We have identified an internal deletion mutation of NDT80 that can completely bypass the pachytene checkpoint, indicating that posttranslational control is the primary regulation for Ndt80. More importantly, we have shown that the pachytene checkpoint controls nuclear localization of Ndt80 in response to recombination or synapsis defects.
Abstract
In budding yeast, the Ndt80 protein is a meiosis-specific transcription factor that is essential for the exit of pachytene and progression into nuclear divisions and spore formation. The pachytene checkpoint responds to defects in meiotic recombination and chromosome synapsis and negatively regulates the activity of Ndt80. The activity of Ndt80 was suggested to be regulated at both transcriptional and posttranslational levels; however, the mechanism for posttranslational regulation of Ndt80 was unclear. From a study of ndt80 in-frame deletion mutations, we have identified a dominant mutation NDT80-bc, which is able to completely bypass the pachytene checkpoint. The NDT80-bc mutation relieves the checkpoint-mediated arrest of the zip1, dmc1, and hop2 mutants, producing spores with low viability. The NDT80-bc mutant provides direct evidence for the posttranslational control of Ndt80 activity. Furthermore, the data presented show that Ndt80 is retained in cytoplasm in the zip1 mutant, whereas Ndt80-bc is found in the nucleus. We propose that the nuclear localization of Ndt80 is regulated by the pachytene checkpoint through a cytoplasmic anchor mechanism.
INTRODUCTION
Progression of the eukaryotic cell division cycle is tightly coordinated through the surveillance mechanisms termed checkpoints to maintain the integrity of genetic information (Hartwell and Weinert, 1989). Checkpoints ensure the proper order of events in the cell cycle by preventing the initiation of late events until earlier events have been successfully completed. Checkpoints cause cells to arrest or delay in response to critical defects in cell-cycle events. Meiosis is a special type of cell division that produces haploid gametes from diploid parental cells. Compared to the mitotic cell cycle, additional meiosis-specific checkpoints are required to ensure the success of the more complicated process of meiosis. In particular, the pachytene checkpoint prevents exit from the pachytene stage of meiotic prophase when meiotic recombination and chromosome synapsis are incomplete (reviewed by Roeder and Bailis, 2000; Hochwagen and Amon, 2006). Recombination and synapsis are necessary for proper chromosome segregation at the first meiotic division. It would be detrimental to cells to enter meiosis I before finishing recombination. The pachytene checkpoint helps to ensure the production of viable haploid products.
In Saccharomyces cerevisiae, mutants that confer defects in recombination or synapsis, such as zip1, dmc1, and hop2, undergo arrest at the pachytene stage. The ZIP1 gene encodes a structural component of the synaptonemal complex; zip1 mutants arrest or delay in meiosis with unsynapsed chromosomes and unresolved Holliday junctions (Sym et al., 1993; Sym and Roeder, 1995; Storlazzi et al., 1996; Tung and Roeder, 1998). The DMC1 gene encodes a homolog of the bacterial RecA strand exchange enzyme; dmc1 mutants arrest or delay in meiosis with unrepaired double-strand breaks (Bishop et al., 1992; Rockmill et al., 1995). The Hop2 protein promotes homologous chromosome pairing in meiosis; hop2 mutants arrest at pachytene with extensive synaptonemal complex formation between nonhomologous chromosomes (Leu et al., 1998; Tsubouchi and Roeder, 2003). The arrest of these mutants is mediated through the pachytene checkpoint and is alleviated by mutations of genes required for the checkpoint (reviewed by Roeder and Bailis, 2000; Hochwagen and Amon, 2006). Several proteins involved in the DNA damage checkpoint that arrest vegetative cells also function in the pachytene checkpoint (Lydall et al., 1996; Hong and Roeder, 2002). The silencing proteins Sir2 and Dot1 are also required for the pachytene checkpoint (San-Segundo and Roeder, 1999, 2000). In addition, the pachytene checkpoint requires certain meiosis-specific proteins, such as Red1, Mek1, and Pch2 (Xu et al., 1997; San-Segundo and Roeder, 1999). Red1 is a component of meiotic chromosome axes (Smith and Roeder, 1997). Mek1 is a meiosis-specific protein kinase (Rockmill and Roeder, 1991). The Pch2 protein colocalizes with Sir2 in the nucleolus, and the nucleolar localization of Pch2 seems to be important for checkpoint function (San-Segundo and Roeder, 1999; Börner et al., 2008).
