Abstract
IL-33/IL-1F11 is a new member of the IL-1 family ligand and provokes T helper-type immune responses. IL-33 is the ligand of ST2 and IL-1 receptor accessory protein (IL-1RAcP) that triggers nuclear factor-κ light chain enhancer of activated B cells (NF-κB) and MAPK signaling. We discovered a novel short splice variant of IL-33 that was termed spIL-33. The new spIL-33 lacks exon 3 containing a proposed caspase-1 cleavage site. We isolated spIL-33 cDNA from the Huh7 human hepatocarcinoma cell line and expressed the recombinant spIL-33 protein in Escherichia coli. The recombinant spIL-33 and pro-IL-33 were not cleaved by caspase-1, unlike IL-18 (IL-1F4). The recombinant spIL-33 was constitutively active, and spIL-33-induced inflammatory cytokine production was caspase-1-independent in HMC-1 and Raw 264.7 cells. The recombinant spIL-33 induced the phosphorylation of IL-1 receptor-associated kinase (IRAK1), NF-κB, p38 MAPK, p44/42 MAPK, and JNK in a time- and dose-dependent manner. Anti-ST2 monoclonal antibody specifically blocked the spIL-33-induced cytokine production. In this study, we identified and characterized a new IL-33 splice variant, which was a constitutively active IL-33 isoform. The existence of constitutively active spIL-33 suggests that the biological activity of IL-33 could be triggered by diverse stimulations during immune responses. Further investigation of the spIL-33 expression pattern may contribute to understanding the involvement of IL-33 in inflammatory disorders.
Keywords: Caspase, Cytokine, Inflammation, NF-kappa B, Signal Transduction
Introduction
IL-33 is a new member of the IL-1 family ligand that was originally discovered as nuclear factor from high endothelial venules (1). IL-33 is considered to be critical in inducing T helper (Th)3 2-type immune responses like immunity against nematodes and allergic diseases with IL-3, IL-4, and/or IL-13 by Th2 cells (2–6), mast cells (4, 7), basophils (8), and eosinophils (5, 9, 10). IL-33 also induces non-Th2 inflammatory cytokines such as TNFα, IL-1β, or IL-6 (11–13). It has also been reported that IL-33 may act as a cytokine and nuclear transcription factor like IL-1β and high mobility group protein B1 (HMGB1) (4, 5, 14–23).
IL-33 is the ligand for ST2 (also known as IL-1RL1, DER4, Fit-1, or T1), which was originally discovered as an orphan receptor (6, 24–28). Thus, the IL-33 receptor complex for signaling is composed of ST2 and IL-1 receptor accessory protein (IL-1RAcP) (2, 29). This receptor complex activates downstream signaling molecules such as nuclear factor-κ light chain enhancer of activated B cells (NF-κB) and activation protein 1 (AP-1) through IL-1 receptor-associated kinase, TNF receptor-associated factor 6 (TRAF6), and/or MAPKs (6). It has been established for several years that ST2 is a selective marker of Th2 cells in mouse and human (30) and is expressed on mast cells (7), eosinophils (9, 10), and even basophils (22) without a known ligand. These cells produce inflammatory cytokines and chemokines, including IL-4, IL-5, IL-6, IL-13, and IL-8 under stimulation of IL-33 (5, 7, 9, 31). Recently, it has been reported that circulating CD34+ hematopoietic progenitors expressed ST2 and responded to IL-33 by releasing high levels of Th2-associated cytokines (32). Such observations suggest potential roles of IL-33 in Th2-associated immune responses, and IL-33 seems to be closely related to allergic inflammatory diseases, including asthma and atopic dermatitis.
IL-1 family members play a variety of pathological roles in autoimmune and inflammatory disorders (33). A blockade of IL-1β with IL-1R antagonist in patients with rheumatoid arthritis (34), ankylosing spondylitis (35), and mutations in the NACHT, LRR, and PYD domain-containing protein 3 (NALP3) gene (36, 37) relieves the symptoms of these diseases. Therefore, IL-1β is used for the immunological marker of inflammatory diseases.
IL-33, similarly to IL-1β and IL-18, is considered to be formed intracellularly as pro-IL-33. This pro-form does not have a signal peptide to be secreted; it is released extracellularly as a mature protein after cleavage (6). It has been known that inflammatory caspases are necessary for the cleavage of IL-1β and IL-18 (38, 39). Although it has been suggested that pro-IL-33 is processed by caspase-1 (6), the exact role of caspases in IL-33 biology still remains controversial (40, 41).
Alternative splicing, which induces several mature mRNAs from a pre-mRNA, is a frequent process in eukaryotes. This is an efficient method for an organism to form various proteins from a single gene (42). Numerous transcripts are formed by cis-regulatory elements such as the promoter, enhancer, and repressor control. Alternative splicing induces structural differences of the translated proteins, which leads to functional changes of produced proteins (43). Alternatively processed mRNAs can even lead to the induction of several diseases, including cancers (44). This process is tightly regulated by various stress conditions, including endoplasmic reticulum stress (45). Furthermore, there are many reports suggesting alternative splicing is efficiently regulated to accomplish specialized tasks (46). Thus, characterizing the biological role of splice variants can help with the understanding of their functional application in human diseases.
In this study, we characterized a newly discovered IL-33 splice variant (spIL-33), which lacks the exon 3 in its coding region. Interestingly, the deleted exon 3 region of spIL-33 contains the proposed caspase-1 cleavage site (6). The existence of a novel spIL-33 and further study on constitutively active spIL-33 in patients with different autoimmune diseases will reveal the precise role of spIL-33 in immune responses.
EXPERIMENTAL PROCEDURES
RT-PCR and Molecular Cloning for E. coli Expression Vectors
Total RNA was isolated with TRI Reagent® (Sigma) from the human A549, HACAT, U937, Huh7, HUVEC, Jurkat, NK, and mouse Raw 264.7 cells. A pair of human IL-33 sense primers, 5′-ACAGAATACTGAAAAATGAAGCC-3′, and reverse primer, 5′-CTTCTCCAGTGGTAGCATTTG-3′; human IL-8 sense primers, 5′-GTGATGACCTGGCCGTCAGG-3′, and reverse primer, 5′-GTGATGACCTGGCCGTCAGG-3′; mouse TNFα sense primer, 5′-CATCTTGGAAATAGCTCCCAG-3′, and reverse primer, 5′-CTGAGCCATAATCCCCTTTCT-3′; mouse macrophage inflammatory protein (MIP-2) sense primer, 5′-ACACTTCAGCCTAGCGCCAT-3′, and reverse primer, 5′-CAGGTCAGTTAGCCTTGCCT-3′; mouse IL-6 sense primer, 5′-GAAACCGCTATGAAGTTCCTC-3′, and reverse primer, 5′-CTTAGGCATAACGCACTAGGT-3′; and β-actin sense primer, 5′-ACCAACTGGGACGACATGGA-3′, and reverse primer, 5′-GTGATGACCTGGCCGTCAGG-3′, was used for the RT-PCR. MMLV-RT (Beams Bio, Korea) was used for converting 2 μg of total RNA to first strand cDNA, and the PCR was performed at 94 °C for 45 s, 70 °C for 2 min, and 59 °C for 1 min for 30 cycles.
