Abstract
To achieve accurate DNA synthesis, DNA polymerases must rapidly sample and discriminate against incorrect nucleotides. Here we report the crystal structure of a high fidelity DNA polymerase I bound to DNA primer-template caught in the act of binding a mismatched (dG:dTTP) nucleoside triphosphate. The polymerase adopts a conformation in between the previously established “open” and “closed” states. In this “ajar” conformation, the template base has moved into the insertion site but misaligns an incorrect nucleotide relative to the primer terminus. The displacement of a conserved active site tyrosine in the insertion site by the template base is accommodated by a distinctive kink in the polymerase O helix, resulting in a partially open ternary complex. We suggest that the ajar conformation allows the template to probe incoming nucleotides for complementarity before closure of the enzyme around the substrate. Based on solution fluorescence, kinetics, and crystallographic analyses of wild-type and mutant polymerases reported here, we present a three-state reaction pathway in which nucleotides either pass through this intermediate conformation to the closed conformation and catalysis or are misaligned within the intermediate, leading to destabilization of the closed conformation.
Keywords: DNA Polymerase, DNA Synthesis, Enzyme Kinetics, Enzyme Mechanisms, X-ray Crystallography
Introduction
DNA polymerases replicate DNA with remarkably lower error rates than would be expected solely on the basis of the free energy differences between correct and incorrect base pairing (1). Polymerases, therefore, actively contribute to the discrimination against mismatched bases. Four mechanisms by which DNA polymerases enhance intrinsic base pairing specificity to achieve high fidelity have been proposed; 1) hydrogen bonding between correctly paired bases, 2) water exclusion from the active site, 3) geometric selection of correct size and shape, and 4) conformational changes associated with dNTP binding (for review, see Ref. 2).
X-ray crystal structures of high fidelity DNA polymerases have revealed a large conformational change that is associated with binding to the complementary dNTP. The structure of DNA polymerases has been likened to a hand with a “fingers” subdomain encoding a solvent-exposed dNTP binding site and the “palm” and “thumb” grasping the nascent DNA chain (Fig. 1A) (3, 4). For family A DNA polymerases, the template base is flipped out of the DNA base stack and resides in a “pre-insertion site” before encountering a dNTP. Upon binding a complementary dNTP, the fingers subdomain transitions from an open to a closed conformation, enclosing the nucleotide opposite the template base (Fig. 1B) (3, 5–8). A concerted dislocation of a highly conserved tyrosine in the O helix of family A enzymes enables the template to pair with the complementary dNTP in a newly formed “insertion site” and positions the triphosphate for in-line attack by the 3′-hydroxyl of the extending DNA strand. Although much is known about the conformational changes associated with binding of complementary nucleotides, mismatched substrates have been structurally less well characterized.
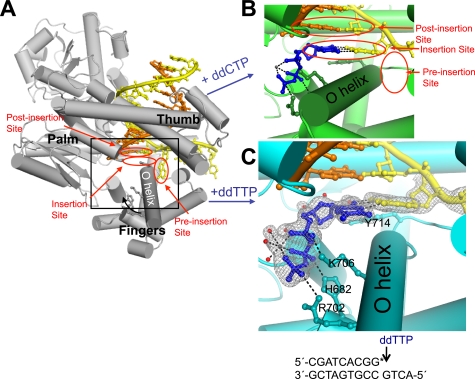
FIGURE 1.
DNA polymerase I structures preceding phosphodiester bond formation. Complementary nucleotide addition is accompanied by the transition from an open (A; PDB code 1L3S (3)) to the closed conformation (B; 2HVI (15)). This conformational change is characterized by rotation of the O helix (curved black arrow) in the fingers subdomain, thereby enclosing the correct ddCTP (stick and balls, blue) within the active site cleft. Red ovals, sites for key base pair interactions; yellow DNA, template strand; orange DNA, primer strand; gray spheres, Mg2+. The addition of a mismatched nucleotide results in an alternate structure (C) in which the template G pairs with the ddTTP (blue sticks and balls) in the insertion site but the fingers do not close. Residues that interact with the dG:ddTTP pair in the active site cleft are shown in sticks and labeled. Red spheres, active-site bound water molecules; black dashes, hydrogen bonds and magnesium-oxygen bonds; gray mesh, simulated-annealing omit map at 1.0 σ. The lower portion of panel C shows the sequence of DNA used in the mismatch crystallization experiment. The asterisk denotes the dideoxy-termination of the primer strand. Panels B and C are close-up views of the region of BF·DNA complex shown inside the black box in A. All models were generated using PyMol.
In the pool of all four dNTPs, DNA polymerases continually probe nucleotides and select the single complementary pair. It has been suggested that conformational changes play a critical role in the selection process. One model proposes that a dynamic equilibrium between the known open and closed conformations is sufficient to account for selectivity (9, 10); only the complementary dNTP can shift the equilibrium toward the closed conformation. A second model posits that an additional, uncharacterized state enables the template to “preview” the incoming dNTP in the open ternary complex (9, 11, 12); only the complementary dNTP progresses beyond this putative conformation to the catalytically competent, closed conformation.
Here we report x-ray crystal structures of the large fragment of DNA polymerase I from Bacillus stearothermophilus (Bacillus fragment (BF))3 bound to DNA and a mismatched nucleoside triphosphate in the active site cleft. We used 2′,3′-dideoxy-terminated primers to prevent nucleotide incorporation, similar to other studies (3, 5, 6, 8, 13). These structures reveal a previously unobserved intermediate conformation between the open and closed states that allows the template base to position the incoming nucleotide relative to the active site. In this conformation, mismatches are distinguished from Watson-Crick base pairs by a combination of effects from hydrogen bonding, active site water molecules, and geometric selection.
