrosines that are phosphorylated upon insulin stimulation. Tyrosines 1158, 1162, and 1163, located in the activation loop of the β subunit, appear to be crucial for the activation of the receptor, the fully active form being the Tris-phosphorylated form, whereas the mono- and bis-phosphorylated forms are much less active (4, 5, 35).
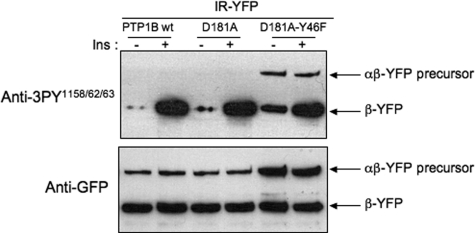
To determine whether the tyrosine phosphorylation of the IR precursor observed with the PTP1B-D181A-Y46F mutant occurred in the activation loop, YFP-tagged insulin receptors were cotransfected with untagged versions of WT-PTP1B, PTP1B-D181A, or PTP1B-D181A-Y46F (Fig. 6). 48 h after transfection, cells were stimulated with insulin for 5 min. After cell lysis, insulin receptors were immunoprecipitated using an anti-GFP antibody and submitted to Western blotting. Hybridization with an antibody that specifically recognizes the Tris-phosphorylated form of the activation loop of the IR (Fig. 6) indicated that a substantial amount of the IR precursor had indeed reached the maximally activated Tris-phosphorylated form when trapped by PTP1B-D181A-Y46. No tyrosine phosphorylation signal could be detected at the level of the IR precursor in cells transfected with the WT-PTP1B or PTP1B-D181A mutant. Reprobing the membrane with an anti-GFP antibody confirmed that the amount of uncleaved precursor was higher in cell expressing the PTP1B-D181A-Y46F mutant than in cells expressing wild-type or PTP1B-D181A mutant.
FIGURE 6.
The insulin receptor precursor can reach the fully activated Tris-phospohorylated form in an insulin-independent manner. HEK-293 cells were transfected with IR-YFP and untagged versions of WT-PTP1B, PTP1B-D181A, or PTP1B-D181A-Y46F. 48 h after transfection, cells were incubated for 5 min in presence (+) of 100 nm insulin (Ins) and lysed. The IR was immunoprecipitated using an anti-GFP antibody and immunoblotted with an antibody directed against the Tris-phosphorylated form of the activation loop of the kinase domain. The membranes were then reprobed with an anti-GFP antibody. Results are representative of four independent experiments.
Interaction with PTP1B Is Similar for IRA (- Exon 11) and IRB (+ Exon 11) Isoforms of the Insulin Receptor
In insulin target cells, the insulin receptor is expressed as two isoforms, resulting from alternative splicing of exon 11, that code for 12 amino acids localized at the C-terminal end of the α-chain. Previous studies have suggested that the tyrosine kinase activity of the IR precursor may be different depending on the presence or absence of exon 11 (36). To determine whether these isoforms differ for their interaction with PTP1B, we have evaluated the basal and the insulin-stimulated BRET signals in cells transfected with either IRA or IRB (supplemental Fig. S3). We observed that upon insulin stimulation, the dynamics of interaction of IRA with PTP1B mutants closely resemble that of IRB. Moreover, the basal BRET signals were also very similar for both IR isoforms. These results suggested that there are no major differences between the two IR isoforms in their interaction with PTP1B, neither during their biosynthesis nor after insulin stimulation (supplemental Fig. S3). This result was further confirmed by immunoprecipitation experiments, which indicated that similar amounts of fully activated uncleaved precursor were recovered with both IR isoforms in cells expressing the D181A-Y46F mutant (supplemental Fig. S4).