The PCR product of spIL-33 and pro-IL-33 was ligated into T&A cloning vector (RBC, Taiwan) for DNA sequencing. The cDNA of pro-IL-33, spIL-33, and mature IL-33 was amplified by PCR with the same reverse primer (5′-GGTTGGTACCTCAGTGTTCTTTAGGCC-3′) and sense primers for pro-IL-33 and spIL-33 (5′-TCCCGAATTCGATTGTGATATTGAA-3′) or mature IL-33 (Ser112– Thr270, 5′-TAAAGAATTCAGTATCACAGGAATTTC-3′). Inserts were digested with EcoRI and KpnI and then transferred into pProEX/HTa (Invitrogen) for recombinant protein expression in Escherichia coli.
Generation of Recombinant Protein
Recombinant pro-IL-33, spIL-33, and mature IL-33 proteins were expressed in E. coli and Rosetta cells (Novagen, Madison, WI) and purified by TALON affinity columns (Invitrogen) by using His6 tag at the N terminus of recombinant proteins. The TALON affinity-purified proteins were subjected to high performance liquid chromatography (GE Healthcare) with a C4 column (Grace Vydac, Hesperia, CA). The two step-purified recombinant proteins were tested for endotoxin levels (below 0.3 enzyme units per μg of recombinant protein) by using the LAL method (Cape Cod, East Falmouth, MA) according to the manufacturer's instructions and then used for experiments.
Caspase-1 Cleavage Test
Recombinant caspase-1 (10 units) from Millipore (Temecula, CA) and recombinant IL-33 (500 ng) protein were incubated in 20 μl of a reaction buffer (25 mm K+HEPES, 1 mm DTT, 1 mm EDTA, 0.1% CHAPS, 10% sucrose, pH 7.5) at 37 °C for 30 min. After the reaction, the sample was subjected to 10% SDS-PAGE for silver staining.
Western Blots
For the detection of His6-tagged recombinant IL-33, HPLC-purified fractions of the protein were loaded on 10% SDS-PAGE. Anti-polyhistidine monoclonal antibody (R&D Systems, Minneapolis, MN) was used for detection. Peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) was used to develop the blots by using Supex (Neuronex, Korea) and LAS-4000 imaging device (Fujifilm, Japan).
For detecting the phosphorylation of signaling molecules (IRAK1, NF-κB, p38 MAPK, p44/42 MAPK, and JNK), HMC-1 and Raw 264.7 cells were stimulated with recombinant spIL-33 at various time points or with several concentrations for 15 min. Cells were lysed with kinase lysis buffer (47) and then subjected to 10% SDS-PAGE. The samples were transferred to nitrocellulose membranes. The membranes were blocked in 3% BSA/TBST (Santa Cruz Biotechnology, Santa Cruz, CA). The membranes were probed first with rabbit anti-phospho-IRAK1, mouse anti-phospho-NF-κB, rabbit anti-phospho-p38 MAPK, mouse anti-phospho-p44/42 MAPK, or rabbit anti-phospho-JNK (Cell Signaling Technology, Beverly, MA). The membranes were reprobed with rabbit anti-NF-κB, p38 MAPK (Santa Cruz Biotechnology), p44/42 MAPK, JNK or IRAK1 (Cell Signaling Technology) for normalization of each protein, and then normalized finally with goat anti-actin (Santa Cruz Biotechnology).
Cell Culture and Bioassay
Human NK cell line was obtained from Dr. Hans Klingerman (48) and cultured in RPMI 1640 medium containing 10% FBS, IL-2 (50 pg/ml), and IL-15 (200 pg/ml) (PeproTech, Rocky Hill, NJ). A549-IL-18Rβ cells were cultured as described previously (49). HUVEC vascular endothelial, A549 lung carcinoma, monocytic U937, Jurkat T cell leukemia, and mouse macrophage Raw 264.7 cell lines were obtained from American Type Culture Collection (ATCC) and maintained according to the instructions. HMC-1 cells were cultured in Iscove's modified Dulbecco's medium enriched with 10% FBS. Huh7 cells were maintained in DMEM with 10% FBS. Bioassays were performed in 96-well plates. Raw cells (5 × 105/ml) and A549-IL-18Rβ cells (2 × 105/ml) were seeded in a 96-well plate and cultured until cells adhered to the plate, and HMC-1 cells (10 × 106/ml) were seeded before assay. For a blockade of ST2, fresh medium (0.2 ml) containing various concentrations of recombinant IL-33 and 0.5 μg of anti-ST2 (R&D Systems) were used. For A549-IL-18Rβ cells assays, the medium was removed and then stimulated with fresh medium (0.2 ml) containing 100 ng/ml IL-18. The plates were placed in a cell culture incubator for 16 h, and cytokines in the supernatant were measured.
Measurement of Cytokine Level
Human IL-8, mouse TNFα, mouse MIP-2, and mouse IL-6 ELISA kits were obtained from R&D Systems. Cytokine levels were measured in culture supernatants by sandwich ELISA according to the manufacturer's instructions.
Statistical Analysis
The data are expressed as means ± S.E. Statistical significance of differences was analyzed by unpaired, two-tailed Student's t test. Values of p < 0.05 were considered statistically significant.
