Abstract
CAPRI is a member of the GAP1 family of GTPase-activating proteins (GAPs) for small G proteins. It is known to function as an amplitude sensor for intracellular Ca2+ levels stimulated by extracellular signals and has a catalytic domain with dual RasGAP and RapGAP activities. Here, we have investigated the mechanism that switches CAPRI between its two GAP activities. We demonstrate that CAPRI forms homodimers in vitro and in vivo in a Ca2+-dependent manner. The site required for dimerization was pinpointed by deletion and point mutations to a helix motif that forms a hydrophobic face in the extreme C-terminal tail of the CAPRI protein. Deletion of this helix motif abolished dimer formation but did not affect translocation of CAPRI to the plasma membrane upon cell stimulation with histamine. We found that dimeric and monomeric CAPRI coexist in cells and that the ratio of dimeric to monomeric CAPRI increases upon cell stimulation with histamine. Free Ca2+ at physiologically relevant concentrations was both necessary and sufficient for dimer formation. Importantly, the monomeric and dimeric forms of CAPRI exhibited differential GAP activities in vivo; the wild-type form of CAPRI had stronger RapGAP activity than RasGAP activity, whereas a monomeric CAPRI mutant showed stronger RasGAP than RapGAP activity. These results demonstrate that CAPRI switches between its dual GAP roles by forming monomers or homodimers through a process regulated by Ca2+. We propose that Ca2+-dependent dimerization of CAPRI may serve to coordinate Ras and Rap1 signaling pathways.
Keywords: Calcium, Membrane Proteins, Protein Translocation, Ras, Signal Transduction, CAPRI, Dual GAP, GAP1, RapGAP, RasGAP
Introduction
The closely related small G proteins (GTPases) Ras and Rap1 are conserved molecular switches that couple extracellular signals to a wide range of cellular responses through different signaling networks (1). Ras plays a central role in cell proliferation and cell survival and is a major oncogene (2). Rap1 was originally identified as a protein that reverts the effects of active Ras, such as the loss of adhesion accompanying cell transformation by oncogenic K-Ras (3) or the activation of ERK and ELK1 (4, 5). However, subsequent studies showed that Rap1 is not a mere anti-Ras protein but has discrete functions, notably in integrin-mediated cell adhesion and spreading, formation of cell/cell contacts (6), superoxide formation, and cAMP-induced neurite outgrowth (1, 7). These distinct physiological roles of Ras and Rap1 are mediated by their differential use of effector proteins and also through differential subcellular localizations of their effectors (8).
As is typical for small G proteins, the biological activities of both Ras and Rap1 are controlled by a GDP/GTP cycle; they are active in their GTP-bound form and inactive in their GDP-bound form (1). Guanine nucleotide exchange factors activate them by promoting the dissociation of GDP, allowing excess free cellular GTP to bind, whereas GAPs2 inactivate them by stimulating their intrinsic GTPase activity. A number of guanine nucleotide exchange factors and GAPs for Ras and Rap have been identified. In general, these are multidomain proteins regulated by receptor-mediated extracellular signals, either through second messengers or localized cues such as protein or lipid binding partners that serve as localization signals or scaffolds, thus controlling their activities in time and space (7, 9, 10). The molecular mechanisms controlling Ras- and Rap1-GEFs have been studied extensively (1), whereas the mechanisms controlling RasGAPs and Rap1GAPs remain comparatively under-investigated.
RasGAPs include p120RasGAP, neurofibromin 1 (NF1), SynGAP, and the GAP1 protein family. Rap1GAPs include RapGAPs I and II, the SPA-1 family (SPA-1, SPAR, SPAL, and E6TP1), and tuberin. There is no detectable sequence homology between RasGAPs and RapGAPs except for similarity in their catalytic GAP-related domains, and although Rap1 is a close homologue of Ras, the mechanisms by which RasGAPs and Rap1GAPs stimulate the GTPase activities of their respective GTPases differ. The catalytic domain of RasGAPs protrudes a catalytic arginine residue (the arginine finger) into the active site of Ras to re-orient Gln-61, which stabilizes the transition state of the GTPase reaction and increases the reaction rate by more than 1,000-fold (11–13). In contrast, the catalytic domain of Rap1GAPs does not possess the arginine finger but instead an asparagine residue (the asparagine thumb) that recapitulates the function of intrinsic Gln (14–17). Despite the fact that RasGAPs and Rap1GAPs act on their small G proteins through distinct mechanisms, some GAPs have dual RasGAP and Rap1GAP activities, namely SynGAP, a synaptic protein important in neuronal development (18), and the following three members of the GAP1 family: GAP1IP4BP, CAPRI, and Rasal (19–21).
The GAP1 family comprises GAP1m, GAP1IP4BP, CAPRI, and Rasal, which share conserved structural modules, i.e. tandem C2 domains, the catalytic GAP-related domain, and a pleckstrin homology domain adjacent to a Bruton tyrosine kinase motif (10). GAP1IP4BP was the first GAP1 family protein found to have dual RasGAP and RapGAP activities (19). The C2 and pleckstrin homology/Bruton tyrosine kinase domains that flank the Ras-GRD are required for Rap1GAP activity but not RasGAP activity, although both Ras and Rap1 bind to the Ras-GRD of GAP1IP4BP (20–22). CAPRI and Rasal are the only members of the GAP1 family that possess five Ca2+-coordinating acidic residues and are known to be regulated by Ca2+ binding (23, 24). Agonist-evoked intracellular Ca2+ mobilization leads to a rapid C2 domain-dependent translocation of CAPRI and Rasal to the plasma membrane. Concomitantly with this plasma membrane translocation, both the RasGAP and Rap1GAP activities of CAPRI increase substantially (20, 25). In contrast to CAPRI, which responds to increases in intracellular Ca2+ with a relatively stable membrane association, Rasal responds to Ca2+ oscillation and acts as a RasGAP only when bound to the membrane through its C2 domains (24). Finally, although membrane association is required for both the RasGAP and RapGAP activities of CAPRI, the RapGAP activity of Rasal is independent of membrane binding (20, 22).
In this study, we have investigated the mechanism that switches CAPRI between its RasGAP and Rap1GAP activities. We describe that CAPRI forms homodimers in a Ca2+-dependent manner and that the dimeric and monomeric forms of CAPRI display differential GAP activities toward Ras and Rap1 in vivo.
EXPERIMENTAL PROCEDURES
Reagents
JetPei was purchased from Autogen Bioclear UK Ltd. (Holly Ditch Farm, Mile ELM, UK). All restriction enzymes, TaqDNA polymerase, and T4 DNA ligase were purchased from Roche Applied Science unless otherwise stated. Rabbit anti-CAPRI serum was custom-made by Harlan SERA-LAB (Leicestershire, UK) using a synthetic peptide CDGAPEDSLAQLLRVLQ corresponding to amino acids (AA) 760–776 of mouse CAPRI. The anti-CAPRI antibody was purified by an affinity column conjugated with the synthetic peptide. Glutathione-Sepharose 4B beads were from Amersham Biosciences Bioscience (Cambridge, UK). ATP, histamine, and protein A-Sepharose beads were purchased from Sigma. Cell culture media and laboratory plastics were purchased from Invitrogen. All other reagents were of standard analytical grade.
Cell Culture and Transfection
HeLa, CHO, HEK293, and COS-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 100 units of penicillin, and 100 μg/ml streptomycin as described previously (23, 26). The cells were transfected with JetPei essentially as recommended by the manufacturer.
Construction of Expression Vectors
Mouse CAPRI cDNA was obtained from Geneservice (MRC, UK). DNA oligomer 5′GCTTCGTGTGTCGCAAGACCATCGAGAAGC3′ was used to generate the CAPRI mutant SH-CAPRI in which the leucines at positions 775 and 778 were replaced by serine and histidine, respectively. The C-terminally deleted CAPRI (ΔcCAPRI) was generated by digesting CAPRI cDNA with SacI, followed by blunting and self-ligation. This deletion removed the 41 most C-terminal AA of CAPRI containing the helix motif. The GFP fusion constructs GFP-CAPRI, GFP-SH-CAPRI, and GFP-ΔcCAPRI were made by cloning the corresponding CAPRI cDNAs into the EcoRI and XhoI sites of the peGFP-C2 vector (Clontech). The red fluorescent protein fusion construct RFP-CAPRI was made by replacing the enhanced GFP of the GFP-CAPRI vector with a Cherry fluorescent protein gene generated by PCR (red fluorescent protein was a gift from Roger Tsien, University of Berkeley, Berkeley, CA). The same cloning strategy was applied to construct the RFP-SH-CAPRI and RFP-ΔcCAPRI expression vectors. The YN and YC vectors were made by replacing the GFP gene in the peGFP-C2 vector with a PCR fragment containing the N-terminal (1–154 AA) (YN) or C-terminal (155–238 AA) (YC) parts of the YFP, respectively. YN-CAPRI, YN-ΔcCAPRI, YN-SH-CAPRI, YC-CAPRI, YC-ΔcCAPRI, and YC-SH-CAPRI vectors were made by cloning the corresponding CAPRI genes into the 3′ end of YN and YC according to Hu and Kerppola (27). Myc-, EE-, and HA-tagged CAPRI constructs (myc-CAPRI, EE-CAPRI, and HA-CAPRI) were generated by cloning CAPRI into the EcoRI and XhoI sites 3′ of the Myc, EE, or HA tag sequences in pcDNA3 (Stratagene). RFP-RBD and GFP-RBD were made as described previously (26). The bacterial expression vector for the C-terminal 48 AA of CAPRI containing the helix motif (His-cCAPRI) was made by cloning the SacI and XhoI DNA fragment into the BamHI and XhoI sites of pET25 (Invitrogen). The same cloning strategy was used to construct the bacterial expression vector for the CAPRI construct with mutations of the leucines at positions 775 (L775S) and 778 (L778H) (His-SH-cCAPRI). GST-RBD and GST-RalGDS expression vectors were made as described previously (20, 23).