DISCUSSION
All members of the PTPase family are characterized by the presence of a signature motif containing a cysteinyl residue (Cys-215 in PTP1B) that is involved in the catalysis of the dephosphorylation reaction. The signature motif, which forms the phosphate recognition site, is located at the base of a cleft. The sides of the cleft are formed by three motifs: the Q loop, which contains Gln-262, the WDP loop, which contains the invariant Asp residue (Asp-181), and the pTyr loop, which contains the conserved Tyr-46. Tyr-46 is believed to define the depth of the catalytic cleft and to contribute to the absolute specificity of PTP1B for phosphotyrosine-containing substrates because the smaller phosphoserine and phosphothreonine residues would not reach down to the phosphate binding site (23). Several lines of evidence indicate that upon insulin or growth factor treatment, the production of reactive oxygen species such as H2O2 induces transient inactivation of PTPases by reversible oxidation of the active site cysteine (37). Structural studies have shown that following PTP1B oxidation by H2O2, a covalent bound is formed between the sulfur atom of Cys-215 and the main chain nitrogen of the adjacent residue, Ser-216, resulting in the formation of a new sulfenyl-amide species. This is accompanied by a profound modification of the architecture of the active site, involving displacement of Tyr-46 to a solvent-exposed position, and results in inhibition of substrate binding. This mechanism is believed to protect PTP1B from the formation of irreversibly oxidized sulfenic acid and to facilitate reactivation of PTP1B by biological thiols (38, 39).
To evaluate the effect of Tyr-46 on the interaction between the IR and PTP1B in living cells, we have mutated this tyrosine into phenylalanine. We observed that this mutation converts PTP1B-D181A into a much more efficient substrate-trapping mutant, as demonstrated by the marked increase in the amount of tyrosine-phosphorylated proteins that could be coprecipitated with PTP1B-D181A-Y46F, both in the absence and presence of insulin (Fig. 1C). The IR is biosynthesized in the endoplasmic reticulum as an uncleaved αβ-precursor, with the kinase domain of the β-chain located on the cytosolic side of the ER, i.e. in the vicinity of the catalytic domain of PTP1B. In a previous study (18), we have demonstrated that PTP1B-D181A interacted with the insulin receptor precursor during its biosynthesis in the ER, resulting in a high basal, insulin-independent BRET signal. In this study, we show that the mutation Y46F on PTP1B-D181A markedly increases the efficiency of the interaction of PTP1B with the IR precursor and thereby reduces the expression of the mature receptor at the plasma membrane. This view is supported by results obtained with cells expressing the PTP1B-D181A-Y46F mutant which displays 1) an increased basal BRET signal, 2) a decreased insulin-induced BRET signal, 3) decreased cell surface I125 insulin binding, and 4) an increased amount of uncleaved IR precursor. In addition, our results demonstrate that the IR precursor can reach the fully active, Tris-phosphorylated form in the absence of insulin. This strongly supports the notion that PTP1B may play a crucial role in the regulation of the activity of the immature IR precursor by precluding uncontrolled autoactivation of the precursor and subsequent activation of intracellular insulin signaling pathways in an autonomous way (18, 34).
It may at first sight appear surprising that in cells transfected with PTP1B-D181A-Y46F, a substantial effect of insulin on tyrosine phosphorylation of the mature IR-β was observed (Figs. 5B and 6), whereas the insulin effect on BRET was markedly reduced in these conditions (Figs. 3, A and B). However, it must be kept in mind that the BRET technique only detects receptors that interact with PTP1B, both in the basal state and upon insulin stimulation. In contrast, in wheat germ lectin (Fig. 5B) or immunoprecipitation (Fig. 6) experiments, all receptors, including those that are not engaged in an interaction with PTP1B, will be detected with anti-phosphotyrosine antibodies. Therefore, although significant amounts of IR precursor interact with the PTP1B-D181A-Y46F mutant (thereby reducing the expression of the mature receptor at the plasma membrane, Fig. 5A), Fig. 5B and Fig. 6 demonstrate that substantial amounts of mature receptors are still present at the cell surface and are capable of undergoing insulin-induced autophosphorylation.
Interestingly, PTP1B-D181A-Y46F appeared to be hyperphosphorylated on tyrosine residues. This hyperphosphorylation is largely due to the kinase of the IR, as demonstrated by its inhibition by AG1024 (Fig. 2B). Several publications have indicated that PTP1B is phosphorylated on tyrosines 66, 152, and 153 by the IR and other tyrosine kinases (20–22). Interestingly, phosphorylation of PTP1B on tyrosine 66 appeared to increase its catalytic activity (21, 40). This insulin-induced tyrosine phosphorylation of PTP1B by stimulating its catalytic activity may be part of a negative feedback loop in insulin signaling (40, 41). Because mutation Y46F increases the substrate-trapping efficiency of PTP1B-D181A, it is conceivable that the resulting prolonged interaction of PTP1B with the IR leads to hyperphosphorylation of the D181A-Y46F double mutant. Moreover, it has been shown previously both in vitro and in intact cells that PTP1B catalyzes its own dephosphorylation through an intermolecular reaction (40), and dimerization of PTP1B was more recently demonstrated using a fluorescent complementation assay (42). Therefore, a highly efficient substrate-trapping mutant may also protect PTP1B from autodephosphorylation.