The pachytene checkpoint controls the progression from pachytene into meiosis I through both protein activation and gene expression. Two downstream targets of the pachytene checkpoint have been identified: Cdc28 and Ndt80. The Cdc28 and Ndt80 pathways cooperate to promote meiotic cell-cycle progression. Cdc28 is the major cyclin-dependent kinase in budding yeast, and its activity is required for both mitotic and meiotic cell-cycle progression (Shuster and Byers, 1989; Xu et al., 1997). Cdc28 activity is regulated by the pachytene checkpoint through two separate mechanisms: phosphorylation of Cdc28 and the availability of its activator for meiotic divisions, Clb1. The Swe1 protein kinase phosphorylates Cdc28 and thereby inactivates Cdc28 (Booher et al., 1993). The Swe1-mediated phosphorylation of Cdc28 is required for the pachytene arrest (Leu and Roeder, 1999). In addition, the activation of CLB1 expression is blocked by the pachytene checkpoint through the inactivation of Ndt80 (Chu and Herskowitz, 1998; Hepworth et al., 1998).
Ndt80 is a meiosis-specific protein, and its activity is required for the exit from pachytene (Xu et al., 1995). It binds to the middle sporulation elements (MSEs) and activates transcription of a large set of genes required for both meiotic nuclear divisions and spore formation, including most B-type cyclin genes and NDT80 itself (Chu and Herskowitz, 1998; Hepworth et al., 1998). The activity of Ndt80 was suggested to be regulated at both transcriptional and posttranslational levels. The pachytene checkpoint may repress the transcription of NDT80 through the transcription repressor Sum1 (Lindgren et al., 2000; Pak and Segall, 2002a). Sum1 binds to MSEs and represses the expression of NDT80 and other middle sporulation genes (MSGs) during vegetative growth (Xie et al., 1999). The level of Sum1 decreases transiently during meiosis, and the decrease is blocked in checkpoint-arrested cells (Lindgren et al., 2000). Mutations of SUM1 alleviate pachytene arrest in dmc1 cells (Lindgren et al., 2000; Pak and Segall, 2002b). All together, it was suggested that the pachytene checkpoint controls the stability of Sum1, and the progression of sporulation is controlled through the competition between Sum1 and Ndt80 for MSE binding (Lindgren et al., 2000; Pak and Segall, 2002b; Pierce et al., 2003). But the expression of MSGs and spore formation in dmc1 cells are not completely restored by the sum1 mutation (Lindgren et al., 2000; Pak and Segall, 2002a, 2002b; Pierce et al., 2003), indicating that transcriptional repression by Sum1 is not the sole regulatory mechanism on Ndt80. The Ndt80 protein itself also could be regulated by the pachytene checkpoint.
One likely mechanism for the regulation of Ndt80 activity is protein phosphorylation. Ndt80 is extensively phosphorylated during meiosis in wild type but not in cells arrested at the pachytene checkpoint, suggesting that phosphorylation of Ndt80 is critical for meiotic progression (Tung et al., 2000). The pachytene checkpoint may block the critical phosphorylation of Ndt80. More recent studies showed that the meiosis-specific kinase Ime2 may phosphorylate Ndt80 and that Ime2-dependent phosphorylation may be required for the maximal or specific activity of Ndt80 (Sopko et al., 2002; Benjamin et al., 2003; Shubassi et al., 2003). The exact function of phosphorylation on Ndt80 activity remains unclear.