Gel Electrophoresis and Western Blotting
SDS-PAGE was performed with 9 or 15% polyacrylamide resolving gels, in the presence or absence of DTT or β-mercaptoethanol, as indicated. Native gels at 15% were prepared and run as described previously (28). Gels were either stained with Coomassie Brilliant Blue or electroblotted onto Immobilon P (Millipore) for immunodetection. Western blotting was performed using the following primary antibodies: purified rabbit anti-CAPRI antibodies (see above); anti-GFP monoclonal antibody (G6539, Sigma); anti-Ras (sc-520) or anti-Rap1 (sc-65) polyclonal and anti-Ras monoclonal antibody (sc-35) (all from Santa Cruz Biotechnology, Cambridge, UK); anti-Myc or anti-HA monoclonal and anti-GFP polyclonal antibodies (all produced in-house). Secondary antibodies against mouse and rabbit IgGs and ECL reagents were purchased from Amersham Biosciences.
Immunoprecipitations
Transfected cells were lysed on ice in lysis buffer containing 1% (v/v) Triton X-100, 150 mm NaCl, 10 mm Tris/HC, pH 7.5, 1 mm NaF, 0.02% NaN3, and proteinase inhibitors after washing three times with cold PBS. CaCl2 or EGTA were added, and immunoprecipitations were performed with the appropriate antibodies as detailed for each individual set of experiments. Purification of EE-CAPRI from HEK293 cells was carried out using anti-EE antibody conjugated to Sepharose. Before the purification, the beads were blocked with 10% bovine serum for 30 min. Cell lysates in 150 mm NaCl, 10 mm Tris/HCl, pH 7.5, 2 mm Ca2+, and 1 μm DTT were incubated with the blocked anti-EE-Sepharose at 4 °C for 2 h washed in the same buffer before elution of the EE-tagged protein with 0.1 m glycine buffer, pH 2.5, and collected into 1/9 volume of 1 m Tris, pH 8.0 (4 °C). Immediately an equal volume of cell lysis buffer with 2 mm Ca2+ was added to each collection.
FPLC Gel Filtration
Gel filtration of bacterially derived purified His-cCAPRI and His-SH-cCAPRI was carried out at 4 °C on an FPLC system and a Superose 12 HR 10/30 column (GE Healthcare) equilibrated in 150 mm NaCl, 100 mm imidazole, 10 mm NaH2PO4, pH 8.0, following the manufacturer's instructions at a flow rate of 150 μl/min. Purified protein (50 μl) was injected into the column and 50-μl fractions were collected. Fractions were analyzed by SDS-PAGE followed by Western blotting. Gel filtration of purified full-length EE-CAPRI was done under the same conditions, except using the following buffer: 150 mm NaCl, 10 mm Tris/HCl, pH 8, and 1 mm CaCl2. Gel filtration of lysates from COS-7 cells expressing myc-CAPRI was carried out in 150 mm NaCl, 10 mm Tris/HCl, pH 7.5, 0.5 mm CaCl2, 1 μm DTT, and 1% Thesit buffer. Post-nuclear supernatant (5 mg/ml; 50 μl) was injected. Standard curves were obtained using calibration kit protein standards (GE Healthcare) and drawn by plotting the elution volume values on the x axis and log molecular masses on the y axis. The molecular masses of the different forms of CAPRI were determined in relation to the known molecular mass markers. Protein standard column runs were performed using the same buffers and conditions as those used for each respective experimental sample.
Purification of GFP-CAPRI and in Vitro Dimerization Assay
Purification of GFP-CAPRI from overexpressing HEK293 cells was carried out using anti-GFP polyclonal antibodies and protein A-Sepharose. Beads were washed three times for 30 min in lysis buffer containing 1 mm EGTA and then divided equally into two tubes. One tube was washed three times in 0.1 m glycine buffer, pH 2.5, and then three times in 0.1 m glycine buffer, pH 9, and supernatants (eluted GFP-CAPRI) were pooled. Eluted GFP-CAPRI was concentrated in Centricon columns and reconstituted in 150 mm NaCl, 10 mm Tris/HCl, pH 7.5. The other tube containing immobilized GFP-CAPRI was blocked with bovine serum for 1 h and then washed three times in 150 mm NaCl, 10 mm Tris/HCl, pH 7.5, and the beads were aliquoted for the dimerization experiment. Beads were washed three more times in 150 mm NaCl, 10 mm Tris/HCl, pH 7.5, and, for each tube, with a different concentration of Ca2+. Then the same amount of purified GFP-CAPRI was added to each tube, and the samples were incubated under constant agitation at 4 °C for 3 h before bound GFP-CAPRI was eluted off the beads with buffer containing 1 mm EGTA.
Ras Pulldown Assay
Ras-RBD was purified as described previously (23). CHO cells in 12-well dishes were transiently transfected with a total of 2 μg of DNA of a mixture of H-Ras cDNA with either empty vector (as control) or with a vector encoding the CAPRI variants (CAPRI or ΔcCAPRI) at a molar ratio of 1:1. 24 h after transfection, the cells were serum-starved for 5 h at 37 °C in serum-free DMEM prior to experiments. Then the cells were stimulated for 2 min with 50 μm ATP and lysed in extraction buffer as described previously (23). Post-nuclear supernatants were incubated with GST-RBD on glutathione-Sepharose beads at 4 °C for 30 min on ice. Beads were washed three times in ice-cold PBS, 0.1% Triton X-100, 10 mm MgCl2. Ras proteins were separated by SDS-PAGE and visualized by immunoblotting on Immobilon-P (Millipore) using anti-HA antibodies and enhanced chemiluminescence.
Rap1 Pulldown Assay
The Rap-GTP binding domain of human RalGDS (GST-RalGDS) was used to pull down endogenous active Rap1. The GST-RalGDS fusion protein was purified as described previously (20). 12-Well dishes of CHO cell were transiently transfected with CAPRI, ΔcCAPRI, or empty vector DNA (2 μg of DNA per well). 24 h after transfection, the cells were serum-starved for 5 h at 37 °C in serum-free DMEM prior to stimulation for 2 min with 50 μm ATP and lysis in extraction buffer. Rap1-GTP pulldown was carried out as described previously (20), and Rap1-GTP was detected by Western blotting with pan-Rap1 antibody.
Imaging
Live cell imaging was performed using a CellR (Biosystems) imaging Station (Olympus) equipped with an MT20 epifluorescence illumination system and a Hamamatsu Orca ER camera. For TIRF microscopy, the same system was used with a dual-port TIRF illuminator and 488 nm/561 nm solid-state lasers, as described previously (26). For TIRF analysis, the fluorescence intensity (F) of a region of interest on the cell surface, representing at least 10% of the surface area, was measured for each cell in a series of images. After background fluorescence was subtracted from the image series, the ratio of the fluorescence intensity at each time point (Ft) and the minimum fluorescence intensity of the same area (Fmin at T = 0) were calculated using the relative translocation parameter 1 − Fmin/Ft.
Analysis
Densitometric analysis of Western blots was performed using ImageJ (National Institutes of Health). A t test or ANOVA were used for statistical analysis, as appropriate.