PTP1B has recently emerged as a major therapeutic target. Indeed, a large number of data now indicate that PTP1B not only plays a negative role in the regulation of insulin and leptin signaling (15, 16, 43) but also has a positive effect on cancer cell growth and migration (44–47). Identification of PTP1B substrates constitutes an important challenge for the treatment of these pathologies. Several substrate-trapping mutants have been developed to identify and study PTP1B substrates by modifying invariant residues in the catalytic domain. Among them, cysteine 215 in PTP1B is positioned to act as a nucleophile to attack the phosphorus atom of the phosphotyrosyl residue of the substrate (23). A first generation of substrate-trapping mutants in which the catalytic cysteine was replaced by an alanine has been largely used for the study of PTP1B and other PTPases substrates (48–50). Mutation of the invariant aspartic acid 181, also involved in the catalysis, was then developed by the group of Tonks (51) as a more efficient substrate-trapping mutant of PTP1B. This observation was then extended to other PTPases in which mutation of the equivalent aspartic residue in the catalytic site also resulted in substrate-trapping enzymes (26). Additional efforts to develop an improved substrate-trapping mutant have been performed by Xie et al. (52). It was shown that mutation of glutamine 262 into an alanine resulted in some improvement of the efficiency of the PTP1B-D181A-Q262A double mutant compared with the conventional D181A single mutant for trapping of the EGF receptor (52). However, trapping of other additional phosphotyrosine-containing proteins was hardly detectable by Western blotting in these experiments (52). Moreover, in a quantitative proteomics approach using the D181A and D181A-Q262A mutants to pull down PTP1B substrates, Mertins et al. (53) found that these mutants interacted with a similar number of tyrosine-phosphorylated proteins. In contrast, our new double mutant turned out to be much more efficient than PTP1B-D181A, as demonstrated by the much larger number and higher intensity of the bands of tyrosine-phosphorylated proteins precipitated by the D181A-Y46F mutant (Fig. 1C). Therefore, the D181A-Y46F mutant will constitute a valuable tool for the identification of new PTP1B substrates using proteomic approaches. In addition, because both Asp-181 and Tyr-46 are highly conserved throughout the PTPase family, our work should permit the development of new, highly efficient trapping mutants for most PTPases.
In summary, our work strongly supports the view that PTP1B not only regulates the activation state of the mature IR but also plays an important role in controlling the autonomous activity of the IR precursor during its biosynthesis. Moreover, we provide a highly efficient tool to isolate new PTP1B substrates.
Supplementary Material
This work was supported by the Institut de Recherche Servier, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and the Association pour la Recherche sur le Cancer and the Ligue contre le Cancer, Comité de Paris.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PTPase
- protein tyrosine phosphatase
- IR
- insulin receptor
- PTP1B
- protein tyrosine phosphatase 1B
- ER
- endoplasmic reticulum
- Rluc
- Renilla luciferase
- BRET
- bioluminescence resonance energy transfer.