RESULTS
Discovery of a Splice Variant of IL-33 mRNA
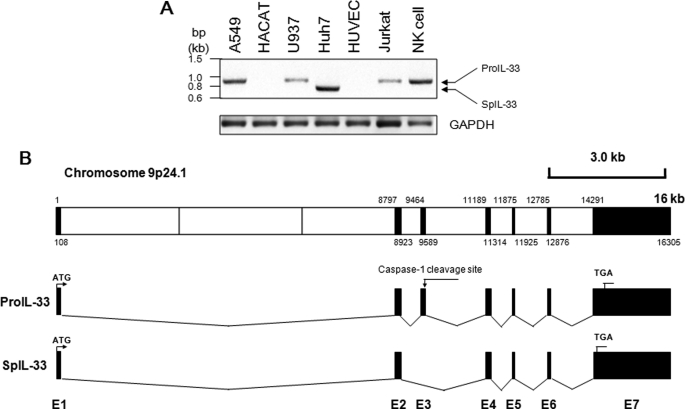
We performed RT-PCR of IL-33 from several human cell lines; these were A549 lung epithelial, HACAT keratinocyte, U937 monocyte, Huh7 hepatocyte, HUVEC vascular endothelial, Jurkat T, and NK cell lines. A549, Huh7, and NK cells expressed relatively high levels of IL-33 mRNA compared with U937 or Jurkat cells but were not expressed in HACAT and HUVEC cells. Interestingly, the IL-33 RT-PCR product from Huh7 cells seemed ∼100 bp smaller than that of other cell lines (Fig. 1A). We performed TA cloning with the PCR product from Huh7, and the sequence was analyzed. The sequencing result revealed that the IL-33 cDNA of Huh7 possesses 126 nucleotides shorter than previously known pro-IL-33. The spIL-33 lacks exon 3 due to alternative splicing of the known pro-IL-33 (Fig. 1B; GenBankTM accession number HQ641439).
FIGURE 1.
Identification of a new splice variant of IL-33 from Huh7 hepatic carcinoma cells. A, RT-PCR of IL-33 was performed with several human cell lines. IL-33 mRNA was detected in A549, U937, Huh7, Jurkat, and NK cells but not in HACAT and HUVEC cells. Interestingly, the PCR product of IL-33 from Huh7 cells was ∼100 bp smaller than that of other cell lines. The data represent one of two independent experiments. B, PCR product was sequenced, and the DNA sequencing result revealed the deletion of 126 bp in previously known pro-IL-33. The mRNA of spIL-33 was alternatively spliced and lacked the exon 3 where the proposed caspase-1 cleavage site exists as indicated (GenBankTM accession No. HQ641439).
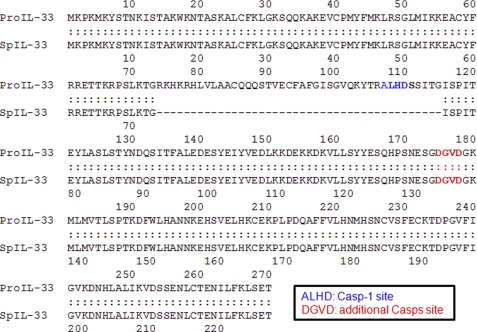
The amino acid sequence of pro-IL-33 and spIL-33 was aligned, and the translated amino acid sequence of spIL-33 possesses 42 amino acids less than pro-IL-33. The proposed caspase-1 cleavage site (Ala-Lys-His-Asp) is deleted in spIL-33 (6); however, the other proposed caspase-1, -3, or -7 cleavage site (Asp-Gly-Val-Asp) (40, 41) remains in this splice variant (Fig. 2).
FIGURE 2.
Alignment between pro-IL-33 and translated amino acid sequence of spIL-33. Amino acid sequence of pro-IL-33 and spIL-33 was aligned. The translated spIL-33 has 228 amino acids, which has 42 amino acids less than pro-IL-33. “ALHD” was originally proposed as a caspase-1 cleavage site, which is deleted in spIL-33; however, the other predicted caspase-1, -3, or -7 cleavage site “DGVD” remains in this splice variant.
Generation of Recombinant IL-33
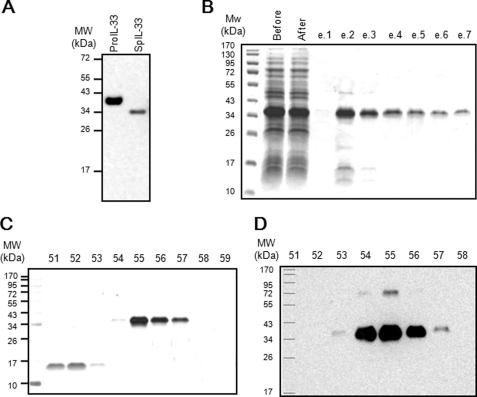
We produced recombinant spIL-33 in E. coli to characterize the newly identified IL-33 variant. The molecular size of His6-tagged recombinant spIL-33 was ∼35 kDa. It was 5 kDa smaller than that of His6-tagged recombinant pro-IL-33 when the molecular size was analyzed using Western blotting with His6 (Fig. 3A). Following the production of recombinant protein, recombinant spIL-33 was first purified by TALON metal affinity chromatography. The purified protein was visualized by 10% SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 3B). Primarily purified 35-kDa recombinant spIL-33 protein was additionally purified using high performance liquid chromatography and visualized by silver staining and Western blotting (Fig. 3, C and D). Recombinant pro-IL-33 (Met1–Thr270) and mature IL-33 proteins (Ser112–Thr270) were also expressed and purified like spIL-33 (data not shown).
FIGURE 3.
Expression of recombinant pro-IL-33 and spIL-33. Recombinant spIL-33 was expressed in E. coli. A, molecular size of recombinant spIL-33 was ∼5 kDa smaller than that of pro-IL-33. B, recombinant spIL-33 was first purified by a TALON metal affinity chromatography. The purified protein was visualized by 10% SDS-PAGE and Coomassie Brilliant Blue staining. The purified (35 kDa) recombinant spIL-33 protein was further purified by HPLC and visualized by silver staining (C) and Western blotting (D). The data represent one of four independent experiments.
Caspase-1 Does Not Affect Pro-IL-33 or SpIL-33
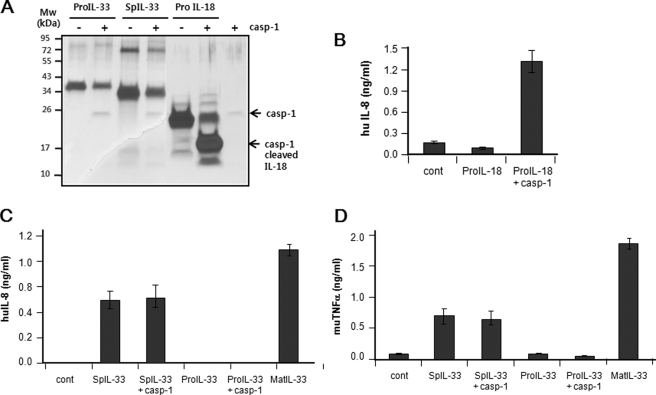
Recombinant pro-IL-33, spIL-33, or pro-IL-18 was digested with caspase-1 to determine whether the caspase-1 cleavage site exists in IL-33. Following treatment of caspase-1 protein to pro-IL-33 and spIL-33, both recombinant proteins were not cleaved, unlike caspase-1-digested pro-IL-18, and produced a mature 18-kDa protein band as described previously (Fig. 4A). In order to confirm the activity of IL-18, pro-IL-18 was cleaved with 10 IU/ml caspase-1. The caspase-1-cleaved pro-IL-18 or intact pro-IL-18 was treated to A549-IL-18Rβ cells overnight. Caspase-1-treated pro-IL-18 induced IL-8 production, but the intact one did not (Fig. 4B). Interestingly, spIL-33 was constitutively active, producing IL-8 in HMC-1 and TNFα in Raw 264.7 cells, but pro-IL-33 was not active in these cell lines (Fig. 4, C and D). The spIL-33 sufficiently induced inflammatory cytokines, although the level was lower than that of mature IL-33. The biological activity of pro-IL-33 and spIL-33 was not changed by caspase-1.