RESULTS
CAPRI Harbors an Evolutionarily Conserved Helix Motif in Its C-terminal Tail
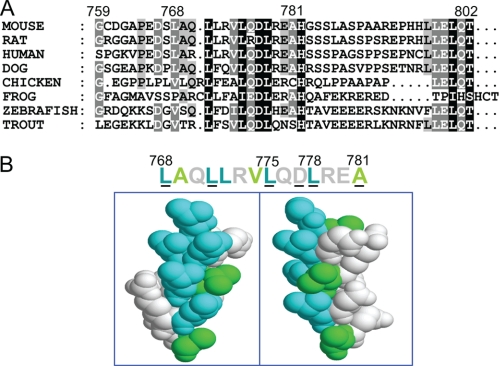
We aimed to investigate the mechanism of CAPRI switching between its RasGAP and Rap1GAP activities, which are catalyzed through the same GAP-related domain. To search for potential functional motifs that may play a role in the switch between the dual GAP functions, we examined the AA sequences of CAPRI proteins from different vertebrate species, ranging from fish to humans. Apart from the already characterized domains and conserved residues in CAPRI, i.e. its tandem C2 domains, catalytic Ras-GRD domain, pleckstrin homology domain, and Bruton tyrosine kinase motif, we identified a region stretching from AA 768 to 781 in the extreme C-terminal tail of the mouse CAPRI sequence that was conserved among CAPRI proteins across all species examined (Fig. 1A). Modeling of this highly conserved region using the Swiss Model option in DEEPVIEW revealed that it forms a simple helix (Fig. 1B). A clear hydrophobic face was present along one surface in this model. We speculated that the helix motif might possibly mediate dimerization of CAPRI through this hydrophobic face. Further computer simulations predicted that mutations of the two leucine residues at positions 775 and 778 might reduce the hydrophobicity of this interface sufficiently to abolish the predicted dimer formation of CAPRI.
FIGURE 1.
A conserved helix motif in the C-terminal tail of CAPRI. A, sequence alignment of the C terminus of CAPRI proteins from different vertebrate species. Mouse sequence LAQLLRVLQDLREA was used for alignment with those of the other species. The intensity of shading indicates the level of conservation of residues between different species. B, AAs 768–781 of the mouse CAPRI sequence were loaded into the Swiss Model option in DEEPVIEW, which automatically displayed the sequence as a simple helix. The conserved residues are underlined. The motif forms a clear hydrophobic face. All residues were space filled and colored according to the linear sequence above.
C Terminus of CAPRI Forms Homodimers through the Predicted Hydrophobic Face of the Helix Motif
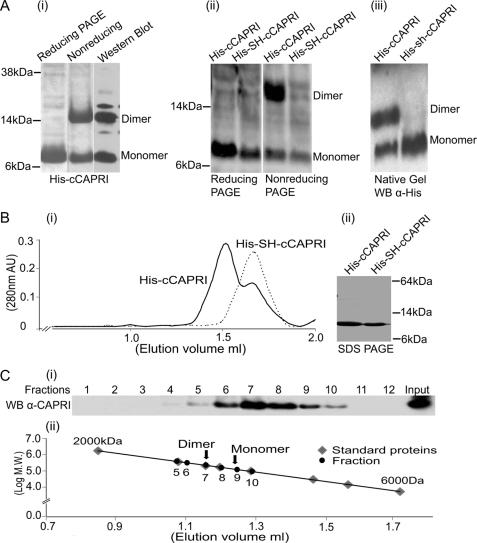
To investigate the possibility that CAPRI may form dimers through the C-terminal helix motif, as suggested by the computer modeling, we expressed the 41 most C-terminal AA of CAPRI with an N-terminal His tag (His-cCAPRI) in bacteria and purified the protein using an nickel-nitrilotriacetic acid column. The purified protein was subjected to SDS-PAGE under both reducing and nonreducing conditions to examine its dimerization state. Coomassie staining detected two bands at the 7- and 14-kDa positions in the nonreducing SDS-PAGE and a sole band at the 7-kDa position in the reducing SDS-PAGE (Fig. 2A, panel i). The lower bands migrated at the expected molecular weight of the monomeric protein fragment, and the upper band in the nonreducing gel was approximately twice the size. Western blots were performed using both anti-His tag monoclonal (Fig. 2A, panel i) and anti-CAPRI polyclonal antibodies (data not shown) to confirm that both bands contained the CAPRI protein. These results indicated that His-cCAPRI might form dimers in bacteria, as predicted by the modeling.
FIGURE 2.
Dimerization of the helix motif in the C-terminal tail of CAPRI. A, panel i, SDS-PAGE of purified His-cCAPRI. His-cCAPRI samples were subjected either to reducing conditions (the protein was treated with loading buffer containing DTT before heating to 94 °C for 5 min and running on SDS-PAGE) or to nonreducing conditions (DTT-free loading buffer). Gels were Coomassie-stained. Nonreducing gels were also Western-blotted using anti-His tag antibody. Panel ii, dimerization of His-cCAPRI was abolished by mutation of leucines at position 775 and 778 to Ser and His. Total lysates of bacteria transformed with His-tagged constructs of both wild type (His-cCAPRI) and mutant CAPRI (His-SH-cCAPRI) were run on reducing or nonreducing SDS-PAGE. Western blotting was performed with anti-His tag antibody. The blots shown are representative of at least three independent experiments. Panel iii, bacterially derived purified His-cCAPRI and His-SH-cCAPRI were analyzed by native PAGE followed by Western blotting with anti-His tag antibody. The blots shown are representative of three independent experiments. B, gel filtration analysis of dimerization of His-cCAPRI. Panel i, aliquots of purified His-cCAPRI or His-SH-cCAPRI were loaded onto a Superose 12 column. Samples were eluted at a flow rate 0.1 ml/min, and fractions were collected and submitted to SDS-PAGE and Western blotting with anti-His tag antibody. The UV absorption at 280 nm is shown and shows that purified His-cCAPRI eluted as two peaks whereas His-SH-cCAPRI eluted as single peak. Panel ii, Coomassie-stained SDS-PAGE showing the purity of His-cCAPRI and His-SH-cCAPRI proteins used for gel filtration analysis. C, gel filtration analysis of full-length CAPRI dimerization. Purified full-length EE-CAPRI was loaded onto Superose 12 column, and samples were eluted at a flow of 0.1 ml/min with UV absorption at 280 nm being measured. Fractions were analyzed by SDS-PAGE followed by Western blotting with anti-CAPRI antibodies. Panel i, Western blot of the fractions. Panel ii, plot showing the molecular masses of the proteins from the column fractions (black) and those of the standard proteins (gray) from the calibration curve. Arrows indicate the positions of the dimeric and monomeric forms of EE-CAPRI within the standard curve.
Next, we tested the prediction that mutation of the two leucine residues at positions 775 and 778 in the hydrophobic motif might be sufficient to abolish dimer formation. We mutated the leucines to serine and histidine, respectively (His-SH-cCAPRI). His-SH-cCAPRI was expressed in Escherichia coli, and dimerization was analyzed by reducing and nonreducing SDS-PAGE and Western blotting as above. In the nonreducing SDS-PAGE, both bands were present in samples containing His-cCAPRI as expected, but the 14-kDa upper band was absent from His-SH-cCAPRI samples, as well as under reducing conditions (Fig. 2A, panel ii). Hence, the replacement of the two leucines with serine or histidine was sufficient to abolish formation of the upper band. This result further supports the possibility that His-cCAPRI forms dimers and suggests that leucine residues 775 and 778 are essential for this dimerization.
To analyze the possibility of His-cCAPRI dimerization further, we used native PAGE. His-cCAPRI and His-SH-cCAPRI proteins were resolved by native PAGE, and the native gels were Western-blotted with anti-His tag monoclonal antibody. This revealed two bands of ∼8 and 17 kDa in the His-cCAPRI sample but only a single band around of 8 kDa for His-SH-cCAPRI (Fig. 2A, panel iii). Although it was difficult to conclude with certainty from the native gels that the upper band is a dimer and lower band is a monomer, these data showed unequivocally that the C-terminal CAPRI forms oligomers that are dependent on leucines 775 and 778.
To evaluate further the molecular mass of the His-cCAPRI oligomers, we carried out gel filtration chromatography. The purity of His-cCAPRI and His-SH-cCAPRI was analyzed by SDS-PAGE before performing the gel filtration (Fig. 2B, panel ii). His-cCAPRI eluted as two peaks from the column (Fig. 2B, panel i), the first peak at an elution volume of 1.52 ml and the second peak at an elution volume of 1.6 ml, whereas His-SH-cCAPRI eluted as a single peak at an elution volume of 1.6 ml. Their molecular masses were determined from a standard graph. The first His-cCAPRI peak had a molecular mass of 19 kDa, equivalent to a dimeric form, whereas the second His-cCAPRI peak and the single His-SH-cCAPRI peak had a molecular mass of 9 kDa, equivalent to the monomeric form. The presence of His-cCAPRI or His-SH-cCAPRI in the fractions was confirmed by Western blotting (data not shown). These results clearly demonstrate that His-cCAPRI forms dimers under native conditions. Moreover, this dimerization is mediated by the leucines 775 and 778 in the C-terminal helix motif.