EXPERIMENTAL PROCEDURES
Materials
Expression and purification of a Bacillus fragment D598A/F710Y double mutant were performed as previously described (3, 14, 15). The mutations facilitate crystallization of trapped ternary complexes. The D598A mutations destabilize crystal contacts of a crystal form in which BF adopts the open conformation and the F710Y mutation allows the incorporation of a dideoxynucleoside monophosphate to the growing DNA strand to prevent further nucleotide addition (15). The V713P and D598A/V713P mutant BFs were generated from wild-type and D598A mutant BF expression vectors using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as described by the manufacturer's instructions. The D329A/Y714S BF double mutant was generated from an expression plasmid for the Asp-329 to Ala BF mutant (3). Protein was expressed and purified as previously described (14). Oligonucleotides template (5′-ACTGCCGTGATCG-3′), T2 (5′-ACTGGCGTGATCG-3′), primer (5′-CGATCACG-3′), 2APTemplate (5′-GACTXGCGTGATCGCA-3′, where X = 2-aminopurine), and PrimerddC (5′-CGATCACGY-3′, where Y = 2′,3′-dideoxycytosine monophosphate) were synthesized by Midland Certified Reagent Co., Inc. (Midland, TX). FluorPrimer (5′-CGAZCACGY-3′, where Z = fluorescein-dT) was synthesized by Operon Biotechnologies, Inc. (Huntsville, AL). High quality nucleotides were purchased from Promega Corp. (Madison, WI) and U. S. Biochemical Corp.
Crystallization and Structure Determination of Bacillus Fragment Bound to DNA and a Mismatched Nucleotide
The BF ternary complexes were crystallized in a similar fashion as described previously for a BF(D598A/F710Y)·DNA(dG)·ddCTP-Mg2+ complex (15). For the BF(D598A/F710Y)·DNA(dG)·ddTTP-Mg2+ crystal, annealed template and primer DNA were mixed with purified D598A/F710Y BF, 2 mm ddGTP, 5 mm ddTTP, and 20 mm MgSO4. Insertion of ddGMP terminates the primer strand to trap the ddTTP in the active site. The use of a 2′,3′-deoxyribonucleoside substrate was intended to complement the F710Y mutation. Crystals were grown by the hanging drop vapor diffusion at 17 °C from a MES (pH 5.8)-buffered solution of ∼50% saturated ammonium sulfate (3) and were transferred into cryoprotectant solutions with increasing concentrations of sucrose before plunging in liquid nitrogen. Our attempts to grow crystals with a chemically dideoxy-terminated primer, BF D598A, and dTTP resulted in an empty and open active site after soaking in cryoprotectant in the presence of dTTP (results not shown). Diffraction data were collected using 1.0 Å synchrotron radiation at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). For crystals of the V713P mutant BF, annealed T2 and PrimerddC were mixed with purified D598A/V13P BF, 5 mm dCTP or dTTP, and 20 mm MgSO4. Diffraction data were collected using 0.972 Å synchrotron radiation at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA). Diffraction data were indexed, integrated, and scaled using XDS (16). All data sets were initially phased by rigid body refinement of the BF·DNA(dG)·ddCTP complex (PDB code 2HVI (15)). Crystal structures were refined using REFMAC5 (17) and rebuilt in Coot (18). Stereochemistry analysis using Molprobity (19) indicated that 98.0 and 2.0% of all residues in BF(D598A/F710Y)·DNA(dG)·ddTTP-Mg2+ and BF(D598A/V713P)·DNA(dG)·dCTP-Mg2+ were in the favored and allowed regions of the Ramachandran plot. No backbone torsions were in the disallowed region.
To crystallize BF(D329A/Y714S)·DNA(dG)·dTTP-Mg2+, complementary oligonucleotides T2 and PrimerddC were annealed and mixed with purified D329A/Y714S BF double mutant, 5 mm dTTP, and 20 mm MgSO4. Crystals were grown as described above. Diffraction data were processed using XDS and then initially phased by molecular replacement using Phaser (20) using a binary BF·DNA complex (PDB code 1L3U (3)) as the search model. After refinement using REFMAC5, the N and O helices of the fingers subdomain in the BF(D598A/Y714S)·DNA(dG)·dTTP-Mg2+ structure showed positive difference electron density near the modeled atoms due to a partial occupancy of the open conformation. The occupancy of the ajar conformation containing the dTTP substrate was estimated to be ∼80% judging by the quality of the Fo − Fc difference electron density maps; the remaining ∼20% was attributed to BF in the open conformation, devoid of substrate. Stereochemistry analysis using Molprobity (19) indicated that 97.8 and 2.2% of all residues in BF(D329A/Y714S)· DNA(dG)·dTTP-Mg2+ were in the favored and allowed regions of the Ramachandran plot. No backbone torsions were in the disallowed region.
Structural Analysis
The open conformation (PDB code 1L3U) and closed conformation (PDB code 2HVI) models of Bacillus fragment were aligned to the ajar conformation model using PyMol (Delano Scientific, Palo Alto, CA). Root mean square distances were calculated using PyMol. Solvent exposure of substrates and hydrophobic residues in the fingers subdomain (residues 678–744) was calculated by determining the solvent accessible surface area (21) using StrucTools. A probe radius of 1.4 Å was used.
2-Aminopurine Fluorescence Spectroscopy
Annealed complementary oligonucleotides 2APTemplate and PrimerddC were mixed with an equimolar amount of wild-type BF (described below) and diluted in a cuvette to 500 nm in reaction buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol). Fluorescence measurements were taken as dCTP or dTTP were titrated into the cuvette at 25 °C (λex = 313 or 315 nm, λem = 330–460 nm). Fluorescence from protein and buffer alone were subtracted from all spectra to measure fluorescence from the template 2-aminopurine (2AP). All spectra were normalized for the small volume changes due to the addition of dNTPs. Fluorescence was measured in an Aminco-Bowman Series 2 Luminescence Spectrometer (Spectronic Instruments, Rochester, NY).