FIGURE 4.
Caspase-1 does not cleave recombinant IL-33 and affects the function of IL-33. A, recombinant pro-IL-33, spIL-33, or pro-IL-18 was digested with caspase (casp)-1. Caspase-1 protein cleaved neither pro-IL-33 nor spIL-33 clearly, unlike the enzyme-digested IL-18. B, caspase-1 cleaves and activates pro-IL-18 properly. A549-IL-18Rβ cells were treated with the caspase-1-cleaved or intact IL-18 overnight. IL-8 induction in A549-IL-18Rβ cells was caspase-1-dependent. C and D, biological activity of recombinant pro-IL-33 and spIL-33 was not affected by caspase-1. Unlike pro-IL-33, spIL-33 and mature IL-33 (Ser112–Thr270) were constitutively active and induced inflammatory cytokines. The data represent one of two independent experiments. Cont, control.
SpIL-33 Stimulates IRAK1, NF-κB, p38 MAPK, p44/42 MAPK, and JNK Signal Pathways
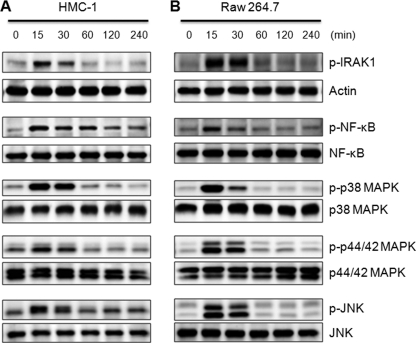
Signal transduction of spIL-33 was also studied. The phosphorylation of IRAK1, NF-κB, p38 MAPK, ERK, and JNK was examined with spIL-33-treated HMC-1 and Raw 264.7 cells because IL-33 is known to activate these inflammatory signal pathways. As shown in Fig. 5, spIL-33 stimulation promptly phosphorylated IRAK1 in a time-dependent manner, reached the maximal level at 15 min after exposure, and decreased dramatically at 60 min. The phosphorylation of NF-κB was similar; however, its phosphorylation in HMC-1 cells was sustained longer than that of other signaling molecules (Fig. 5A, 2nd lane). The phosphorylation pattern of p38 MAPK, p42/44 MAPK, and JNK was similar to IRAK1, although there was a difference in the density of activation. This result confirms that spIL-33 induces inflammatory cytokines through stimulation of IRAK1, NF-κB, p38 MAPK, p42/44 MAPK, and JNK signal pathways.
FIGURE 5.
spIL-33 induced the phosphorylation of IRAK1, NF-κB, p38 MAPK, p44/42 MAPK, and JNK. Human mast HMC-1 cells (A) and mouse macrophage Raw 264.7 cells (B) were treated with 50 ng/ml human spIL-33 as indicated time points. The phosphorylation of IRAK1, NF-κB, p38 MAPK, p44/42 MAPK, and JNK was significantly increased at 15 min and then drastically decreased at 60 min. However, phospho-NF-κB remained until 240 min in HMC-1 cell. The bottom of each panel exhibits the expression level of nonphosphorylated signaling molecule in cell lysates to show an equal amount of protein sample was loaded in each lane. The data represent one of three independent experiments.
Examination of Chemokines and Inflammatory Cytokine Transcripts
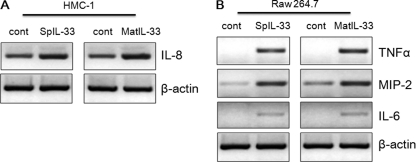
HMC-1 cells and Raw 264.7 cells were stimulated with spIL-33 or mature IL-33, and then the induction of chemokines and inflammatory cytokine transcript was assessed by RT-PCR. Both spIL-33 and mature IL-33 induced the transcriptions of chemokines (IL-8 and MIP-2) and inflammatory cytokines (TNFα and IL-6) compared with the untreated control cells (Fig. 6). SpIL-33 and mature IL-33 sufficiently increased mRNA level of the chemokines and cytokines, although the induction of transcripts varied. Chemokines (IL-8 and MIP-2) exhibited the basal level of transcripts in untreated control cells (Fig. 6A).
FIGURE 6.
Recombinant spIL-33 induces the transcription of inflammatory cytokines. HMC-1 cells (A) and Raw 264.7 cells (B) were treated with spIL-33 or mature IL-33, and the transcription level of inflammatory cytokine was assessed by RT-PCR. Both spIL-33 and mature IL-33 induced chemokines (IL-8 and MIP-2) and inflammatory cytokines (TNFα and IL-6) compared with nontreated control cells. The data represent one of three independent experiments. cont, control.
Dose-dependent Phosphorylation of spIL-33-induced Signaling Molecules
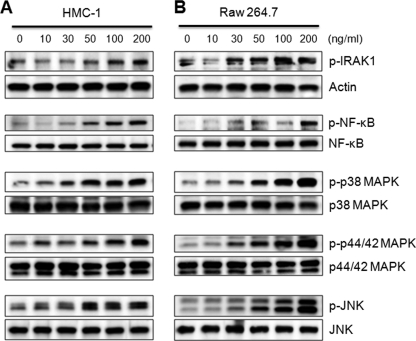
In addition, we determined whether spIL-33 phosphorylates signaling molecules in a dose-dependent manner. HMC-1 and Raw 264.7 cells were stimulated with various concentrations of spIL-33 for 15 min because the phosphorylation of signaling molecules exhibited the highest activation (Fig. 5). The phosphorylation of IRAK1, NF-κB, p38 MAPK, p44/42 MAPK, and JNK was augmented along with the increased concentrations of spIL-33 (Fig. 7). The activation of p38 MAPK and p44/42 MAPK in HMC-1 was initiated at 10 ng/ml concentration of spIL-33, whereas the other signaling molecules were phosphorylated at 30 or 50 ng/ml concentration of spIL-33 (Fig. 7, A and B).