Full-length CAPRI Forms Dimers
To determine whether full-length CAPRI is able to form dimers, we purified full-length EE-CAPRI from HEK293 cells as detailed under “Experimental Procedures” and subjected the purified protein to gel filtration. Eluted fractions were analyzed by SDS-PAGE followed by Western blotting. As shown in Fig. 2C, EE-CAPRI mainly eluted between fraction 6 and 9. The peak of eluted EE-CAPRI was in fraction 7, close to the 200-kDa marker, and equivalent to size of dimeric EE-CAPRI (192 kDa). Some EE-CAPRI was also present in fraction 9, which is equivalent to the size of the monomer. Hence, it is likely that a proportion of CAPRI was present in monomeric form, although the conditions used here did not provide sufficient resolution to separate the large dimer peak from fraction 9, which could be due to a proportion of CAPRI dimers dissociating during the gel filtration process (but see also Fig. 5D for more data on dimer and monomer ratios). Furthermore, EE-CAPRI was barely detectable in fractions 5 or below (protein mass of 400 kDa or more), so although we cannot categorically rule out that some higher oligomerization states also exist, they would be present in minor quantities at best. In summary, this gel filtration assay clearly showed that full-length CAPRI is capable of forming dimers and that the dimeric form is the main form in solution.
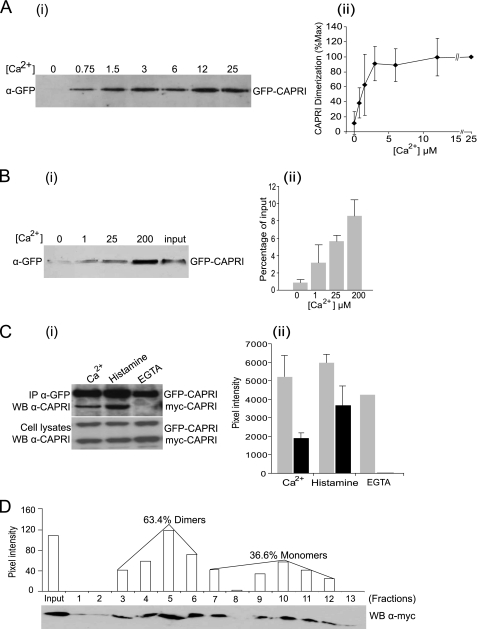
FIGURE 5.
Ca2+ dependence of CAPRI dimerization. A, CAPRI dimerization requires the presence of Ca2+. GFP-CAPRI was overexpressed in COS-7 cells. The cells were lysed in buffer containing 2 mm Ca2+. Immunoprecipitation was carried out with anti-GFP polyclonal antibody and protein A-Sepharose beads, and the beads were washed in buffers containing the indicated concentrations of Ca2+. Dimerized GFP-CAPRI was eluted with lysis buffer containing 1 mm EGTA and subjected to Western blotting with anti-GFP monoclonal antibody. Panel i, Representative Western blot. Panel ii, quantification by densitometric scanning of Western blots. Data are mean ± S.D. of three independent experiments. B, Ca2+ is sufficient to drive dimerization of purified GFP-CAPRI in vitro. Monomeric GFP-CAPRI was prepared as detailed under “Experimental Procedures,” and one-half was kept in solution, and the other was immobilized on beads. Aliquots of immobilized and free GFP-CAPRI were recombined in the presence of the indicated concentrations of Ca2+. Dimerized GFP-CAPRI was eluted with buffer containing 1 mm EGTA and subjected to Western blotting with anti-GFP monoclonal antibody. Panel i, representative Western blot. Input, 5% of the total GFP-CAPRI in each reaction. Panel ii, quantification of dimerized GFP-CAPRI as the percentage of total input. Data are mean ± range of two independent experiments. C, dimerization of CAPRI can be induced by histamine stimulation of HeLa cells. GFP-CAPRI and myc-CAPRI were co-overexpressed in HeLa cells. The cells were either lysed in buffer containing 1 mm Ca2+ or stimulated with 100 μm histamine before lysis in Ca2+ buffer or lysed in Ca2+-free buffer containing 0.5 mm EGTA, as indicated. Immunoprecipitation was carried out with anti-GFP monoclonal antibody, and the blots were probed with anti-CAPRI antibody. Panel i, representative Western blot (WB). Panel ii, quantification by densitometric scanning. Data are mean ± range of two independent experiments, except for EGTA, which are from one experiment. D, quantification of CAPRI monomers and dimers by gel filtration. myc-CAPRI was overexpressed in Cos-7 cells, and the cells were lysed in buffer with 500 μm Ca2+. The lysates were subjected to FPLC Superose 12 gel filtration column as detailed under “Experimental Procedures.” Column fraction aliquots (30 μl/fraction) were subjected to SDS-PAGE and Western blotting with anti-Myc monoclonal antibody. Quantification of the monomeric and dimeric forms of CAPRI was obtained by densitometric scanning. One-tenth input proteins were loaded in the input lane. The standard protein markers are blue dextran 2000 kDa, apoferritin 443 kDa, β-amylase 200 kDa, alcohol dehydrogenase 150 kDa, BSA 66.2 kDa, carbonic anhydrase 29 kDa, cytochrome c 12.4 kDa, and aprotinin 6500 Da.
Full-length CAPRI Protein Forms Dimers through Its C-terminal Helix Motif in Mammalian Cells
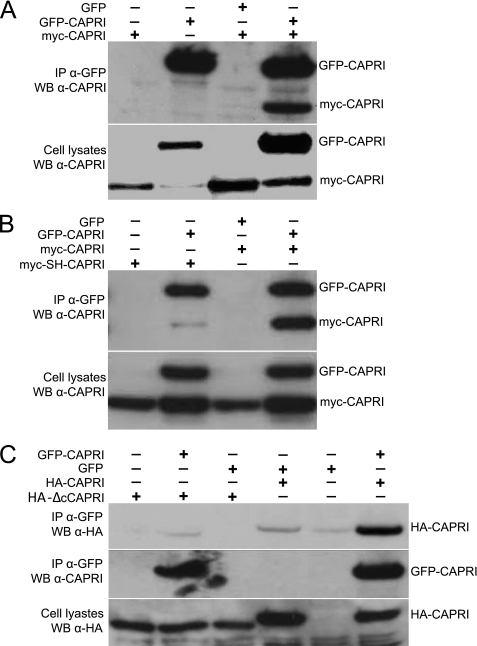
We further investigated the dimerization of full-length CAPRI in mammalian cells. We cotransfected COS-7 cells with GFP-CAPRI and myc-CAPRI constructs and used anti-GFP antibody for immunoprecipitation from total cell lysates. Western blotting showed that myc-CAPRI coimmunoprecipitated with GFP-CAPRI from cotransfected cells but not from cells transfected with myc-CAPRI or GFP-CAPRI alone or those cotransfected with GFP and myc-CAPRI constructs (Fig. 3A). This result suggests that full-length CAPRI forms homodimers in mammalian cells.
FIGURE 3.
Dimerization of full-length CAPRI in cells requires the helix motif in the C-terminal tail. A, myc-CAPRI coimmunoprecipitates with GFP-CAPRI. Cos-7 cells were transfected to co-overexpress GFP-CAPRI (or GFP as control) with myc-CAPRI. Anti-GFP monoclonal antibody was used for immunoprecipitation (IP) and anti-CAPRI antibody for Western blotting (WB). B, mutation of the two leucines in the helix motif of the C-terminal tail of CAPRI abolishes the dimerization of full-length CAPRI. Cos-7 cells were transfected to co-overexpress GFP-CAPRI (of GFP as control) with myc-CAPRI or myc-SH-CAPRI. Anti-GFP antibody was used for immunoprecipitation and anti-CAPRI antibody for Western blotting. myc-SH-CAPRI protein failed to coimmunoprecipitate with GFP-CAPRI, whereas myc-CAPRI did. C, deletion of the C-terminal tail of CAPRI abolishes the dimerization of full-length CAPRI. HA-tagged CAPRI mutant with a 41-AA C-terminal deletion, HA-ΔcCAPRI, was co-overexpressed with GFP-CAPRI or GFP. Immunoprecipitation was performed using anti-GFP antibody, and blotting was done with anti-HA and GFP antibodies. HA-ΔcCAPRI failed to coimmunoprecipitate with GFP-CAPRI. The blots shown are representative of at least three independent experiments.