REFERENCES
- 1. Combettes-Souverain M., Issad T. (1998) Diabetes. Metab. 24, 477–489 [PubMed] [Google Scholar]
- 2. Virkamäki A., Ueki K., Kahn C. R. (1999) J. Clin. Invest. 103, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellis L., Clauser E., Morgan D. O., Edery M., Roth R. A., Rutter W. J. (1986) Cell 45, 721–732 [DOI] [PubMed] [Google Scholar]
- 4. White M. F., Shoelson S. E., Keutmann H., Kahn C. R. (1988) J. Biol. Chem. 263, 2969–2980 [PubMed] [Google Scholar]
- 5. Issad T., Combettes M., Ferre P. (1995) Eur. J. Biochem. 234, 108–115 [DOI] [PubMed] [Google Scholar]
- 6. King M. J., Sharma R. P., Sale G. J. (1991) Biochem. J. 275(Pt 2), 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldstein B. J., Ahmad F., Ding W., Li P. M., Zhang W. R. (1998) Mol. Cell Biochem. 182, 91–99 [PubMed] [Google Scholar]
- 8. Cheng A., Dubé N., Gu F., Tremblay M. L. (2002) Eur. J. Biochem. 269, 1050–1059 [DOI] [PubMed] [Google Scholar]
- 9. Galic S., Klingler-Hoffmann M., Fodero-Tavoletti M. T., Puryer M. A., Meng T. C., Tonks N. K., Tiganis T. (2003) Mol. Cell. Biol. 23, 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lacasa D., Boute N., Issad T. (2005) Mol. Pharmacol. 67, 1206–1213 [DOI] [PubMed] [Google Scholar]
- 11. Frangioni J. V., Beahm P. H., Shifrin V., Jost C. A., Neel B. G. (1992) Cell 68, 545–560 [DOI] [PubMed] [Google Scholar]
- 12. Cicirelli M. F., Tonks N. K., Diltz C. D., Weiel J. E., Fischer E. H., Krebs E. G. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5514–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahmad F., Li P. M., Meyerovitch J., Goldstein B. J. (1995) J. Biol. Chem. 270, 20503–20508 [DOI] [PubMed] [Google Scholar]
- 14. Kenner K. A., Anyanwu E., Olefsky J. M., Kusari J. (1996) J. Biol. Chem. 271, 19810–19816 [DOI] [PubMed] [Google Scholar]
- 15. Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., Loy A. L., Normandin D., Cheng A., Himms-Hagen J., Chan C. C., Ramachandran C., Gresser M. J., Tremblay M. L., Kennedy B. P. (1999) Science 283, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 16. Klaman L. D., Boss O., Peroni O. D., Kim J. K., Martino J. L., Zabolotny J. M., Moghal N., Lubkin M., Kim Y. B., Sharpe A. H., Stricker-Krongrad A., Shulman G. I., Neel B. G., Kahn B. B. (2000) Mol. Cell. Biol. 20, 5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Issad T., Boute N., Boubekeur S., Lacasa D., Pernet K. (2003) Diabetes Metab. 29, 111–117 [DOI] [PubMed] [Google Scholar]
- 18. Boute N., Boubekeur S., Lacasa D., Issad T. (2003) EMBO Rep. 4, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romsicki Y., Reece M., Gauthier J. Y., Asante-Appiah E., Kennedy B. P. (2004) J. Biol. Chem. 279, 12868–12875 [DOI] [PubMed] [Google Scholar]
- 20. Bandyopadhyay D., Kusari A., Kenner K. A., Liu F., Chernoff J., Gustafson T. A., Kusari J. (1997) J. Biol. Chem. 272, 1639–1645 [DOI] [PubMed] [Google Scholar]
- 21. Liu F., Chernoff J. (1997) Biochem. J. 327, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tao J., Malbon C. C., Wang H. Y. (2001) J. Biol. Chem. 276, 29520–29525 [DOI] [PubMed] [Google Scholar]
- 23. Tonks N. K. (2003) FEBS Lett. 546, 140–148 [DOI] [PubMed] [Google Scholar]
- 24. Nouaille S., Blanquart C., Zilberfarb V., Boute N., Perdereau D., Burnol A. F., Issad T. (2006) Biochem. Pharmacol. 72, 1355–1366 [DOI] [PubMed] [Google Scholar]
- 25. Blanquart C., Achi J., Issad T. (2008) Biochem. Pharmacol. 76, 873–883 [DOI] [PubMed] [Google Scholar]
- 26. Blanquart C., Boute N., Lacasa D., Issad T. (2005) Mol. Pharmacol. 68, 885–894 [DOI] [PubMed] [Google Scholar]