FIGURE 7.
spIL-33 induces signal transduction in a dose-dependent manner. HMC-1 (A) and Raw 264.7 (B) cells were stimulated with various concentrations of recombinant spIL-33 for 15 min as indicated at the top. The phosphorylated or total signaling protein was detected with anti-phosphoprotein or total signaling protein as indicated at the right of each panel. The data represent one of three independent experiments.
Dose-dependent Induction of Inflammatory Cytokines by spIL-33
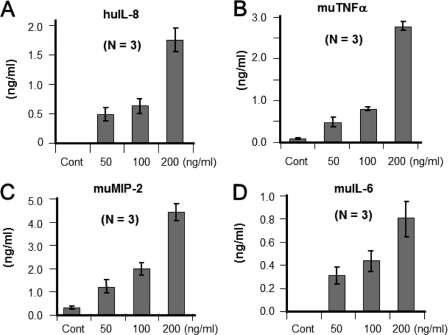
We decided to assess whether spIL-33 induces inflammatory cytokines by a dose-dependent manner. HMC-1 and Raw 264.7 cells were stimulated with several concentrations of spIL-33. spIL-33 induced chemokine (IL-8 and MIP-2) productions in a dose-dependent manner from HMC-1 and Raw 264.7 cells, respectively (Fig. 8, A and C). In addition, the induction of inflammatory cytokines (TNFα and IL-6) was similar to chemokine production (Fig. 8, B and D). This phenomenon was also repeated in the study with mature IL-33 proteins (data not shown).
FIGURE 8.
Dose-dependent induction of inflammatory cytokines by spIL-33. Several concentrations of spIL-33 were treated to HMC-1 or Raw 264.7 cells overnight. A, spIL-33 increased chemokine IL-8 in a dose-dependent manner from HMC-1 cells. B–D, inflammatory cytokines (TNFα and IL-6) and chemokines (MIP-2) were also induced by spIL-33 in mouse Raw 264.7 cells in a dose-dependent manner. The data represent one of two independent experiments. Cont, control; hu, human; mu, murine.
IL-33 Induces Inflammatory Cytokines through ST2 Receptors
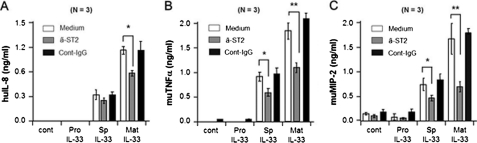
We examined whether spIL-33 induces chemokines and inflammatory cytokines via ST2 on the cell surface of HMC-1 and Raw 264.7 cells. These cells were pretreated with an anti-ST2-neutralizing antibody, and then pro-IL-33, spIL-33, or mature IL-33 was used to stimulate HMC-1 and Raw 264.7 cells. The culture supernatant was harvested after 16 h, and secreted IL-8, TNFα, and MIP-2 were measured by ELISA. The anti-ST2 antibody-pretreated HMC-1 cells (gray bar) produced less IL-8 compared with nontreated cells (open bar) and control antibody (black bar) in Fig. 9. Although spIL-33 or mature IL-33-induced IL-8 production was decreased, statistical significance was observed only in mature IL-33 (Fig. 9A). In mouse Raw 264.7 cells, TNFα and MIP-2 productions were sufficiently reduced by the anti-ST2 antibody, and the results exhibited were statistically significant in both spIL-33 and mature IL-33 (Fig. 9, B and C).
FIGURE 9.
IL-33 activates inflammatory effects via ST2. HMC-1 cells (A) and Raw 264.7 cells (B) were pretreated with anti-ST2 antibody or control IgG and then pro-IL-33 (Pro), spIL-33 (Sp), or mature (Mat) IL-33 was added to cells. The culture supernatant was harvested, and secreted IL-8 (A), TNFα (B), or MIP-2 (C) level was measured. Mean ± S.E. A, huIL-8; B, muTNFα; C, muMIP-2. *, p < 0.05; **, p < 0.01 (n = 3 per group). The data represent one of three independent experiments. Cont, control; hu, human; mu, murine.
DISCUSSION
In this study, we first demonstrated a novel isoform of human IL-33 splice variant (spIL-33), and spIL-33 has been characterized as a constitutively active IL-33 isoform. The discovery of spIL-33 was carried out while we attempted to isolate cDNA of IL-33 from various human cell lines. We found spIL-33 in Huh7 cells and revealed the lack of exon 3 at the predicted caspase-1 cleavage site (Figs. 1B and 2) (6).
The molecular structure, signal transduction, and the biological activity of spIL-33 were studied and compared with that of pro-IL-33 and mature IL-33. The previously known IL-33 was reported as a Th2-associated inflammatory cytokine (6). Although spIL-33 lacks the 42 amino acids (Arg74–Gly115) of the known pro-IL-33 (Fig. 2), its ability to activate the typical inflammatory signal pathways resulted in the induction of inflammatory cytokines such as human IL-8 in HMC and mouse TNFα, IL-6, and MIP-2 in mouse Raw 264.7 cells (Figs. 4–8).
The role of caspase-1 in the activation of IL-33 was controversial in several previous studies. The first study of IL-33 suggested that caspase-1 cleaves the amino acid residue (Asp110–Ser111) on human pro-IL-33, which was predicted by the incubation of in vitro translated IL-33 rather than recombinant pro-IL-33 protein (6). Interestingly, our results in Fig. 4A showed that recombinant pro-IL-33 and spIL-33 proteins were not cleaved by caspase-1, but recombinant pro-IL-18 protein was specifically processed by caspase-1 and became an active mature 18-kDa IL-18 (Fig. 4, A and B). Moreover, the recombinant spIL-33 and mature IL-33 stimulated HMC-1 and Raw 264.7 cells and induced inflammatory cytokines, which were independent of caspase-1 cleavage on spIL-33. The recombinant pro-IL-33 was not cleaved by caspase-1 (Fig. 4A) and was also not active in inducing inflammatory cytokines (Fig. 4, C and D).