Fluorescence Resonance Energy Transfer (FRET)
The wild-type Bacillus fragment open reading frame was mutated using the QuikChange multi-site-directed mutagenesis kit (Stratagene) in three locations to change cysteines 388 and 845 to serines and glutamine 691 to cysteine as directed by the manufacturer's instructions. The BF triple mutant (C388S/C845S/Q691C) was expressed and purified as previously described (14). Concentrated protein was then treated with tris-(2-carboxethyl)phosphine to reduce the lone cysteine. Cy3-maleimide (Amersham Biosciences) was dissolved in dimethylformamide and mixed with reduced protein for 2 h at room temperature, then overnight at 4 °C. Excess dye was separated from Cy3-labeled protein (Cy3BF) using a desalting column. Absorbance measurements at 280 and 550 nm indicated that 82% of the protein was labeled (data not shown). FRET was observed by comparing the fluorescence spectrum (λex = 475 nm, λem = 490–640 nm) of a 500 nm mixture of Cy3BF and annealed FluorPrimer and T2 oligonucleotides (FlDNAddG) with the spectra of Cy3BF alone and FlDNAddG alone (see supplemental Fig. 3). Fluorescence measurements were taken as dCTP or dTTP were titrated into a cuvette containing 500 nm Cy3BF and 100 nm FlDNAddG in reaction buffer.
Presteady State Kinetics of BF V713P Mutant
Complementary oligonucleotides T75 (5′- TTACTTGACCAGATACACTGTCTTTGACACGTTGATGGATTAGAGCAATCACATCCAAGACTGGCTATGCACGAA-3′) and P67 (5′-(6-carboxyfluorescein)-TCGTGCATAGCCAGTCTTGGATGTGATTGCTCTAATCCATCAACGTGTCAAAGACAGTGTATCTGGT-3′) were synthesized (Operon Biotechnologies, Inc., Huntsville, AL) then dissolved and annealed as described above. P67 contains a fluorescent 6-carboxyfluorescein dye at the 5′ end. Pilot experiments showed that mixing annealed T75:P67 DNA with BF and dCTP resulted in the extension of the fluorescently labeled P67 strand by one nucleotide, which could be separated from P67 and quantitated using capillary electrophoresis (data not shown). T75:P67 DNA and BF wild type or BF V713P were diluted with reaction buffer to 0.1 and 0.5 μm, respectively. The BF:DNA complex was then mixed with an equal volume of dCTP or dTTP at various concentrations at room temperature and then quenched using 4 reaction volumes 95 °C formamide. Reactions too fast to capture on the benchtop were performed in a KinTek RQF-3 Rapid Quench Flow instrument and quenched using 95% formamide, 25 mm EDTA. Reaction products were separated and quantitated using an ABI3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). The fraction of P67 that had been extended was plotted for each time point, and the data were fitted to a hyperbolic curve to determine the rate at each dNTP concentration. Kinetic parameters were derived from curve fits of rates versus dNTP concentration plots.
Accession Codes
Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 3HP6 (BF·DNA(dG)·ddTTP-Mg2+), 3HPO (BF(Y714S)·DNA(dG)·dTTP-Mg2+), and 3HT3 (BF(V713P)·DNA(dG)· dCTP-Mg2+.
RESULTS
The dG:ddTTP mismatch was captured in the BF polymerase double mutant F710Y/D598A by co-crystallization with a 3′-recessed double-stranded DNA containing a deoxyguanosine (dG) in the template overhang together with 2′,3′-dideoxythymidine triphosphate (ddTTP) as the mismatched incoming nucleotide. The Phe-710 to Tyr mutation promotes incorporation of dideoxynucleotides (22, 23); the Asp-598 to Ala mutation destabilizes a crystal contact that predisposes BF to crystallize in the open conformation (3, 15). To capture reaction intermediates, the primer strand was terminated by the incorporation of a ddGMP. Using this DNA substrate, we observed a misaligned dG:ddTTP complex in which the O helix adopts a previously unreported, bent conformation that has characteristics in between the fully open and closed state, which we term the ajar conformation.
Structure of a Distinct Intermediate Conformation in the Bacillus Fragment
We crystallized a ternary complex of Bacillus fragment bound to a 2′,3′-dideoxythymidine triphosphate opposite a template guanine and solved its structure to 1.81 Å resolution (BF(D598A/F710Y)·DNA(dG)·ddTTP-Mg2+; r = 21.0, Rfree = 24.6; Fig. 1C and Table 1). The asymmetric unit contains two polymerases that are structurally indistinguishable from each other (r.m.s.d. (Cα) = 0.40 Å). In these mismatched ternary complexes, we observed the thymine base of the incoming ddTTP directly interacting with the template guanine by forming specific hydrogen bonds. The dG:ddTTP mismatch forms a wobble pair (Fig. 2A) with two hydrogen bonds between the bases. A water molecule positioned approximately in the base pair plane completes the hydrogen-bonding potential between the two bases (Fig. 2A). This wobble pairing displaces the incoming ddTTP by 2.8 Å (root mean square deviation) relative to a complementary dG:ddCTP pair and thereby misaligns the α-phosphate for attack by the 3′-hydroxyl of the primer terminus (Fig. 2B). Seven water molecules fill the void resulting from this displacement (Fig. 2, B and C); ddTTP is, therefore, more solvated than the complementary ddCTP (solvent-accessible surface areas of 121 and 37 Å2, respectively) in the BF active site. Three of the extra water molecules in the ddTTP complex together with three triphosphate oxygen atoms chelate a magnesium (II) in an octahedral geometry (Fig. 1C). The non-bridging phosphate oxygens that are not coordinated by Mg(II) interact with His-682, Arg-702, and Lys-706 (Fig. 1C). We did not observe a second metal ion common in the active sites of polymerase enzymes (24) using anomalous scattering in the presence of manganese instead of magnesium (results not shown), likely due to the missing 3′-OH in the primer strand that chelates the metal ion. The template guanine displaces the conserved Tyr-714 from the insertion site and occupies a nearly identical position in both the complementary and mismatched nucleotide structures (Fig. 2A).
TABLE 1.