It has been observed that overproduction of Ndt80 suppresses checkpoint-arrested mutants, raising the possibility that the pachytene checkpoint elevates the abundance or activity of an inhibitor to interact with Ndt80 and block its function (Tung et al., 2000). Excess Ndt80 protein resulting from overproduction might titrate out the inhibitor and allow the Ndt80 protein to be activated. To test this possibility, we constructed and analyzed a set of in-frame deletion mutations of NDT80. One of these deletion mutations, NDT80-bc, can bypass the pachytene checkpoint, supporting a direct protein inhibition mechanism by the pachytene checkpoint. In addition, we show that nuclear localization of Ndt80 is blocked by the pachytene checkpoint. We propose that the exclusion of Ndt80 from the nucleus is an essential piece of the regulatory mechanism, whereby the pachytene checkpoint controls progression of meiosis in budding yeast.
RESULTS
The NDT80Δ346–402 deletion mutation suppresses the zip1 sporulation defect
To define the functional domains of Ndt80, especially those responsible for the posttranslational regulation by the pachytene checkpoint, we constructed a set of ndt80 in-frame deletion mutations and tested them for their ability to suppress the zip1 mutant sporulation defect. These mutations were generated based on the availability of in-frame fusion sites or protein hydrophobicity plots. For the purpose of protein detection, these mutant alleles were tagged by the hemagglutinin (HA) epitope. These in-frame deletion mutations carried on high-copy plasmids were transformed into the ndt80 null mutant to determine whether they were functional. Also, they were transformed into the zip1 cells to assess their ability to suppress the sporulation defect when overexpressed. As expected, most deletions failed to sporulate, and those ndt80 alleles that retained the ability for sporulation also suppressed zip1 sporulation deficiency when overexpressed (Supplemental Figure S1). Among these deletion mutations, the NDT80Δ346–402 mutation, in which the amino acid residues from 346 to 402 are deleted, retained apparently full Ndt80 function. Interestingly, the NDT80Δ346–402 mutant suppressed the zip1 mutant better than the wild-type NDT80 did when overexpressed (sporulation frequency: 28%, compared to 19% for NDT80). We suspected that the NDT80Δ346–402 mutation might bypass the control of the pachytene checkpoint; therefore, we renamed the NDT80Δ346–402 allele as NDT80-bc (for bypassing checkpoint).
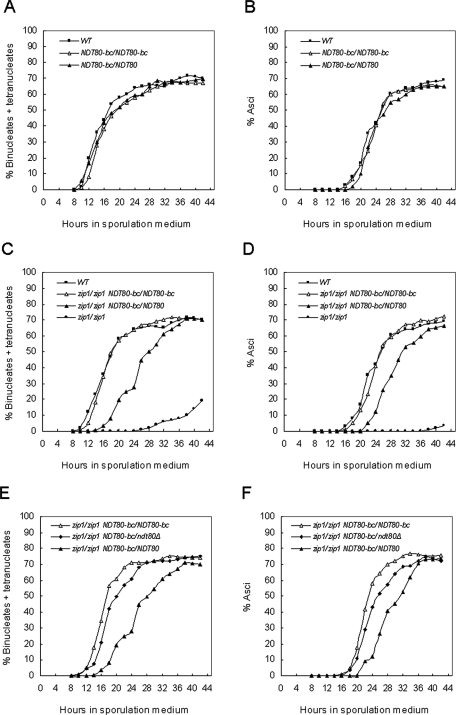
To determine whether the NDT80-bc allele required overexpression to suppress the zip1 sporulation defect, strains were constructed in which the chromosomal NDT80 loci were replaced with the NDT80-bc allele (without the HA tag). The NDT80-bc cells sporulated as well as the wild-type cells in both the BR2495 and SK1 strains (Table 1). The NDT80-bc mutation did not affect spore viability, recombination, or the kinetics of sporulation (Table 1; Figure 1, A and B), indicating that the Ndt80-bc protein is apparently fully functional. The NDT80-bc mutation, however, completely bypassed the checkpoint-mediated arrest of zip1 in both the BR2495 and SK1 backgrounds (Table 1). The kinetics of the nuclear divisions and asci formation in zip1 NDT80-bc cells were also equivalent to those of the wild-type cells (Figure 1, C and D). The zip1 pch2 cells were included in the experiment as a comparison for zip1 cells undergoing bypass of the pachytene checkpoint. Pch2 is required for the pachytene checkpoint–mediated arrest of zip1 (San-Segundo and Roeder, 1999). The zip1 NDT80-bc cells were comparable to the zip1 pch2 cells for sporulation (Table 1). The zip1 defects in spore viability and recombination in the SK1 background were not suppressed by the NDT80-bc mutation (Table 1). The map distances for zip1 NDT80-bc appeared to be greater than those from zip1 NDT80, but the difference is not statistically significant. The NDT80/NDT80-bc heterozygote was capable of suppressing the zip1 mutation (Table 1), indicating that the NDT80-bc mutation is dominant to the wild-type NDT80. These observations support our hypothesis that NDT80-bc bypasses the control of the pachytene checkpoint.