We then investigated if mutation of the two leucines at position 775 and 778 in the C-terminal tail of CAPRI containing the helix motif would abolish dimerization between full-length myc-CAPRI and GFP-CAPRI in cells. In contrast to wild-type myc-CAPRI, which coprecipitated with GFP-CAPRI from control samples, myc-SH-CAPRI did not coprecipitate with GFP-CAPRI (Fig. 3B). A similar result was obtained when we cotransfected cells with a CAPRI mutant that lacked the 41 most C-terminal AA (HA-ΔcCAPRI). Again, wild-type HA-CAPRI, but not HA-ΔcCAPRI, coimmunoprecipitated with GFP-CAPRI (Fig. 3C). Combined, these results show that full-length CAPRI forms homodimers in mammalian cells and that dimerization is dependent on the helix motif in its C-terminal tail.
CAPRI Forms Dimers in Living Mammalian Cells
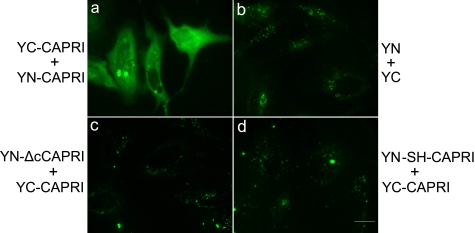
To confirm through a different approach that full-length CAPRI forms dimers in mammalian cells (and to exclude the possibility that CAPRI dimers could form artifactually during protein isolation), we studied CAPRI dimerization in living cells using bimolecular fluorescence complementation (BiFC). BiFC provides a sophisticated imaging tool to study direct protein-protein interactions (29). The approach is based on the complementation of two nonfluorescent reporter molecule fragments being able to reconstitute a native fluorescent reporter molecule. Here, we used the nonfluorescent N-terminal (YN) and C-terminal (YC) fragments of YFP, fused C-terminally to full-length CAPRI (YN-CAPRI and YC-CAPRI), reasoning that dimerization of CAPRI should bring both YFP fragments into sufficiently close spatial proximity for native YFP to form, thus leading to detectable fluorescence. We cotransfected HeLa cells with YN-CAPRI and YC-CAPRI or other pairs of CAPRI and control constructs, and Western blotting was performed to ascertain that all proteins were expressed at comparable levels. Imaging of HeLa cells cotransfected with YN-CAPRI and YC-CAPRI constructs revealed a strong fluorescence signal in the cytoplasm (Fig. 4A). In contrast, no fluorescence signal was detectable in cells cotransfected with YN and YC controls (Fig. 4B). These results demonstrate that full-length CAPRI forms dimers in living cells, in agreement with our findings from the coimmunoprecipitation experiments.
FIGURE 4.
Detection of CAPRI dimerization in living cells using BiFC. To detect CAPRI dimers in living cells, CAPRI constructs were fused N-terminally to either the N-terminal (YN) or the C-terminal (YC) fragment of YFP protein, and HeLa cells were transfected to coexpress the following: either YN-CAPRI and YC-CAPRI (panel a) or YN and YC only (panel b); YC-CAPRI with a YN-mutant of CAPRI that lacks the 41 C-terminal AA (YN-ΔcCAPRI) (panel c) or YC-CAPRI with YN-mutant L775S/L778H of CAPRI (YN-SH-CAPRI) (panel d). Fluorescence signals were only detectable when CAPRI dimerization brought both halves of YFP into sufficiently close proximity to reconstitute the mature YFP fluorophore. In each group, over 200 cells were examined. Fluorescence signals were absent in the cells transfected with the constructs of panels b–d, respectively. 77% of cells transfected with the constructs of panel a were fluorescence-positive. The photographs shown are representative of three independent experiments. Scale bars, 20 μm.
To test whether dimerization of full-length CAPRI is mediated through the helix motif in the C-terminal tail in living cells, we fused YN or YC either to the L775S/L775H-mutated CAPRI (YN-SH-CAPRI and YC-SH-CAPRI) or to the C-terminal 41-AA deletion mutant (YN-ΔcCAPRI and YC-ΔcCAPRI). In contrast to coexpression of YC-CAPRI with wild-type YN-CAPRI, coexpression of YC-CAPRI with YN-ΔcCAPRI (Fig. 4C) or YN-SH-CAPRI (Fig. 4D) led only to background fluorescence. Similarly, the inverse combinations (coexpression of YN-CAPRI with YC-ΔcCAPRI or YC-SH-CAPRI) also produced only background fluorescence (data not shown). These results show that the dimerization of CAPRI is mediated through the helix motif in the C-terminal tail in living cells, which further confirms the coimmunoprecipitation results.
Dimerization of CAPRI Is Mediated by Free Ca2+ in Vivo
CAPRI is a sensor for the amplitude of the initial increase in intracellular [Ca2+] in response to receptor stimulation. Hence, we investigated if Ca2+ regulates the dimerization of full-length CAPRI in cells. GFP-CAPRI was immunoprecipitated from HEK cell lysates in the presence of 2 mm CaCl2 using anti-GFP antibody and protein A-Sepharose beads. Aliquots of the beads were each washed in a buffer containing a different concentration of Ca2+ ranging from 0 to 25 μm. Dimerized GFP-CAPRI was eluted using 1 mm EGTA and subjected to SDS-PAGE and Western analysis (Fig. 5A, panel i). Dimerization of GFP-CAPRI was dependent on the concentration of Ca2+ in the wash buffer. It was almost undetectable after washing in Ca2+-free buffer but increased dose-dependently and reached a plateau at 3 μm Ca2+ (Fig. 5A, panel ii). The EC50 was estimated to be 0.8 μm. This result demonstrates that dimerization of full-length CAPRI requires Ca2+ in a concentration-dependent manner in vivo. Importantly, the concentrations of Ca2+ required were within the relevant high nanomolar to low micromolar physiological range. In contrast to the full-length protein, the isolated C-terminal fragment of CAPRI used in Fig. 2 did not require Ca2+ for dimerization. This is presumably because Ca2+ induces a conformational change in the full-length protein to enable dimerization, whereas the dimerization site is already exposed in the C-terminal fragment.
Ca2+ Is Sufficient to Drive CAPRI Dimerization in Vitro
We investigated whether Ca2+ is sufficient for dimerization of purified full-length GFP-CAPRI in vitro. GFP-CAPRI monomers were purified from HEK cells. One-half was immobilized on protein A-Sepharose beads and the other half was kept in solution as detailed under “Experimental Procedures.” Aliquots of immobilized GFP-CAPRI and free GFP-CAPRI were combined. Each tube was incubated and washed with a different concentration of Ca2+, ranging from 0 to 200 μm. Free GFP-CAPRI that had bound to the immobilized GFP-CAPRI was eluted with 1 mm EGTA and analyzed by SDS-PAGE and Western blot (Fig. 5B, panel i). Densitometric analysis showed that less than 1% of GFP-CAPRI formed dimers in the absence of Ca2+, but the presence of Ca2+ was sufficient to drive CAPRI dimer formation. CAPRI dimerization was Ca2+ dose-dependent, such that 3.2, 5.6, or 8.6% of the total input was dimerized in the presence of 1, 25, or 200 μm Ca2+, respectively (Fig. 5B, panel ii). This result shows that Ca2+ is not only necessary but sufficient for driving CAPRI dimerization directly.
Monomeric and Dimeric Forms of CAPRI Coexist within the Cell
We next addressed the relative abundance of monomeric and dimeric forms of CAPRI in cells. The first approach was similar to the in vivo dimerization assay. Cells cotransfected with GFP-CAPRI and myc-CAPRI constructs were equally divided into three groups and subjected to different treatments. The first group was lysed directly in buffer containing 1 mm CaCl2; the second group was treated with 100 μm histamine for 30 s before being lysed in the same buffer, and the third group was treated like the second group except that the cells were lysed in a Ca2+-free buffer containing 500 μm EGTA. Immunoprecipitation with anti-GFP antibody and Western blotting with anti-CAPRI were carried out, and the amount of myc-CAPRI that dimerized with GFP-CAPRI was quantified by densitometric analysis. There was no detectable dimerization of myc-CAPRI with GFP-CAPRI in the absence of Ca2+, as expected. In contrast, CAPRI dimers were detected in unstimulated cells in the presence of Ca2+, and the amount of myc-CAPRI that dimerized with GFP-CAPRI increased almost 2-fold upon stimulation with histamine (Fig. 5C). These results demonstrated again that monomers and dimers of CAPRI coexist within the cell, consistent with the result of the BiFC assays. Importantly, they furthermore showed that cell stimulation with histamine, which causes an intracellular elevation of Ca2+, induces an increase in CAPRI dimers, suggesting that Ca2+-dependent dimerization of CAPRI is indeed a physiologically relevant process.