Crystallographic data collection and refinement statistics
| BF(D598A/F710Y)·DNA(dG)·ddTTP-Mg2+ | BF(D329A/Y714S)·DNA(dG)·dTTP-Mg2+ | BF(D598A/V713P)·DNA(dG)·dCTP-Mg2+ | |
|---|---|---|---|
| Chain terminator | ddGMP incorporation | Synthetic preterminated oligonucleotide | Synthetic preterminated oligonucleotide |
| Data collection | |||
| Space group | P 212121 | P 212121 | P 212121 |
| Cell dimensions a, b, c (Å) | 93.2, 108.7, 152.2 | 90.2, 93.7, 105.5 | 93.3, 108.8, 152.1 |
| No. complexes in asymmetric unit | 2 | 1 | 2 |
| Resolution (Å)a | 42.9-1.81 (1.91-1.81) | 46.0-1.74 (1.85-1.74) | 47.0-1.70 (1.74-1.70) |
| Rsym (%)a | 5.8 (51.0) | 8.4 (40.9) | 6.0 (56.1) |
| I/σIa | 15.5 (3.0) | 10.7 (2.9) | 14.4 (2.5) |
| Completeness, %a | 98.9 (98.2) | 90.6 (77.9) | 99.8 (99.8) |
| Redundancya | 5.0 (5.0) | 5.0 (4.5) | 3.9 (3.9) |
| Refinement | |||
| Resolution (Å) | 42.8-1.81 | 46.0-1.75 | 47.0-1.70 |
| No. reflections | 133,362 | 79,376 | 162,156 |
| Rwork/Rfreeb | 21.0/24.6 | 20.1/24.1 | 21.0/24.7 |
| No. atoms | |||
| Protein | 9,376 | 4,685 | 9,299 |
| DNA | 848 | 387 | 848 |
| Ligands and ions | 119 | 58 | 162 |
| Water | 869 | 724 | 996 |
| B-factor (Å2) | |||
| Protein | 29.2 | 23.5 | 22.5 |
| DNA | 32.1 | 26.2 | 27.6 |
| Ligands and ions | 32.9 | 23.6 | 27.2 |
| Water | 35.9 | 36.6 | 30.2 |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.012 | 0.008 | 0.010 |
| Bond angles (°) | 1.43 | 1.18 | 1.30 |
| PDB code | 3HP6 | 3HPO | 3HT3 |
a The highest resolution shell is shown in parentheses.
b Test setup to 1.98 Å resolution was inherited from PDB code 2HVI (15). 5% of reflections from 1.98-1.70 Å were randomly selected to complete the set.
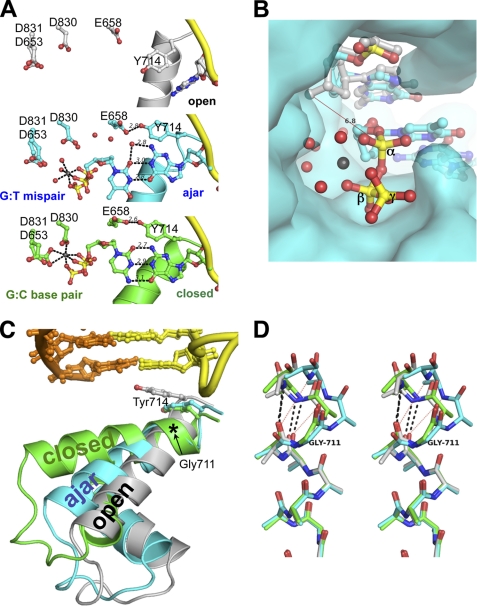
FIGURE 2.
Structures of a dG:ddTTP mismatch in the ajar Bacillus fragment active site. A, shown is a comparison between the binary BFO·DNA complex (gray; top panel), the dG:ddTTP mismatch (cyan; center panel), and the complementary dG:ddCTP base pair (gold; bottom panel). Red spheres, water molecules; gray spheres, Mg2+. B, the triphosphate of the mismatched nucleotide is misaligned for attack by the primer 3′-OH for catalysis. The open binary complex was superimposed onto the BF·DNA(dG)·ddTTP-Mg2+ crystal structure. A red line connects the modeled 3′-hydroxyl of the primer strand in the open conformation with the α-phosphate of the mismatched ddTTP. The interatomic distance is 6.8 Å. The BF polymerase is shown as its solvent-accessible surface. Red spheres, water molecules; gray sphere, Mg2+; yellow, α, β, and γ phosphorus atoms. C, the N and O helices of the fingers subdomain of BF polymerase in the open (gray), ajar (cyan), and closed (green) conformations is shown in ribbons. Tyr-714 and the post-insertion site base pair are shown in sticks. The location of Gly-711 is denoted by an asterisk. Incoming dNTPs from the ajar and closed structures have been omitted for clarity. D, structural superposition of the residues 700–710 of the O helix (in stereo) shows breakage of backbone hydrogen bonds starting at Gly-711 in the ajar conformation. Only backbone and Cβ atoms are shown. Hydrogen bonds between the backbone carbonyl oxygens of Asn-709 and Phe-710 and nitrogens of Val-713 and Tyr-714 are shown in black dashed lines for the open (gray) and closed (green) conformations. Red dotted lines show the broken hydrogen bonds in the ajar (cyan) conformation.
The misaligned base pair is bound to Bacillus fragment in an ajar conformation of the fingers subdomain, which is intermediate between the open and closed conformations and contains elements of both (Fig. 2C). In this ajar conformation, most of the residues in the mobile α-helices N, O, and O1 (residues 681–725) are nearer to the open conformation than the closed conformation (root mean square deviation (681–725, Cα)mismatch-open = 3.2 Å, root mean square deviation (681–725, Cα)mismatch-closed = 4.8 Å). However, the C-terminal helical turn of the O helix (residues 710–715; see supplemental Fig. 1) superimposes well with the closed conformation; it is distorted due to the displacement of Tyr-714 from the insertion site (Fig. 2A). To accommodate the template base in the insertion site, the hydrogen bonds between the backbone carbonyl oxygens of Asn-709 and Tyr-710 and the backbone amide nitrogens of Val-713 and Tyr-714, respectively, are broken, thereby unraveling the end of the O helix (Fig. 2D). In contrast, the O helix is intact in the open and closed conformations. The O helices of all three conformations pivot at a highly conserved glycine (711 in BF; Fig. 2, C and D), which is a strong breaker of α-helices (25). The breakage of the O helix at Gly-711 appears to ease the displacement of Tyr-714 from the insertion site without forcing the rest of the O helix to rotate to the closed conformation.