TABLE 1:
Sporulation, spore viability, and crossing over in zip1 NDT80-bc cells.
| Strains | BR2495 background | SK1 background | ||||
|---|---|---|---|---|---|---|
| Sporulation efficiency, % | Spore viability, % | Sporulation efficiency, % | Spore viability, % | Crossing over, cM | ||
| MAT/CEN3 | CEN3/HIS4 | |||||
| Wild type | 64.0 | 92.2 | 97.5 | 90.0 | 14.5 | 33.3 |
| NDT80-bc/NDT80-bc | 61.0 | 94.8 | 97.0 | 92.2 | 11.2 | 30.8 |
| NDT80/NDT80-bc | 66.0 | 96.0 | — | — | — | — |
| zip1/zip1 | 0 | — | 52.0 | 51.5 | 4.8 | 2.7 |
| zip1/zip1 NDT80-bc/NDT80-bc | 65.0 | 56.6 | 94.5 | 41.8 | 7.6 | 5.0 |
| zip1/zip1 NDT80/NDT80-bc | 60.5 | 64.0 | — | — | — | — |
| zip1/zip1 pch2/pch2 | 64.5 | 47.0 | — | — | — | — |
Sporulation frequencies represent the averages of two independent cultures for each strain, with 200 cells counted for each culture. For BR2495 spore viability data, at least 44 tetrads were dissected for each strain. For spore viability and crossover frequencies in SK1 background strains, at least 119 tetrads were dissected for each strain.
FIGURE 1:
NDT80-bc suppresses the zip1 sporulation defect. Nuclear divisions and spore formation were monitored and compared among wild type, NDT80-bc/NDT80-bc, and NDT80-bc/NDT80 (A and B); wild type, zip1, zip1 NDT80-bc/NDT80-bc, and zip1 NDT80-bc/NDT80 (C and D); zip1 NDT80-bc/NDT80-bc, zip1 NDT80-bc/ndt80Δ, and zip1 NDT80-bc/NDT80 (E and F). These experiments were repeated with similar results.
The NDT80-bc mutant suppresses the dmc1 and hop2 mutants for sporulation
If the NDT80-bc mutant provides a full bypass of the pachytene checkpoint, then the suppression should not be limited to zip1 mutants. To determine whether the suppression by NDT80-bc is specific to the zip1 mutation or is more general, NDT80-bc was examined in dmc1 and hop2 cells. Both mutants undergo checkpoint-mediated arrest at the pachytene stage of meiosis (Bishop et al., 1992; Rockmill et al., 1995; Leu et al., 1998). In the BR2495 strain background, the dmc1 mutant sporulated at low levels and the hop2 mutant did not sporulate. Similar to the suppression in zip1 cells, both the dmc1 NDT80-bc and the hop2 NDT80-bc cells sporulated to the level of the wild type (Table 2), although nuclear divisions were delayed in the hop2 NDT80-bc cells, and spore formation was delayed in both the dmc1 NDT80-bc and the hop2 NDT80-bc cells (Supplemental Figure S2). These results are consistent with previous observations suggesting that the meiotic defect in hop2 is more severe than in zip1 (Leu et al., 1998). There were only occasional viable spores generated from the dmc1 NDT80-bc and the hop2 NDT80-bc tetrads (Table 2), suggesting that the suppression by NDT80-bc was due to a direct bypass of the checkpoint without repairing the recombination defects. An analysis of Rad51 foci confirmed that the double-strand breaks remained unrepaired in the dmc1 NDT80-bc and the hop2 NDT80-bc, but not in the wild-type cells after the pachytene stage (Supplemental Figure S3).