We used gel filtration analysis of cell lysates as a second approach to quantify the monomeric and dimeric forms of CAPRI within the cell. COS-7 cells expressing myc-CAPRI were lysed in a buffer containing 500 μm CaCl2. Post-nuclear supernatants were loaded onto an FPLC Superose 12 gel filtration column and eluted as fractions (Fig. 5D). The fractions were subjected to SDS-PAGE and Western-blotted with anti-Myc monoclonal antibody. Two peaks of myc-CAPRI were detected as follows: one in fractions 3–6 and the other in fractions 7–12. Comparison with the molecular mass of protein standards allowed us to conclude that fractions 3–6 contained dimeric myc-CAPRI and fractions 7–12 contained monomeric myc-CAPRI. Densitometry showed that CAPRI monomers and dimers were present in the total cell lysate at 32.6 and 67.4%, respectively. Therefore, the dimeric form of CAPRI is two times more abundant than the monomeric form when overexpressed in Cos-7 cells. Although these data suggest that myc-CAPRI is present in the cell both as a monomer and dimer, they do not rule out the presence of small amounts of higher order oligomers and might show, in addition, complex formation of CAPRI with other proteins (e.g. Rac1 and Cdc42 are known to interact with CAPRI in vivo). We attempted to analyze the dimerization of endogenous CAPRI using similar methods as with the overexpressed protein, but the low abundance of endogenous CAPRI in cells combined with the limited affinities of available anti-CAPRI antibodies prevented us from detecting a convincing signal.
Wild-type and Monomeric Forms of CAPRI Have Different GAP Activities toward Ras and Rap1
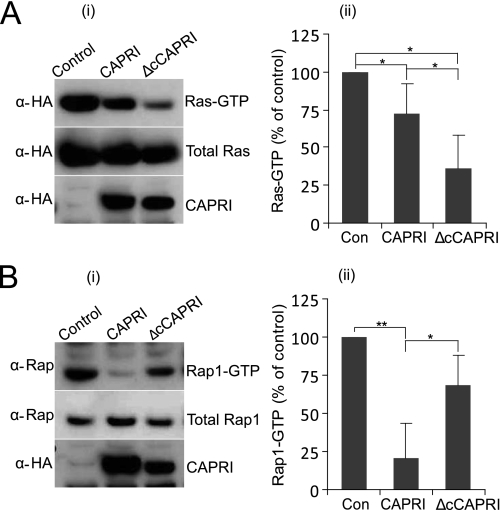
Both GAP activities of CAPRI reside within its Ras-GRD domain, although the Rap1GAP activity is influenced by the flanking regions (20, 22). This suggests that protein interactions and/or conformational changes may act as mechanisms controlling CAPRI activity toward Ras or Rap1 selectively. Accordingly, we examined the RasGAP and Rap1GAP activities of monomeric and WT-CAPRI. Previous work has shown that, in serum-starved cells, a significant proportion of H-Ras and Rap1 are in the GTP-bound state and that CAPRI has insignificant basal RasGAP and Rap1GAP activities. However, treatment of cells with ATP stimulates both the RasGAP and Rap1GAP activities of CAPRI, leading to a reduction in Ras-GTP and Rap1-GTP (20, 23). For RasGAP assays, we transiently cotransfected CHO cells with HA-Ras and HA-CAPRI or HA-ΔcCAPRI, whereas control cells were transfected with HA-Ras and empty vector (Fig. 6A). For Rap1GAP assays, endogenous Rap1 activity was analyzed in CHO cells transfected with HA-CAPRI, HA-ΔcCAPRI, or empty vector as control (Fig. 6B). Data from both Ras and Rap1 pulldown assays were normalized to control cells.
FIGURE 6.
Wild-type and monomeric CAPRI display different RasGAP and Rap1GAP activities. A, RasGAP assays (n = 5). CHO cells were cotransfected with myc-H-Ras and either HA-CAPRI or HA-ΔcCAPRI, and RasGAP assays were performed with GST-RBD pulldown as detailed under “Experimental Procedures” with anti-Myc and anti-HA antibodies to detect Ras and CAPRI, respectively. B, Rap1GAP assays (n = 4). CHO cells were transfected with empty vector DNA as control (Con), full-length HA-CAPRI, or HA-ΔcCAPRI, and RapGAP assays were performed as detailed under “Experimental Procedures” using GST-RalGDS to pull down Rap1-GTP. Anti-HA and anti-Rap1 antibodies were used to detect CAPRI and Rap1, respectively. Panel i show Western blots from one representative experiment; panel ii shows mean ± S.E. of five (A) and four (B) independent experiments. Quantification of Ras-GTP and Ras-GTP levels is expressed as percentage of levels in control cells. Statistics are analysis of variance. *, p ≤ 0.05; **, p ≤ 0.001.
First, we analyzed Ras-GTP levels in ATP-treated cells, and we found that expression of HA-CAPRI reduced Ras-GTP levels to 72% (p = 0.047) (Fig. 6A, panel ii), as expected from published results. However, expression of HA-ΔcCAPRI reduced the level of Ras-GTP significantly more, to 35.9% (p = 0.014). Importantly, HA-ΔcCAPRI reduced the level of Ras-GTP significantly further than wild type HA-CAPRI (to 35.9% versus 72%; p = 0.028), showing that HA-ΔcCAPRI has stronger RasGAP activity than HA-CAPRI. Expression levels of wild type HA-CAPRI and HA-ΔcCAPRI varied somewhat, but across different experiments, there was no significant correlation between these variations and GAP activities. Hence, we conclude that monomeric CAPRI possesses a stronger RasGAP activity than WT-CAPRI.
When we analyzed Rap1-GTP levels in ATP-stimulated cells, we found that HA-CAPRI expression significantly reduced the level of Rap1-GTP to 20.5% of the control (p = 0.007) (Fig. 6B, panel ii). In contrast, HA-ΔcCAPRI expression reduced Rap1-GTP levels to 68.2% of the control, and Rap1-GTP levels were significantly lower in cells expressing HA-CAPRI than those expressing HA-ΔcCAPRI (p = 0.023). We conclude that WT-CAPRI displays a stronger Rap1GAP activity than monomeric CAPRI. As the HA-ΔcCAPRI mutant shows increased RasGAP activity compared with HA-CAPRI, its lower RapGAP activity is likely to be a specific consequence of the loss of its dimerization ability. We also used the leucine double point mutant SH-CAPRI and the deletion mutant instead of the deletion mutant throughout these GAP assays, and we obtained similar results (data not shown). Combined, the results from the pulldown experiments showed that the wild type, dimerization-competent CAPRI possesses a stronger GAP activity for Rap1 than Ras, whereas monomeric CAPRI possesses a stronger GAP activity for Ras than Rap1 in vivo.
Wild-type and Monomeric CAPRI Translocate to the Plasma Membrane Similarly, but Monomers Are Stronger RasGAPs at the Membrane Level
Given that WT-CAPRI and monomeric CAPRI have different RasGAP and Rap1GAP activities, we investigated their subcellular localizations and their abilities to translocate to the membrane and to act as RasGAPs at the plasma membrane level.
First, we transiently coexpressed RFP-CAPRI with GFP-ΔcCAPRI in CHO cells and analyzed their subcellular localizations by wide field epifluorescence microscopy (supplemental Fig. 1). In basal cells, both appeared to be largely cytosolic, and there was no obvious difference in the localization of GFP-ΔcCAPRI and RFP-CAPRI. Next, we investigated the dynamics of ΔcCAPRI and WT-CAPRI translocation to the plasma membrane upon cell stimulation with histamine. RFP-CAPRI and GFP-ΔcCAPRI were transiently coexpressed in CHO cells and analyzed by TIRF microscopy. This showed that monomeric CAPRI translocated to and was retained at the membrane with similar dynamics to WT-CAPRI upon histamine stimulation (supplemental Fig. 2 and supplemental Movie 1). The same results were obtained when RFP-ΔcCAPRI was coexpressed with GFP-CAPRI (supplemental Movie 2).