To further probe the role of the tyrosine 714 steric gate in nucleotide discrimination, we solved the crystal structure of a D329A/Y714S double mutant of Bacillus fragment bound to a dG:dTTP mismatch. The homologous Y766S mutant of the Escherichia coli DNA polymerase I Klenow fragment (KF) is error-prone (26) and favors misinserting T opposite a template G (27). Like D598A, D329A destabilizes a crystal contact that predisposes BF to crystallize in the open conformation (3). The cocrystallization of the Y714S mutant with a dG·dTTP mismatch was performed with a synthesized oligonucleotide primer strand that contained a dideoxy nucleoside monophosphate at the terminus and thus did not require the F710Y mutation. The structure of BF(D329A/Y714S)·DNA(dG)·dTTP-Mg2+ was solved to 1.75 Å resolution in a crystal form with only one molecule in the asymmetric unit and a smaller unit cell (r = 20.1, Rfree = 24.1; Table 1) and is nearly identical to the BF(D598A/F710Y)·DNA(dG)·ddTTP-Mg2+ complex in the ajar conformation (see supplemental Fig. 2). There is no obvious difference between the structures of mismatches with a Tyr-714 or a smaller Ser-714 that would explain the increase in misinserting nucleotides by the mutant. On the other hand, the similarity of these two crystal structures indicates that the crystal packing and the location of a single hydroxyl group, whether on the incoming nucleotide or on residue 710, do not have any appreciable impact on the structure of the mismatch or the enzyme active site.
The intermediate ajar conformation of BF observed in both crystals had not been observed when the complementary nucleotide is bound. It was, therefore, not clear if the ajar conformation arises only in the presence of a G·T mismatch or if it is an intermediate between the open and closed conformations in a more general mechanism for nucleotide binding and discrimination. We asked if the enzyme could be trapped in the ajar conformation in the presence of the complementary nucleotide. To capture the selection for Watson-Crick base pairing in the ajar conformation, we mutated valine 713 to proline, which should selectively stabilize the ajar conformation relative to the closed states. The proline mutation blocks the formation of a hydrogen bond between the carbonyl oxygen of residue 709 and the backbone nitrogen of residue 713. It also sterically hinders the formation of a contiguous O helix observed in the closed conformation. The cocrystallization of the V713P mutant was also performed with a synthesized oligonucleotide primer strand that contained a dideoxy nucleoside monophosphate at the terminus and thus did not require the F710Y mutation. As expected, a mismatched dG:dTTP pair induces the ajar conformation in the V713P mutant BF polymerase that is nearly identical to the BF(D598A/F710Y)·DNA(dG)·ddTTP-Mg2+ complex (data not shown). The crystal structure of a complementary dG:dCTP pair in the V713P mutant BF polymerase (BF(D598A/V713P)·DNA(dG)·dCTP-Mg2+; r = 21.0%, Rfree = 24.1%; Table 1) shows a complementary dCTP making a Watson-Crick base pair in the ajar conformation (Fig. 3A). Instead of enclosing the complementary nucleotide, the O helix of the V713P mutant BF is bent at Gly-711, and the bound dCTP is solvent-exposed (Fig. 3B). The triphosphate moiety is held far away from the active site, which is likely to slow catalysis.
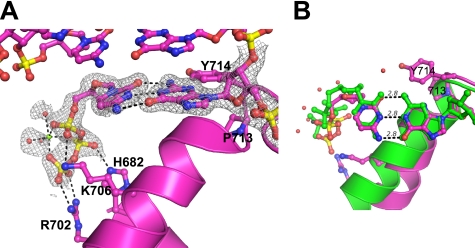
FIGURE 3.
Structure of complementary base pair in the insertion site trapped in the ajar conformation. The structure of the V713P mutant BF bound to a dG:dCTP pair (magenta) is shown from the side (A) and from the top (B). The N and O helices are represented by ribbons (A). The closed conformation (PDB code 2HVI) is shown in green. Gray sphere, magnesium ion; red spheres, nearby water molecules; gray mesh, simulated-annealing omit map at 1.0σ; black lines, hydrogen bonds and magnesium-oxygen bonds.
The V713P Mutant Slows Catalysis
If the ajar conformation is indeed an intermediate in the enzymatic mechanism, obstacles to transitions from the ajar conformation to the closed conformation should affect the rates of catalysis for complementary nucleotides. We measured solution kinetics of complementary (dC) and incorrect (dT) nucleotide incorporation into an oligonucleotide by wild-type or V713P BF (Table 2), which should have a relatively unstable closed conformation. In comparison to wild-type BF, the V713P mutant BF exhibits a similar affinity for the complementary nucleotide but an ∼10-fold slower rate of polymerization (kpol). The kpol for incorrect nucleotide incorporation is slowed over 100-fold by the mutation. As a result, the V713P mutant BF polymerase is generally slower than wild-type BF, whether binding the correct or incorrect nucleotide.
TABLE 2.
Pre-steady state kinetic parameters for nucleotide insertion opposite a template guanine in wild-type BF and BF V713P mutant
See supplemental Fig. 4 online for experimental data and curve fits.
| BF | Nascent base pair | KD | Kpol | Kpol/KD | Relative efficiencya |
|---|---|---|---|---|---|
| μm | s−1 | μm−1s−1 | |||
| Wild type | dG:dCTP | 21 | 52 | 2.3 | |
| dG:dTTP | 830 | 0.55 | 0.00066 | 2.6 × 10−4 | |
| V713P | dG:dCTP | 12 | 2.8 | 0.24 | |
| dG:dTTP | 220 | 0.0036 | 0.000017 | 7 × 10−5 |
a Ratio of kpol/KD values for dTTP vs dCTP incorporation opposite dG.