TABLE 2:
Sporulation and spore viability in dmc1 NDT80-bc and hop2 NDT80-bc cells.
| Strains | Sporulation efficiency, % | Spore viability, % |
|---|---|---|
| Wild type (BR2495) | 53 | 87.8 |
| dmc1/dmc1 | 8 | — |
| dmc1/dmc1 NDT80-bc/NDT80-bc | 55.5 | 1.9 |
| hop2/hop2 | 0 | — |
| hop2/hop2 NDT80-bc/NDT80-bc | 54 | 0 |
Sporulation frequencies represent the averages of two independent cultures for each strain, with 200 cells counted for each culture. For spore viability data, at least 44 tetrads were dissected for each strain.
The wild-type Ndt80 interferes with the ability of Ndt80-bc to suppress checkpoint arrest
The suppression ability of the NDT80-bc deletion mutation provides strong evidence for the posttranslational regulation of Ndt80. In zip1 cells, the Ndt80 activity is inhibited by the checkpoint; in contrast, the deletion in Ndt80-bc allows checkpoint bypass. Although the NDT80-bc allele is dominant, it is possible that the presence of wild-type Ndt80 protein could influence the suppression efficiency of Ndt80-bc. Examination of zip1/zip1 NDT80/NDT80-bc and zip1/zip1 NDT80-bc/NDT80-bc strains revealed no significant difference in the final sporulation frequencies (Table 1). The zip1/zip1 NDT80/NDT80-bc heterozygote, however, displayed a delay in nuclear divisions and asci formation, compared to the zip1/zip1 NDT80-bc/NDT80-bc homozygote and the wild type (Figure 1, C and D). This delay in meiotic progression did not occur in the ZIP1/ZIP1 NDT80/NDT80-bc strain (Figure 1, A and B), indicating that the delay is checkpoint dependent.
One possible cause for this delay in the zip1/zip1 NDT80/NDT80-bc heterozygote is that wild-type Ndt80 might interfere with the processing or functioning of Ndt80-bc. Alternatively, the delay could be due to a dosage effect of the Ndt80-bc. To distinguish between these two possibilities, we constructed the isogenic zip1/zip1 ndt80Δ/NDT80-bc strain, which contains only one copy of NDT80-bc, but no NDT80. The kinetics of meiosis in the zip1/zip1 ndt80Δ/NDT80-bc was faster than that in the zip1/zip1 NDT80/NDT80-bc heterozygote, and was similar to the zip1/zip1 NDT80-bc/NDT80-bc homozygote (Figure 1, E and F). The deletion of NDT80 from the zip1/zip1 NDT80/NDT80-bc cells alleviated the delay in meiosis, indicating that the delay is caused by the presence of Ndt80. The result is consistent with the possibility that the Ndt80 protein interferes with Ndt80-bc processing or functioning in zip1 mutants. The coexistence of checkpoint-responsive Ndt80 and checkpoint-bypassing Ndt80-bc provides a good tool for the study of regulatory mechanism.
The expression patterns of Ndt80 and Ndt80-bc are similar in zip1/zip1 NDT80/NDT80-bc cells
The results just described suggest that the posttranslational control of Ndt80 plays a critical role in regulating meiotic progression, yet the expression of Ndt80 is also regulated (Chu and Herskowitz, 1998; Hepworth et al., 1998). The posttranslational regulation of Ndt80 activity has been difficult to study due to low levels of accumulated protein in pachytene-arrested mutants (Tung et al., 2000). The NDT80-bc mutant, which bypasses the pachytene checkpoint, should induce high levels of Ndt80 in the zip1/zip1 NDT80/NDT80-bc cells, providing an opportunity to study the posttranslational control of Ndt80.