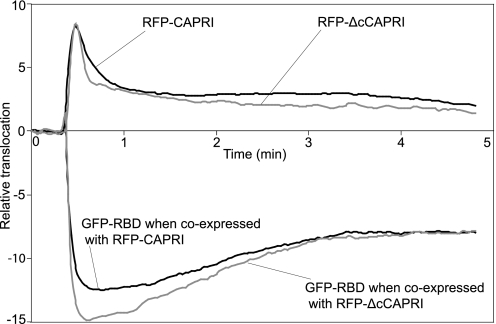
Finally, we compared the RasGAP activities of monomeric CAPRI and WT-CAPRI at the plasma membrane level. It has previously been shown that the isolated Ras-GTP binding domain of B-Raf localizes to the plasma membrane in a Ras GTP-dependent manner and relocates from the membrane to the cytosol upon hydrolysis of Ras GTP to GDP (23). Here, we transiently cotransfected CHO cells with H-Ras and GFP-RBD, together with either RFP-CAPRI or RFP-ΔcCAPRI. We tested both for the histamine-stimulated association of the red fluorescent protein-tagged proteins with the plasma membrane and for the dissociation of GFP-RBD from the plasma membrane by TIRF microscopy. Upon histamine stimulation, both RFP-CAPRI and RFP-ΔcCAPRI were rapidly recruited to the plasma membrane, and this was mirrored by a concomitant dissociation of the GFP-RBD from the plasma membrane (Fig. 7; and supplemental Movies 3 and 4). The dissociation of GFP-RBD from the plasma membrane indicated that membrane-associated Ras-GTP was converted to Ras-GDP both by WT-CAPRI and ΔcCAPRI. However, whereas the extent of membrane translocation of RFP-CAPRI and RFP-ΔcCAPRI was similar, the amplitude of GFP-RBD dissociation from the membrane was larger in cells expressing RFP-ΔcCAPRI than in those expressing RFP-CAPRI (ratio of ΔcCAPRI to WT-CAPRI = 15:12.5 (ΔF/F); p = 0.053). This result indicates that, upon histamine stimulation, RFP-ΔcCAPRI induced a bigger decrease in Ras-GTP levels at the plasma membrane than RFP-CAPRI. Together, these data show that both WT-CAPRI and monomeric CAPRI can act as RasGAPs at the plasma membrane, but they also suggest, as did the pulldown assays, that monomeric CAPRI has stronger RasGAP activity than WT-CAPRI.
FIGURE 7.
Wild-type CAPRI and monomeric CAPRI have different RasGAP activities in vivo. CHO cells were cotransfected with GFP-RBD and RFP-CAPRI or with GFP-RBD and RFP-ΔcCAPRI and analyzed by TIRF microscopy. The relative translocations to and from the plasma membrane of GFP-RBD versus RFP-CAPRI or GFP-RBD versus RFP-ΔcCAPRI induced by 100 μm histamine stimulation were compared. The black and gray traces in the upper panel represent the membrane association of RFP-CAPRI (mean of 33 cells) and RFP-ΔcCAPRI (mean of 30 cells), respectively, in CHO cells stimulated with 100 μm histamine. The gray and black traces in the lower panel represent the concomitant translocation of GFP-RBD away from the plasma membrane in the same cells as above.
DISCUSSION
In this study, we demonstrated that CAPRI forms dimers in a Ca2+-dependent manner. Dimerization results from a direct interaction between CAPRI proteins, and free Ca2+ is both necessary and sufficient for dimer formation. Importantly, WT-CAPRI and monomeric CAPRI display different GAP activities toward Ras and Rap1 GTPases. Hence, our findings describe the mechanism that enables CAPRI to switch between its RasGAP and Rap1GAP activities in the cell.
Through deletion and point mutations, we showed that dimerization of CAPRI is achieved through the helix motif in the C-terminal tail of CAPRI. We have examined the AA sequence of other members of the GAP1 family for the existence of possible dimerization motifs and did not find any motifs equivalent to the helix motif in CAPRI, not even in the most closely related family member Rasal. We therefore suggest that dimerization is unique to CAPRI within the GAP1 family.
We have shown that the dimerization of full-length CAPRI not only requires the presence of Ca2+ but also occurs within the range of physiological concentrations of Ca2+ (30), in a dose-dependent manner. We further demonstrated that dimerization of CAPRI occurs in living cells under normal physiological conditions by using BiFC microscopy, which reveals that dimerization of CAPRI is a relevant physiological process. Importantly, stimulation of cells with histamine, which induces intracellular Ca2+ mobilization, increased the level of dimerization of CAPRI. These observations echo the function of CAPRI as the amplitude sensor of the free Ca2+ evoked by agonists in cells. Although the precise molecular mechanism of regulation of CAPRI dimerization through Ca2+ is still unknown, we hypothesize that the two C2 domains of CAPRI may serve as Ca2+-binding modules, similar to the roles of the two C2 domains in synaptotagmin I (31). In this model, the number of Ca2+ ions associated with the two C2 domains depends on the physiological condition. When the C2 domains are depleted of Ca2+, the C terminus of CAPRI containing the helix motif may be embedded inside the CAPRI protein, thus preventing exposure of the helix motif and blocking dimerization. Upon a partial association of the C2 domains with Ca2+ (unsaturated with Ca2+), a conformational change might occur in CAPRI that exposes the C terminus, thus initiating the formation of homodimers. Agonist stimulation of cells leading to a rapid elevation of intracellular Ca2+ would enable this mobilized Ca2+ to quickly bind to the C2 domains until they are saturated with Ca2+, thus inducing a further conformational change in CAPRI and triggering membrane translocation. The proportions of CAPRI existing as monomers and dimers would be dependent both on the Ca2+ elevation evoked by extracellular signaling and on the localization of CAPRI within the cell relative to these Ca2+ fluxes (32).
We have shown that the monomeric and dimeric forms of CAPRI coexist within the cell in a Ca2+-dependent manner. It seems likely that the monomers and dimers are subject to further control through their localization and complex formation with other proteins within the cell. Ca2+ microdomains exist in the immediate vicinity of the cytosolic mouth of a variety of Ca2+ channels (30). In such microdomains, the free Ca2+ concentration can rise above 100 μm at a distance of 50–150 nm from the channel (33). Also, it has been demonstrated that CAPRI can localize to the endoplasmic reticulum (32). Therefore, it is possible that some of the dimerization of CAPRI will take place at these Ca2+ microdomains or the endoplasmic reticulum, although we did not observe any particular subcompartmental cytoplasmic localization of CAPRI even in the BiFC experiments. Equally, it is possible that CAPRI binding to other interactors, such as Rac1 or Cdc42 (34), may prevent some of the CAPRI dimer formation in the cell.
Ras is a key small G protein and is activated by a large range of extracellular signaling events, so-called “outside-in” signaling, whereas Rap1 often plays opposite roles to Ras signaling, referred “inside-out” signaling (35). Ca2+, as a second messenger, regulates dynamic and highly versatile signals that control a plethora of cellular processes (36). There are many examples where the balance of the signaling between Ras and Rap1 determines important cell fate decisions, such as cell proliferation and survival (35). Regulation of the dual RasGAP and Rap1GAP activities of CAPRI by Ca2+, as described in this paper, provides a new mechanism that could enable Ca2+ to coordinate Ras and Rap1 signaling pathways. One can envisage that, by modulating the RasGAP versus RapGAP activities of CAPRI through Ca2+, the cell may coordinate the relative activation profile of these two small GTPases, similar to the example of SynGAP in neurons, where the strength of long term potentiation versus long term depression is determined by Ca2+ acting through calcium/calmodulin-dependent kinase II (37).
Monomeric CAPRI has a strong RasGAP activity, as demonstrated by pulldown assays and TIRF microscopy. We cannot rule out that CAPRI dimers also possess RasGAP activity, as cells expressing WT-CAPRI contained a mixture of CAPRI monomers and dimers and because of the difficulty specifically expressing the dimeric form of CAPRI. However, two lines of evidence argue against the dimeric form of CAPRI having RasGAP activity. First, cells expressing monomeric CAPRI showed twice as much RasGAP activity as those expressing WT-CAPRI. Second, two-thirds of the CAPRI molecules could form dimers in the cell. Therefore, it seems likely that the RasGAP activity observed in WT-CAPRI-expressing cells could be derived from the present CAPRI monomers. We are confident that dimeric CAPRI possesses at best weak RasGAP activity compared with monomeric CAPRI. In contrast, WT-CAPRI possessed a much stronger Rap1GAP activity than monomeric CAPRI. Indeed, monomeric CAPRI displayed weak Rap1GAP activity, not significantly different from control. We speculate that, structurally, monomeric CAPRI may be partially accessible to Rap1 but cannot form a stable catalytic complex. CAPRI may use the mechanism of dimerization to form a stable catalytic site for Rap1.
Mutation of the arginine finger in the GAP domain of CAPRI inactivates both RasGAP and Rap1GAP activities (20, 23), which is similar to the effects of mutating the arginine finger of GAP1IP4BP (20) or SynGAP (11, 38). This observation indicates that the Rap1-binding site within the catalytic domain of CAPRI is the same as for Ras. However, mutation of the asparagine thumb or glutamine residues within the GAP domain in GAP1IP4PB failed to inhibit Rap1GAP activity (21). In contrast, mutation of the corresponding asparagine in SynGAP significantly reduced its Rap1GAP activity but did not affect binding affinity (38). These data imply that each of these dual Ras- and Rap-GAPs employs its own mechanism to act on Rap1. Apparently, the flanking regions of the GAP domain of GAP1IP4BP and Rasal promote a different orientation of switch II and catalytic residues in the active sites of Ras and Rap. Therefore, Gln-63 of Rap adopts the role normally played by Gln-61 of Ras (22). In agreement with this hypothesis, the crystal structure of SynGAP revealed that the C2 domain is in the vicinity of the nucleotide-binding region of the G protein target to assist GTP hydrolysis (38). In addition, it was suggested that the dual GAP activity of SynGAP is modulated through protein phosphorylation (39).