Conformational States in Solution Probed by Fluorescence
We used FRET to probe whether complementary and mismatch base pairs induce different conformational changes (10, 12). A fluorescein donor fluorophore was attached to the n-6 position of the primer strand of a dideoxy-terminated substrate DNA, and Cy3 acceptor was attached to a cysteine introduced at position 691 in the BF O helix (see supplemental Fig. 3). Consistent with previous work on homologous KF and T7 DNA polymerases that were fluorescently labeled in a similar location near the O helix (12, 28), titration of dCTP (complementary nucleotide) or dTTP (mismatch) to this labeled complex resulted in a decrease and increase respectively in donor fluorescence emission intensity (Fig. 4, A and B). Because of the lack of knowledge about the relative orientations of the donor and acceptor fluorophores, we cannot relate distances between the fluorophores in the presence of the correct or incorrect nucleotides. The opposite directions of fluorescence intensity changes, however, indicate that the two nucleotides elicit distinct conformational changes in the enzyme, consistent with the inability of the mismatch to form the closed state. The data fit to a single-site binding isotherm (Fig. 4C) revealed that the mismatch binds ∼30-fold more weakly than the complementary base.
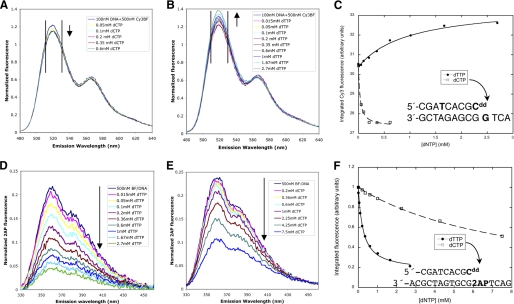
FIGURE 4.
Solution fluorescence studies of BF bound to double-stranded DNA containing a dideoxy-terminated primer strand. A–C, fluorescence resonance energy transfer between a fluorescent donor attached to the n-6 position of the DNA primer strand (green) and a Cy3 acceptor attached to Cys691 in the finger domain of BF wild type. Donor emission decreases upon titration with the complementary dCTP (A) but increases upon titration with a dTTP mismatch (B) as indicated by arrows. Emission intensities were integrated over the 511–530-nm interval (black vertical lines) and fit to a single-site binding isotherm (C) with Kd values of 29 mm for dCTP and 1.0 mm for dTTP. The DNA sequence is shown in the inset. D–F, change in emission intensity of 2-aminopurine placed in the template acceptor base position (magenta) upon titration of BF wild type with the complementary dTTP (D) and a dCTP mismatch (E) were integrated over the 330–460-nm interval and fit to single-site binding isotherms with Kd,dTTP = 0.29 mm and Kd,dCTP = 11 mm for BF wild type.
The structure of the ajar conformation revealed that the incorrect nucleotide can also induce the template base to move from the preinsertion into the insertion site. This movement can be observed in solution by monitoring the fluorescence intensity emission of 2AP, an environmentally sensitive nucleoside analog, placed in the template acceptor position (29). 2AP, like adenine, can form two hydrogen bonds with thymine that are isosteric with a Watson-Crick base pair. Consistent with previous work on 2AP as the template base in KF (29), titration of dTTP (complementary base) into BF complexed with a 2AP template decreases fluorescence emission intensity (Fig. 4D), presumably as a result of the movement of 2AP into the insertion site. Titration of dCTP (mismatch) also decreases 2AP fluorescence emission intensity to a similar saturation point (Fig. 4E), supporting the hypothesis that a mismatched nucleotide also induces the template base to move into the insertion site. Furthermore, binding isotherms again show that the mismatch binds ∼30-fold more weakly than the complementary nucleotide (Fig. 4F). Together, these equilibrium studies show that, although both mismatched and complementary nucleotides move the template base into the insertion site (Fig. 4, D–F), the dG:dTTP mismatch induces an O helix conformation that is distinct from that observed in the presence of the complementary nucleotide (Fig. 4, A–C). The ajar conformation observed in the crystal structure of BF when bound to a mismatch (Fig. 2, A and C) is consistent with both of these observations in solution phase.
DISCUSSION
Before chemical incorporation, DNA polymerases must test incoming nucleotides against the template base to distinguish between complementary and mismatch nucleotides. Experimental studies (11, 12) suggest that this process involves at least one distinct conformational state that precedes the adoption of the closed state in family A DNA polymerases. We determined the crystal structure of a BF polymerase complex in which the template base has moved into the insertion site and is paired with a mismatched and misaligned nucleotide. Importantly, the template base occupies the same location when bound to a mismatched or a complementary nucleotide, indicating that the template is used to align or misalign incoming nucleotides relative to the active site using hydrogen bonds between bases. The mismatched and misaligned ternary complex appears unable to fully desolvate the incoming nucleotide, suggesting that active site water molecules play a role in nucleotide selection.
A key feature of the protein structure in the mismatched ternary complex is the pronounced kink in the mobile O helix near the active site. In the ajar conformation, the O helix is positioned at an angle intermediate to that adopted in the open or close state. In contrast to the open and closed states, the O helix is bent at Gly-711, allowing the template base to interrogate the incoming nucleotide without enclosing it. In addition, we trapped the enzyme complex bound to a complementary dNTP in the ajar conformation by mutating an O helix residue that inhibits the closed state. This observation demonstrates that the polymerase is capable of adopting the ajar conformation in the presence of a complementary or a noncomplementary nucleotide. Thus, this ajar conformation has the hallmarks expected for the structure of a reaction intermediate involved in previewing nucleotides in the polymerase active site.
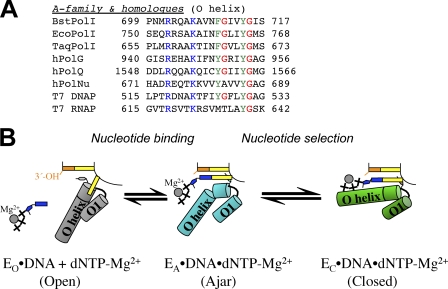
The importance of the glycine hinge is further underlined by its conservation pattern among the members of the family of polymerases. This residue is conserved in T7 DNA polymerase, bacterial DNA polymerase I enzymes, human family A DNA polymerases γ (hPolG), which replicates mitochondrial DNA with high fidelity (30), and θ (hPolQ), a DNA repair DNA polymerase with conflicting reports on its fidelity (31, 32), but not in DNA polymerase υ (hPolNu), a low fidelity repair polymerase with a particular propensity for incorporating a T opposite a template G (33) (Fig. 5A). Low fidelity Y-family DNA polymerases that participate in DNA repair are hypothesized not to employ large-scale conformational changes during nucleotide binding of complementary or mismatched nucleotides (34) nor do they have an O-helix equivalent (35). The juxtaposition of these high and low fidelity family A DNA polymerases suggests that the existence of the ajar conformation contributes to fidelity in DNA replication.