- 27. Blanquart C., Gonzalez-Yanes C., Issad T. (2006) Mol. Pharmacol. 70, 1802–1811 [DOI] [PubMed] [Google Scholar]
- 28. Angers S., Salahpour A., Joly E., Hilairet S., Chelsky D., Dennis M., Bouvier M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Issad T., Boute N., Pernet K. (2002) Biochem. Pharmacol. 64, 813–817 [DOI] [PubMed] [Google Scholar]
- 30. Issad T., Blanquart C., Gonzalez-Yanes C. (2007) Expert. Opin. Ther. Targets 11, 541–556 [DOI] [PubMed] [Google Scholar]
- 31. Liu J. F., Issad T., Chevet E., Ledoux D., Courty J., Caruelle J. P., Barritault D., Crépin M., Bertin B. (1998) Eur. J. Biochem. 258, 271–276 [DOI] [PubMed] [Google Scholar]
- 32. Issad T., Ferré P., Pastor-Anglada M., Baudon M. A., Girard J. (1989) Biochem. J. 264, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Formisano P., Najjar S. M., Gross C. N., Philippe N., Oriente F., Kern-Buell C. L., Accili D., Gorden P. (1995) J. Biol. Chem. 270, 24073–24077 [DOI] [PubMed] [Google Scholar]
- 34. Issad T., Boute N., Boubekeur S., Lacasa D. (2005) Biochimie 87, 111–116 [DOI] [PubMed] [Google Scholar]
- 35. Issad T., Young S. W., Tavaré J. M., Denton R. M. (1992) FEBS Lett. 296, 41–45 [DOI] [PubMed] [Google Scholar]
- 36. Pashmforoush M., Yoshimasa Y., Steiner D. F. (1994) J. Biol. Chem. 269, 32639–32648 [PubMed] [Google Scholar]
- 37. Goldstein B. J., Mahadev K., Kalyankar M., Wu X. (2005) Diabetes 54, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Montfort R. L., Congreve M., Tisi D., Carr R., Jhoti H. (2003) Nature 423, 773–777 [DOI] [PubMed] [Google Scholar]
- 39. Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 40. Dadke S., Kusari A., Kusari J. (2001) Mol. Cell. Biochem. 221, 147–154 [DOI] [PubMed] [Google Scholar]
- 41. Byon J. C., Kusari A. B., Kusari J. (1998) Mol. Cell. Biochem. 182, 101–108 [PubMed] [Google Scholar]
- 42. Anderie I., Schulz I., Schmid A. (2007) Exp. Cell Res. 313, 3189–3197 [DOI] [PubMed] [Google Scholar]
- 43. Zabolotny J. M., Bence-Hanulec K. K., Stricker-Krongrad A., Haj F., Wang Y., Minokoshi Y., Kim Y. B., Elmquist J. K., Tartaglia L. A., Kahn B. B., Neel B. G. (2002) Dev. Cell 2, 489–495 [DOI] [PubMed] [Google Scholar]
- 44. Julien S. G., Dubé N., Read M., Penney J., Paquet M., Han Y., Kennedy B. P., Muller W. J., Tremblay M. L. (2007) Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 45. Bentires-Alj M., Neel B. G. (2007) Cancer Res. 67, 2420–2424 [DOI] [PubMed] [Google Scholar]
- 46. Blanquart C., Karouri S. E., Issad T. (2009) Biochem. Biophys. Res. Commun. 387, 748–753 [DOI] [PubMed] [Google Scholar]
- 47. Blanquart C., Karouri S. E., Issad T. (2010) Biochem. Biophys. Res. Commun. 392, 83–88 [DOI] [PubMed] [Google Scholar]
- 48. Bliska J. B., Clemens J. C., Dixon J. E., Falkow S. (1992) J. Exp. Med. 176, 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun H., Charles C. H., Lau L. F., Tonks N. K. (1993) Cell 75, 487–493 [DOI] [PubMed] [Google Scholar]
- 50. Milarski K. L., Zhu G., Pearl C. G., McNamara D. J., Dobrusin E. M., MacLean D., Thieme-Sefler A., Zhang Z. Y., Sawyer T., Decker S. J., et al. (1993) J. Biol. Chem. 268, 23634–23639 [PubMed] [Google Scholar]
- 51. Flint A. J., Tiganis T., Barford D., Tonks N. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie L., Zhang Y. L., Zhang Z. Y. (2002) Biochemistry 41, 4032–4039 [DOI] [PubMed] [Google Scholar]
- 53. Mertins P., Eberl H. C., Renkawitz J., Olsen J. V., Tremblay M. L., Mann M., Ullrich A., Daub H. (2008) Mol. Cell. Proteomics 7, 1763–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.