Although dimerization as a mechanism to promote RapGAP activity seems to be unique to CAPRI, dimerization is common among Rap1-specific GAPs, as revealed by the crystal structures of the GAP domain of Rap1GAP (16) and of the Rap-Rap1GAP complex (17). Although dimerization itself is not critical for its GAP activity (16), Rap1GAP activity does appear to require the presence of the dimerization domain, possibly to support binding of the Rap target. This was concluded from mutational analysis of the residues in the contact area with the catalytic domain (16, 17). We speculate that CAPRI may use dimerization to create an allosteric conformation of the GAP domain that enables it to bind and catalyze the inactivation of Rap1.
The CAPRI gene has been knocked out in mice. Surprisingly, the only phenotype found so far in these mice was a defect in the phagocytic function of macrophages (34). It was suggested by these authors that a primary role for CAPRI may be that of a scaffold protein for Rac1 and Cdc42. In addition, it was revealed that CAPRI may also function as a tumor suppressor (40), probably similar to the role of the GAP DAP2IP in tumors (41). The diverse functions of CAPRI in these systems indicate that CAPRI may play a central role within a range of different cellular signaling pathways.
Supplementary Material
Acknowledgments
We thank Dr. Len Stephens for encouragement and critically reading this manuscript and Dr. Trevor Smith for assistance with the gel filtration assays.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Movies 1–4.
- GAP
- GTPase-activating protein
- AA
- amino acid
- BiFC
- bimolecular fluorescence complementation
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Raaijmakers J. H., Bos J. L. (2009) J. Biol. Chem. 284, 10995–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hancock J. F. (2003) Nat. Rev. Mol. Cell Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 3. Kitayama H., Sugimoto Y., Matsuzaki T., Ikawa Y., Noda M. (1989) Cell 56, 77–84 [DOI] [PubMed] [Google Scholar]
- 4. Cook S. J., Rubinfeld B., Albert I., McCormick F. (1993) EMBO J. 12, 3475–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vossler M. R., Yao H., York R. D., Pan M. G., Rim C. S., Stork P. J. (1997) Cell 89, 73–82 [DOI] [PubMed] [Google Scholar]
- 6. Ohba Y., Ikuta K., Ogura A., Matsuda J., Mochizuki N., Nagashima K., Kurokawa K., Mayer B. J., Maki K., Miyazaki J., Matsuda M. (2001) EMBO J. 20, 3333–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kooistra M. R., Dubé N., Bos J. L. (2007) J. Cell Sci. 120, 17–22 [DOI] [PubMed] [Google Scholar]
- 8. Stork P. J. (2003) Trends Biochem. Sci. 28, 267–275 [DOI] [PubMed] [Google Scholar]
- 9. Bernards A. (2003) Biochim. Biophys. Acta 1603, 47–82 [DOI] [PubMed] [Google Scholar]
- 10. Yarwood S., Bouyoucef-Cherchalli D., Cullen P. J., Kupzig S. (2006) Biochem. Soc. Trans. 34, 846–850 [DOI] [PubMed] [Google Scholar]
- 11. Ahmadian M. R., Stege P., Scheffzek K., Wittinghofer A. (1997) Nat. Struct. Biol. 4, 686–689 [DOI] [PubMed] [Google Scholar]
- 12. Scheffzek K., Lautwein A., Kabsch W., Ahmadian M. R., Wittinghofer A. (1996) Nature 384, 591–596 [DOI] [PubMed] [Google Scholar]
- 13. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 14. Brinkmann T., Daumke O., Herbrand U., Kühlmann D., Stege P., Ahmadian M. R., Wittinghofer A. (2002) J. Biol. Chem. 277, 12525–12531 [DOI] [PubMed] [Google Scholar]
- 15. Chakrabarti P. P., Suveyzdis Y., Wittinghofer A., Gerwert K. (2004) J. Biol. Chem. 279, 46226–46233 [DOI] [PubMed] [Google Scholar]
- 16. Daumke O., Weyand M., Chakrabarti P. P., Vetter I. R., Wittinghofer A. (2004) Nature 429, 197–201 [DOI] [PubMed] [Google Scholar]
- 17. Scrima A., Thomas C., Deaconescu D., Wittinghofer A. (2008) EMBO J. 27, 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim J. H., Lee H. K., Takamiya K., Huganir R. L. (2003) J. Neurosci. 23, 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cullen P. J., Hsuan J. J., Truong O., Letcher A. J., Jackson T. R., Dawson A. P., Irvine R. F. (1995) Nature 376, 527–530 [DOI] [PubMed] [Google Scholar]
- 20. Kupzig S., Deaconescu D., Bouyoucef D., Walker S. A., Liu Q., Polte C. L., Daumke O., Ishizaki T., Lockyer P. J., Wittinghofer A., Cullen P. J. (2006) J. Biol. Chem. 281, 9891–9900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kupzig S., Bouyoucef-Cherchalli D., Yarwood S., Sessions R., Cullen P. J. (2009) Mol. Cell. Biol. 29, 3929–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sot B., Kötting C., Deaconescu D., Suveyzdis Y., Gerwert K., Wittinghofer A. (2010) EMBO J. 29, 1205–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lockyer P. J., Kupzig S., Cullen P. J. (2001) Curr. Biol. 11, 981–986 [DOI] [PubMed] [Google Scholar]
- 24. Walker S. A., Kupzig S., Bouyoucef D., Davies L. C., Tsuboi T., Bivona T. G., Cozier G. E., Lockyer P. J., Buckler A., Rutter G. A., Allen M. J., Philips M. R., Cullen P. J. (2004) EMBO J. 23, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cook S. J., Lockyer P. J. (2006) Cell Calcium 39, 101–112 [DOI] [PubMed] [Google Scholar]
- 26. Liu Q., Walker S. A., Gao D., Taylor J. A., Dai Y. F., Arkell R. S., Bootman M. D., Roderick H. L., Cullen P. J., Lockyer P. J. (2005) J. Cell Biol. 170, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu C. D., Kerppola T. K. (2003) Nat. Biotechnol. 21, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hames B. (1990) in Gel Electrophoresis of Proteins (Hames B., Rockwood D. eds) pp. 1–139, 2nd Ed., Oxford University Press, Oxford [Google Scholar]
- 29. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 30. Bootman M. D., Collins T. J., Peppiatt C. M., Prothero L. S., MacKenzie L., De Smet P., Travers M., Tovey S. C., Seo J. T., Berridge M. J., Ciccolini F., Lipp P. (2001) Semin. Cell Dev. Biol. 12, 3–10 [DOI] [PubMed] [Google Scholar]
- 31. Nalefski E. A., Falke J. J. (1996) Protein Sci. 5, 2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bivona T. G., Pérez De Castro I., Ahearn I. M., Grana T. M., Chiu V. K., Lockyer P. J., Cullen P. J., Pellicer A., Cox A. D., Philips M. R. (2003) Nature 424, 694–698 [DOI] [PubMed] [Google Scholar]
- 33. Cullen P. J. (2006) Curr. Opin. Cell Biol. 18, 157–161 [DOI] [PubMed] [Google Scholar]
- 34. Zhang J., Guo J., Dzhagalov I., He Y. W. (2005) Nat. Immunol. 6, 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinashi T. (2005) Nat. Rev. Immunol. 5, 546–559 [DOI] [PubMed] [Google Scholar]
- 36. Roderick H. L., Cook S. J. (2008) Nat. Rev. Cancer 8, 361–375 [DOI] [PubMed] [Google Scholar]
- 37. De Koninck P., Schulman H. (1998) Science 279, 227–230 [DOI] [PubMed] [Google Scholar]
- 38. Pena V., Hothorn M., Eberth A., Kaschau N., Parret A., Gremer L., Bonneau F., Ahmadian M. R., Scheffzek K. (2008) EMBO Rep. 9, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oh J. S., Manzerra P., Kennedy M. B. (2004) J. Biol. Chem. 279, 17980–17988 [DOI] [PubMed] [Google Scholar]
- 40. Westbrook T. F., Martin E. S., Schlabach M. R., Leng Y., Liang A. C., Feng B., Zhao J. J., Roberts T. M., Mandel G., Hannon G. J., Depinho R. A., Chin L., Elledge S. J. (2005) Cell 121, 837–848 [DOI] [PubMed] [Google Scholar]
- 41. Dote H., Toyooka S., Tsukuda K., Yano M., Ouchida M., Doihara H., Suzuki M., Chen H., Hsieh J. T., Gazdar A. F., Shimizu N. (2004) Clin. Cancer Res. 10, 2082–2089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.