Contrary to the initial report (6), it has been reported that full-length pro-IL-33 is active, and the processing by caspase-1 results in IL-33 inactivation (40). Further study on IL-33 processing by Luthi et al. (41) reported that apoptotic caspases process pro-IL-33 in apoptotic cells resulting in inactivation of pro-IL-33, whereas pro-IL-33 released from necrotic cell is spontaneously active. They suggested that the cleavage site of caspase-1 does not occur at the site initially proposed (Asp110–Ser111) but rather at the amino acid residues Asp178–Gly179, which is the consensus site of cleavage by caspase-3 (40, 41, 50). Apoptotic caspase-3 and -7 destroy the biological activity of pro-IL-33 by cleavage of amino acid residues Asp178–Gly179. Nevertheless, these studies have performed in vitro; the biological activity of the pro-IL-33 was shown by only NFκ-B luciferase assay (41). Unlike this study, our experiments were performed with HPLC-purified recombinant IL-33 proteins (endotoxin level below 0.3 enzyme units per 1 μg).
Another study suggested calpain-dependent processing of pro-IL-33 like IL-1α, which was shown by treatment with calcium ionophore. Calpain processes pro-IL-33, which produces mature IL-33 in human epithelial and endothelial cells without proving biological activity of the calpain-cleaved IL-33 (51). Contrary to this result, Ohno et al. (52) reported that caspase-1/8 and calpain are dispensable for IL-33 release from macrophage and mast cells. Murine IL-33 was released spontaneously, and its secretion was increased by LPS or phorbol 12-myristate 13-acetate plus ionomycin from the peritoneal macrophages of caspase-1-deficient BALB/c mice (52). The discrepancy of these results may be explained by the fact that the experiment of calpain-dependent IL-33 processing was carried out on human cell lines, whereas the conflicting report by Ohno et al. (52) was performed with the peritoneal macrophages of caspase-1-deficient mice. Although there is significant sequence homology between human and mouse IL-33, they share only 55% identity at the amino acid level.
Splice variants are atypical in cytokine genes, yet splice variants exist in some cytokines such as IL-15, IL-1F7, IL-32, and VEGF (49, 53–57). There are two isoforms of a well characterized IL-15, one has a long signal peptide (48 amino acids) and the other one has a short signal peptide (21 amino acids) due to an alternative splicing in exon 5 (58), which is similar to IL-33, but the splicing region of IL-33 (in pro piece) is distinct from IL-15 (signal peptide). Unlike IL-15, the splicing area of spIL-33 is located in a critical region containing the proposed caspase-1 cleavage site (Asp110–Ser111). As shown in Fig. 4, spIL-33 is constitutively active and its activity is not dependent on caspase-1 cleavage (Fig. 4, C and D). Our data showed that we were not able to confirm the cleavage of recombinant spIL-33, pro-IL-33 (Fig. 4A), and mature IL-33 (Ser112–Thr270) (data not shown) from different sources; our recombinant mature IL-33 and mature IL-33 were from R & D Systems. The spIL-33 and mature IL-33 were highly active, but pro-IL-33 was not active in both human HMC-1 and mouse Raw 264.7 cells (Figs. 4–9). This result implies that the mature form of IL-33 (Ser112–Thr270) is processed by other cytokine-processing enzymes.
Although both IL-15 isoforms possess identical biological properties, sharing the same receptors and biological functions, each IL-15 isoform has a distinct regulation and expression pattern. The long signal peptide IL-15 isoform is more efficiently secreted in supernatant (58), than that with the short one. Thus, we chose to down-regulate the activity of spIL-33 with mouse anti-ST2 in order to confirm whether spIL-33 activates inflammatory effects through ST2. The anti-ST2 monoclonal antibody effectively blocks the production of inflammatory cytokines induced by recombinant spIL-33 and mature IL-33 (Fig. 9). This result confirms that both spIL-33 and mature IL-33 share ST2 as a receptor. Although we confirmed that spIL-33 utilizes the same receptor (ST2) and exhibits the same biological functions, further investigation is necessary to characterize the expression pattern and regulation of spIL-33 in human tissues and different organs.
Neutrophil proteinase 3 (PR3), also known as Wegener autoantigen, is a granule serine protease that is localized in neutrophils and monocytes, and it is capable of processing multiple biological substrates (59). PR3 processes several cytokines such as IL-8 (60), TGFβ1 (61), membrane-bound TNFα (62), IL-1β (63), IL-18 (64), and IL-32 (65) and enhances the biological activity of cytokines. Thus, PR3 could be a potential processing enzyme for pro-IL-33, since PR3 processes the proform of IL-1 family ligands such as IL-1β and IL-18 (63, 64).
It is critical to study the effect of spIL-33 in vivo for investigation of the physiological role of the molecule. However, we have not discovered a similar splice variant in mouse IL-33, although we tried RT-PCR with various mouse tissues and organs (supplemental Fig. 1). There is variation in the number of splice variants with the IL-18-binding protein (IL-18BP). For example human IL-18BP exists in four isoforms, and mouse IL-18BP exists in only two isoforms (66).
spIL-33 sufficiently activates inflammatory signal pathways and induces inflammatory cytokines, although the biological activity of spIL-33 is weaker than mature IL-33. It is necessary to perform further studies on the precise structure of spIL-33 in order to address the difference in biological activity of spIL-33 and pro-IL-33. However, spIL-33 has 42 less amino acids in the pro-piece of IL-33, and the deleted region of spIL-33 seems to influence the active site of IL-33. Although the predicted caspase-1 cleavage site (Asp110–Ser111) is controversial, the mature recombinant IL-33 (Ser112–Thr270) was highly active, and the deletion of 42 amino acids in the pro-piece of spIL-33 is structurally similar to mature IL-33 that contributes to the biological activity of spIL-33.
Collectively, we identified a constitutively active spIL-33, which lacks the proposed caspase-1 cleavage site, and characterized the molecular structure, signal transduction, and the biological activity of spIL-33. Further study on the expression pattern of spIL-33 could help us understand the inflammatory disorders of epithelial tissues such as asthma and atopic dermatitis.
Supplementary Material
This work was supported by National Research Foundation Grants WCU R33-2008-000-10022-0 and KRF-2008-313-C00644 funded by the Korean government and the Korea Healthcare Technology R&D Project Grant A100460 from the Ministry of Health & Welfare, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) HQ641439.
- Th
- T helper
- NF-κB
- nuclear factor-κ light chain enhancer of activated B cells
- HUVEC
- human umbilical vein endothelial cell
- NK
- natural killer.