FIGURE 5.
A, shown is structure-based manual sequence alignment of nucleotide binding helices of A family DNA polymerases. Blue, phosphate interaction; green, aromatic residues in the nucleotide binding sites; red, proposed glycine hinges in the helices. BstPolI, EcoPolI, and TaqPolI, DNA polymerase I from B. stearothermophilus, E. coli, and Thermus aquaticus; hPolG, hPolQ, and hPolNu, human DNA polymerases γ, θ, and ν. B, a model for nucleotide sampling and selection is shown. Nucleotides are sampled in the ajar conformation (EA, cyan) and are released if it incorrect, whereupon the enzyme returns to the open conformation (EO, gray) or entrapped in the closed conformation (EC, green) if it is complementary to the template base. Schematic representations of each state are shown in the center.
Mismatch ternary complexes have also been captured in human DNA polymerase β (Pol β), an X-family DNA polymerase, using non-hydrolyzable nucleotide analogues and manganese in place of magnesium (36). Despite the similarity in DNA replication accuracy and the two-metal ion catalytic mechanisms between A- and X-family DNA polymerases, BF and polymerase β recognize mismatches using distinct mechanisms (36). Polymerase β does not contain the glycine hinge. Instead of forming a mismatched base pair as in BF, incorrect nucleotides in polymerase β slow catalysis by displacing the primer terminus and template (36).
Recently, the conformations adopted by the homologous Klenow fragment from E. coli DNA polymerase I (KF) in the presence of DNA and nucleotides were studied by single-molecule FRET between fluorophores on the thumb and fingers subdomains (37). When bound to mismatches (template A with dGTP or template T with dTTP), the apparent FRET efficiency slightly increased to an intermediate level between values for the open and closed conformations, suggesting that the O helix of KF adopted a conformation intermediate of the open and closed conformations. This alternate ternary complex is more similar to the open than the closed conformations and is presented as a candidate for a kinetic checkpoint before the closed conformation, in which the template tests the incoming nucleotide for complementarity. The ajar conformation we describe here using the dG:dTTP mismatch in BF, which is more similar to the open than the closed conformation (Fig. 2C), matches closely with the mismatched ternary complexes observed in KF. It is quite possible that both enzymes use a similar mechanism to distinguish multiple mismatches.
In previously proposed structural reaction pathways, the template base pairs with an incoming dNTP either in the preinsertion site in the open conformation (37) or in the insertion site with the triphosphate bound to the active site aspartic acid residues instead of the O helix (11). Here, we observe deoxynucleoside triphosphates base-paired with the template base in the insertion site in an intermediate conformation with the triphosphate clearly bound to the O helix. In addition, our 2-aminopurine fluorescence quenching experiments indicate that both complementary and mismatched nucleotides induce template movement into the insertion site, which is inconsistent with base pairing in the pre-insertion site. A targeted molecular dynamics simulation of events after phosphodiester bond formation in the BF polymerase showed that the enzyme transitions from the closed state back to the open state through a bent O helix state similar to the ajar conformation during DNA translocation (38), further supporting the ajar conformation as a key intermediate in DNA polymerase mechanism. Based on the intermediate nature of the ajar conformation between open and closed states, we present a reaction pathway by family A high fidelity DNA polymerases.
In our interpretation (Fig. 5B), nucleotide selection operates by a modulation of the free energies of the closed state and relaxation back to the open state from the ajar conformation. A nucleotide binds along the O helix and initially interacts with the template base in the ajar conformation. Our studies leave open the question of whether the incoming nucleotide induces the ajar conformation upon binding or the enzyme exists in equilibrium between the open and ajar states. A complementary dG:dCTP base pair passes through the ajar conformation to the low energy closed conformation (see supplemental Movie S1), stabilized by three hydrogen bonds between bases, proper geometric alignment, and binding of magnesium to the polymerase active site and to the incoming triphosphate. The closed conformation correctly aligns the chemical partners for catalysis. For the dG:dTTP mismatch, the closed state is destabilized due to having two instead of three interbase hydrogen bonds and to misalignment of the incoming nucleotide away from the active site. Misalignment of the mismatched nucleotide appears to impede its desolvation in the active site and may interfere with the proper chelation of catalytic magnesium ions by active site aspartates. The relatively high energy of the mismatched closed state results mostly in relaxation back to the open state and nucleotide release. FRET studies suggest that the closed state is partially occupied by Klenow fragment DNA polymerase in the presence of mismatches (37), and further discrimination of the mismatch may occur in the closed state or in additional states between ajar and closed.
In this model, the transitional ajar conformation is relatively unstable compared with the open and closed states, explaining why we are unable to observe the ajar conformation without the use of BF mutants (F710Y with dG·ddTTP, Y714S with dG·dTTP, or V713P with dG·dCTP) that potentially stabilize the ajar conformation selectively (see “Experimental Procedures”). The observation of the ajar conformation with three different mutations at three different residues suggests that the bending of the O helix is not the result of any single mutation. An additional factor in the instability of the observed ajar conformation is the missing divalent cation at metal “site A” (24). The use of a 2′,3′-dideoxy-terminated primer strand to prevent nucleotide insertion removes a chelating ligand for the site A metal, but it has been observed in the complementary ternary complex of Bacillus fragment (38). Recent solution fluorescence experiments on E. coli KF indicate that the site A metal does not bind until the fingers subdomain have closed around the complementary nucleotide (39). The absence of a site A metal in the BF ajar conformation correlates with the possibility that the ajar conformation precedes the closed conformation during nucleotide selection.