REFERENCES
- 1. Baekkevold E. S., Roussigné M., Yamanaka T., Johansen F. E., Jahnsen F. L., Amalric F., Brandtzaeg P., Erard M., Haraldsen G., Girard J. P. (2003) Am. J. Pathol. 163, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chackerian A. A., Oldham E. R., Murphy E. E., Schmitz J., Pflanz S., Kastelein R. A. (2007) J. Immunol. 179, 2551–2555 [DOI] [PubMed] [Google Scholar]
- 3. Guo L., Wei G., Zhu J., Liao W., Leonard W. J., Zhao K., Paul W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13463–13468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kondo Y., Yoshimoto T., Yasuda K., Futatsugi-Yumikura S., Morimoto M., Hayashi N., Hoshino T., Fujimoto J., Nakanishi K. (2008) Int. Immunol. 20, 791–800 [DOI] [PubMed] [Google Scholar]
- 5. Pecaric-Petkovic T., Didichenko S. A., Kaempfer S., Spiegl N., Dahinden C. A. (2009) Blood 113, 1526–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T. K., Zurawski G., Moshrefi M., Qin J., Li X., Gorman D. M., Bazan J. F., Kastelein R. A. (2005) Immunity 23, 479–490 [DOI] [PubMed] [Google Scholar]
- 7. Allakhverdi Z., Smith D. E., Comeau M. R., Delespesse G. (2007) J. Immunol. 179, 2051–2054 [DOI] [PubMed] [Google Scholar]
- 8. Sullivan B. M., Locksley R. M. (2009) Immunity 30, 12–20 [DOI] [PubMed] [Google Scholar]
- 9. Cherry W. B., Yoon J., Bartemes K. R., Iijima K., Kita H. (2008) J. Allergy Clin. Immunol. 121, 1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suzukawa M., Koketsu R., Iikura M., Nakae S., Matsumoto K., Nagase H., Saito H., Matsushima K., Ohta K., Yamamoto K., Yamaguchi M. (2008) Lab. Invest. 88, 1245–1253 [DOI] [PubMed] [Google Scholar]
- 11. Moulin D., Donzé O., Talabot-Ayer D., Mézin F., Palmer G., Gabay C. (2007) Cytokine 40, 216–225 [DOI] [PubMed] [Google Scholar]
- 12. Smithgall M. D., Comeau M. R., Yoon B. R., Kaufman D., Armitage R., Smith D. E. (2008) Int. Immunol. 20, 1019–1030 [DOI] [PubMed] [Google Scholar]
- 13. Verri W. A., Jr., Souto F. O., Vieira S. M., Almeida S. C., Fukada S. Y., Xu D., Alves-Filho J. C., Cunha T. M., Guerrero A. T., Mattos-Guimaraes R. B., Oliveira F. R., Teixeira M. M., Silva J. S., McInnes I. B., Ferreira S. H., Louzada-Junior P., Liew F. Y., Cunha F. Q. (2010) Ann. Rheum. Dis. 69, 1697–1703 [DOI] [PubMed] [Google Scholar]
- 14. Brint E. K., Xu D., Liu H., Dunne A., McKenzie A. N., O'Neill L. A., Liew F. Y. (2004) Nat. Immunol. 5, 373–379 [DOI] [PubMed] [Google Scholar]
- 15. Carriere V., Roussel L., Ortega N., Lacorre D. A., Americh L., Aguilar L., Bouche G., Girard J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ho L. H., Ohno T., Oboki K., Kajiwara N., Suto H., Iikura M., Okayama Y., Akira S., Saito H., Galli S. J., Nakae S. (2007) J. Leukocyte Biol. 82, 1481–1490 [DOI] [PubMed] [Google Scholar]
- 17. Hoshino K., Kashiwamura S., Kuribayashi K., Kodama T., Tsujimura T., Nakanishi K., Matsuyama T., Takeda K., Akira S. (1999) J. Exp. Med. 190, 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurowska-Stolarska M., Stolarski B., Kewin P., Murphy G., Corrigan C. J., Ying S., Pitman N., Mirchandani A., Rana B., van Rooijen N., Shepherd M., McSharry C., McInnes I. B., Xu D., Liew F. Y. (2009) J. Immunol. 183, 6469–6477 [DOI] [PubMed] [Google Scholar]
- 19. Moussion C., Ortega N., Girard J. P. (2008) PLoS One. 3, e3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rank M. A., Kobayashi T., Kozaki H., Bartemes K. R., Squillace D. L., Kita H. (2009) J. Allergy Clin. Immunol. 123, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Senn K. A., McCoy K. D., Maloy K. J., Stark G., Fröhli E., Rülicke T., Klemenz R. (2000) Eur. J. Immunol. 30, 1929–1938 [DOI] [PubMed] [Google Scholar]
- 22. Suzukawa M., Iikura M., Koketsu R., Nagase H., Tamura C., Komiya A., Nakae S., Matsushima K., Ohta K., Yamamoto K., Yamaguchi M. (2008) J. Immunol. 181, 5981–5989 [DOI] [PubMed] [Google Scholar]
- 23. Townsend M. J., Fallon P. G., Matthews D. J., Jolin H. E., McKenzie A. N. (2000) J. Exp. Med. 191, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bergers G., Reikerstorfer A., Braselmann S., Graninger P., Busslinger M. (1994) EMBO J. 13, 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klemenz R., Hoffmann S., Werenskiold A. K. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5708–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tominaga S. (1989) FEBS Lett. 258, 301–304 [DOI] [PubMed] [Google Scholar]
- 27. Tominaga S., Jenkins N. A., Gilbert D. J., Copeland N. G., Tetsuka T. (1991) Biochim. Biophys. Acta 1090, 1–8 [DOI] [PubMed] [Google Scholar]
- 28. Tominaga S., Yokota T., Yanagisawa K., Tsukamoto T., Takagi T., Tetsuka T. (1992) Biochim. Biophys. Acta 1171, 215–218 [DOI] [PubMed] [Google Scholar]
- 29. Palmer G., Lipsky B. P., Smithgall M. D., Meininger D., Siu S., Talabot-Ayer D., Gabay C., Smith D. E. (2008) Cytokine 42, 358–364 [DOI] [PubMed] [Google Scholar]
- 30. Trajkovic V., Sweet M. J., Xu D. (2004) Cytokine Growth Factor Rev. 15, 87–95 [DOI] [PubMed] [Google Scholar]
- 31. Iikura M., Suto H., Kajiwara N., Oboki K., Ohno T., Okayama Y., Saito H., Galli S. J., Nakae S. (2007) Lab. Invest. 87, 971–978 [DOI] [PubMed] [Google Scholar]
- 32. Allakhverdi Z., Comeau M. R., Smith D. E., Toy D., Endam L. M., Desrosiers M., Liu Y. J., Howie K. J., Denburg J. A., Gauvreau G. M., Delespesse G. (2009) J. Allergy Clin. Immunol. 123, 472–478 [DOI] [PubMed] [Google Scholar]
- 33. Dinarello C. A. (2004) Curr. Opin. Pharmacol. 4, 378–385 [DOI] [PubMed] [Google Scholar]
- 34. Bresnihan B., Alvaro-Gracia J. M., Cobby M., Doherty M., Domljan Z., Emery P., Nuki G., Pavelka K., Rau R., Rozman B., Watt I., Williams B., Aitchison R., McCabe D., Musikic P. (1998) Arthritis Rheum. 41, 2196–2204 [DOI] [PubMed] [Google Scholar]
- 35. Haibel H., Rudwaleit M., Listing J., Sieper J. (2005) Ann. Rheum. Dis. 64, 296–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aksentijevich I., Nowak M., Mallah M., Chae J. J., Watford W. T., Hofmann S. R., Stein L., Russo R., Goldsmith D., Dent P., Rosenberg H. F., Austin F., Remmers E. F., Balow J. E., Jr., Rosenzweig S., Komarow H., Shoham N. G., Wood G., Jones J., Mangra N., Carrero H., Adams B. S., Moore T. L., Schikler K., Hoffman H., Lovell D. J., Lipnick R., Barron K., O'Shea J. J., Kastner D. L., Goldbach-Mansky R. (2002) Arthritis Rheum. 46, 3340–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hawkins P. N., Lachmann H. J., Aganna E., McDermott M. F. (2004) Arthritis Rheum. 50, 607–612 [DOI] [PubMed] [Google Scholar]
- 38. Arend W. P., Palmer G., Gabay C. (2008) Immunol. Rev. 223, 20–38 [DOI] [PubMed] [Google Scholar]
- 39. Dinarello C. A. (1998) Ann. N.Y. Acad. Sci. 856, 1–11 [DOI] [PubMed] [Google Scholar]
- 40. Cayrol C., Girard J. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9021–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lüthi A. U., Cullen S. P., McNeela E. A., Duriez P. J., Afonina I. S., Sheridan C., Brumatti G., Taylor R. C., Kersse K., Vandenabeele P., Lavelle E. C., Martin S. J. (2009) Immunity 31, 84–98 [DOI] [PubMed] [Google Scholar]
- 42. Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T. A., Soreq H. (2005) Gene 344, 1–20 [DOI] [PubMed] [Google Scholar]
- 43. Kriventseva E. V., Koch I., Apweiler R., Vingron M., Bork P., Gelfand M. S., Sunyaev S. (2003) Trends Genet. 19, 124–128 [DOI] [PubMed] [Google Scholar]
- 44. Philips A. V., Cooper T. A. (2000) Cell. Mol. Life Sci. 57, 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schröder M., Kaufman R. J. (2005) Mutat. Res. 569, 29–63 [DOI] [PubMed] [Google Scholar]
- 46. Ast G. (2004) Nat. Rev. Genet. 5, 773–782 [DOI] [PubMed] [Google Scholar]
- 47. Han S. Y., Kim S. H., Heasley L. E. (2002) J. Biol. Chem. 277, 47167–47174 [DOI] [PubMed] [Google Scholar]
- 48. Kim S. H., Eisenstein M., Reznikov L., Fantuzzi G., Novick D., Rubinstein M., Dinarello C. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim S. H., Han S. Y., Azam T., Yoon D. Y., Dinarello C. A. (2005) Immunity 22, 131–142 [DOI] [PubMed] [Google Scholar]
- 50. Lamkanfi M., Dixit V. M. (2009) Immunity 31, 5–7 [DOI] [PubMed] [Google Scholar]
- 51. Hayakawa M., Hayakawa H., Matsuyama Y., Tamemoto H., Okazaki H., Tominaga S. (2009) Biochem. Biophys. Res. Commun. 387, 218–222 [DOI] [PubMed] [Google Scholar]
- 52. Ohno T., Oboki K., Kajiwara N., Morii E., Aozasa K., Flavell R. A., Okumura K., Saito H., Nakae S. (2009) J. Immunol. 183, 7890–7897 [DOI] [PubMed] [Google Scholar]
- 53. Bufler P., Azam T., Gamboni-Robertson F., Reznikov L. L., Kumar S., Dinarello C. A., Kim S. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13723–13728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumar S., Hanning C. R., Brigham-Burke M. R., Rieman D. J., Lehr R., Khandekar S., Kirkpatrick R. B., Scott G. F., Lee J. C., Lynch F. J., Gao W., Gambotto A., Lotze M. T. (2002) Cytokine 18, 61–71 [DOI] [PubMed] [Google Scholar]
- 55. Meazza R., Verdiani S., Biassoni R., Coppolecchia M., Gaggero A., Orengo A. M., Colombo M. P., Azzarone B., Ferrini S. (1996) Oncogene 12, 2187–2192 [PubMed] [Google Scholar]
- 56. Nishimura H., Washizu J., Nakamura N., Enomoto A., Yoshikai Y. (1998) J. Immunol. 160, 936–942 [PubMed] [Google Scholar]
- 57. Pufe T., Petersen W., Tillmann B., Mentlein R. (2001) Arthritis Rheum. 44, 1082–1088 [DOI] [PubMed] [Google Scholar]
- 58. Tagaya Y., Kurys G., Thies T. A., Losi J. M., Azimi N., Hanover J. A., Bamford R. N., Waldmann T. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14444–14449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baggiolini M., Bretz U., Dewald B., Feigenson M. E. (1978) Agents Actions 8, 3–10 [DOI] [PubMed] [Google Scholar]
- 60. Padrines M., Wolf M., Walz A., Baggiolini M. (1994) FEBS Lett. 352, 231–235 [DOI] [PubMed] [Google Scholar]
- 61. Csernok E., Szymkowiak C. H., Mistry N., Daha M. R., Gross W. L., Kekow J. (1996) Clin. Exp. Immunol. 105, 104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robache-Gallea S., Morand V., Bruneau J. M., Schoot B., Tagat E., Réalo E., Chouaib S., Roman-Roman S. (1995) J. Biol. Chem. 270, 23688–23692 [DOI] [PubMed] [Google Scholar]
- 63. Coeshott C., Ohnemus C., Pilyavskaya A., Ross S., Wieczorek M., Kroona H., Leimer A. H., Cheronis J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6261–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sugawara S., Uehara A., Nochi T., Yamaguchi T., Ueda H., Sugiyama A., Hanzawa K., Kumagai K., Okamura H., Takada H. (2001) J. Immunol. 167, 6568–6575 [DOI] [PubMed] [Google Scholar]
- 65. Novick D., Rubinstein M., Azam T., Rabinkov A., Dinarello C. A., Kim S. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3316–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Novick D., Kim S. H., Fantuzzi G., Reznikov L. L., Dinarello C. A., Rubinstein M. (1999) Immunity 10, 127–136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.