Although we have captured a mismatched nucleotide bound in the active site cleft of a high fidelity DNA polymerase, the structure does not answer how mismatches are incorporated. Because of a high energy barrier to incorrect nucleotide incorporation in the open conformation, it was hypothesized that “nucleotide misinsertions may occur from a partially open conformation” (40). The triphosphates of dG:dTTP mismatches we describe in the ajar conformation are distant from the location of the 3′-OH and not properly aligned for attack, suggesting that catalysis of mismatch incorporation in the ajar conformation would be very slow. In the context of our model for nucleotide selection, the observation that the V713P mutant that inhibits the closed state slows mismatched nucleotide incorporation more than complementary nucleotide incorporation (Table 2) suggests that mismatched nucleotides must still transition from the ajar the closed conformation for catalysis.
Supplementary Material
Acknowledgments
We thank H. Hellinga and W. Wang (Duke University Medical Center) for discussion and critically reading this manuscript. Data were collected at SE Regional Collaborative Access Team (SER-CAT) 22-ID (or 22-BM) beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at the SER-CAT website. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38. J. Phillips assisted with X-ray data collection. T. Pohlhaus, S. Armstrong, and C. Nelson assisted with site-directed mutagenesis. S. Langdon (Duke University Medical Center) assisted with kinetics experiments. H. Hellinga and P. Modrich (Duke University Medical Center) graciously allowed use of key instrumentation.
This work was supported, in whole or in part, by National Institutes of Health Grants 5 P01 CA92584 and R01 GM91487 (to L. S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Movie S1.
The atomic coordinates and structure factors (codes 3HP6, 3HPO, and 3HT3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BF
- Bacillus fragment
- 2AP
- 2-aminopurine
- KF
- Klenow fragment
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Goodman M. F. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10493–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kunkel T. A. (2004) J. Biol. Chem. 279, 16895–16898 [DOI] [PubMed] [Google Scholar]
- 3. Johnson S. J., Taylor J. S., Beese L. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y., Kong Y., Korolev S., Waksman G. (1998) Protein Sci. 7, 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Franklin M. C., Wang J., Steitz T. A. (2001) Cell 105, 657–667 [DOI] [PubMed] [Google Scholar]
- 6. Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. (1998) Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 7. Doublié S., Sawaya M. R., Ellenberger T. (1999) Structure 7, R31–R35 [DOI] [PubMed] [Google Scholar]
- 8. Li Y., Korolev S., Waksman G. (1998) EMBO J. 17, 7514–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joyce C. M., Benkovic S. J. (2004) Biochemistry 43, 14317–14324 [DOI] [PubMed] [Google Scholar]
- 10. Rothwell P. J., Mitaksov V., Waksman G. (2005) Mol. Cell 19, 345–355 [DOI] [PubMed] [Google Scholar]
- 11. Rothwell P. J., Waksman G. (2007) J. Biol. Chem. 282, 28884–28892 [DOI] [PubMed] [Google Scholar]
- 12. Joyce C. M., Potapova O., Delucia A. M., Huang X., Basu V. P., Grindley N. D. (2008) Biochemistry 47, 6103–6116 [DOI] [PubMed] [Google Scholar]
- 13. Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. (1994) Science 264, 1891–1903 [PubMed] [Google Scholar]
- 14. Kiefer J. R., Mao C., Hansen C. J., Basehore S. L., Hogrefe H. H., Braman J. C., Beese L. S. (1997) Structure 5, 95–108 [DOI] [PubMed] [Google Scholar]
- 15. Warren J. J., Forsberg L. J., Beese L. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19701–19706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 17. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerstein M., Richards F. M., Chapman M. S., Connolly M. L. (2001) in International Tables for Crystallography (Rossmann M. G. A. E. ed) pp. 531–545, Kluwer Academic Publishers, Norwell, MA [Google Scholar]
- 22. Tabor S., Richardson C. C. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6339–6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Astatke M., Grindley N. D., Joyce C. M. (1998) J. Mol. Biol. 278, 147–165 [DOI] [PubMed] [Google Scholar]
- 24. Steitz T. A. (1998) Nature 391, 231–232 [DOI] [PubMed] [Google Scholar]
- 25. Chou P. Y., Fasman G. D. (1978) Annu. Rev. Biochem. 47, 251–276 [DOI] [PubMed] [Google Scholar]
- 26. Minnick D. T., Bebenek K., Osheroff W. P., Turner R. M., Jr., Astatke M., Liu L., Kunkel T. A., Joyce C. M. (1999) J. Biol. Chem. 274, 3067–3075 [DOI] [PubMed] [Google Scholar]
- 27. Bell J. B., Eckert K. A., Joyce C. M., Kunkel T. A. (1997) J. Biol. Chem. 272, 7345–7351 [DOI] [PubMed] [Google Scholar]
- 28. Tsai Y. C., Johnson K. A. (2006) Biochemistry 45, 9675–9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purohit V., Grindley N. D., Joyce C. M. (2003) Biochemistry 42, 10200–10211 [DOI] [PubMed] [Google Scholar]
- 30. Longley M. J., Nguyen D., Kunkel T. A., Copeland W. C. (2001) J. Biol. Chem. 276, 38555–38562 [DOI] [PubMed] [Google Scholar]
- 31. Maga G., Shevelev I., Ramadan K., Spadari S., Hübscher U. (2002) J. Mol. Biol. 319, 359–369 [DOI] [PubMed] [Google Scholar]
- 32. Arana M. E., Seki M., Wood R. D., Rogozin I. B., Kunkel T. A. (2008) Nucleic Acids Res. 36, 3847–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takata K., Shimizu T., Iwai S., Wood R. D. (2006) J. Biol. Chem. 281, 23445–23455 [DOI] [PubMed] [Google Scholar]
- 34. Vaisman A., Ling H., Woodgate R., Yang W. (2005) EMBO J. 24, 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ling H., Boudsocq F., Woodgate R., Yang W. (2001) Cell 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 36. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2008) Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santoso Y., Joyce C. M., Potapova O., Le Reste L., Hohlbein J., Torella J. P., Grindley N. D., Kapanidis A. N. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Golosov A. A., Warren J. J., Beese L. S., Karplus M. (2010) Structure 18, 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bermek O., Grindley N. D., Joyce C. M. (2011) J. Biol. Chem. 286, 3755–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Florián J., Goodman M. F., Warshel A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6819–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.