We first checked the expression of Ndt80 and Ndt80-bc in the zip1/zip1 NDT80/NDT80-bc heterozygote. Ndt80 and Ndt80-bc proteins were differentially tagged (NDT80–6XHA and NDT80-bc-6XMYC) to detect and distinguish them at the same time in the same cells. The NDT80-HA allele is functional and has been used in our previous study (Tung et al., 2000). The NDT80-bc-MYC allele is also fully functional in sporulation and in suppression of zip1.
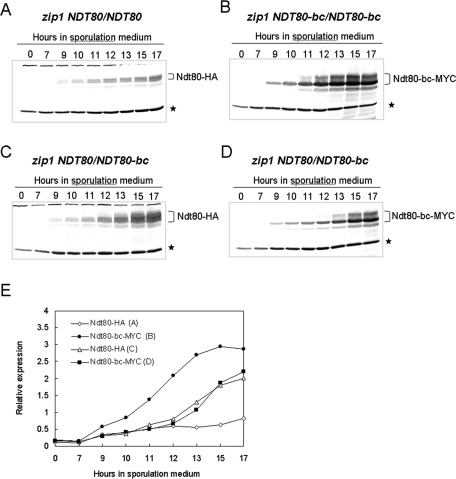
The accumulation of Ndt80 and Ndt80-bc in the zip1/zip1 NDT80/NDT80, zip1/zip1 NDT80-bc/NDT80-bc, and zip1/zip1 NDT80/NDT80-bc cells were monitored throughout meiosis by Western blot analysis. As reported previously, the induction of Ndt80 was inhibited in the zip1/zip1 NDT80/NDT80 cells (Tung et al., 2000; Figure 2A). In contrast, Ndt80-bc rapidly accumulated in the zip1/zip1 NDT80-bc/NDT80-bc cells (Figure 2B). As expected, in the zip1/zip1 NDT80/NDT80-bc cells, the induction pattern of Ndt80 appeared to be similar to that of Ndt80-bc (Figure 2, C and D). The kinetics of protein accumulation were equivalent for Ndt80 and Ndt80-bc in the zip1/zip1 NDT80/NDT80-bc cells (Figure 2E), making it possible to compare the posttranslational differences between Ndt80 and Ndt80-bc without bias from differential expression.
FIGURE 2:
The expressions of Ndt80 and Ndt80-bc are similar in zip1 cells. Production of Ndt80 in zip1 (A) and zip1 NDT80/NDT80-bc (C) cells were monitored through meiosis by Western blot analysis with anti-HA antibodies. Production of Ndt80-bc in zip1 NDT80-bc/NDT80-bc (B) and zip1 NDT80/NDT80-bc (D) cells were analyzed through meiosis with anti-myc antibodies. Tubulin was also analyzed and used as a loading control on each blot (*). The ratios in expression of Ndt80 or Ndt80-bc to tubulin are plotted (E).
Consistent with the delay in progression of meiosis, the accumulation of Ndt80-bc was delayed in the zip1/zip1 NDT80/NDT80-bc cells, compared to the isogenic zip1/zip1 NDT80-bc/NDT80-bc homozygote (Figure 2, B, D, and E). Thus the delay in meiotic progression is likely due to a delay in reaching a threshold amount of the Ndt80-bc protein.
Nuclear localization of Ndt80, but not Ndt80-bc, is inhibited in zip1 cells
Ndt80 is a transcription factor and, as such, is localized to chromosomes in the nucleus. In the zip1/zip1 NDT80/NDT80 cells, we did not detect signals of Ndt80 concentrated in nuclei (Supplemental Figure S4). Because of the low amount of Ndt80 expression (Figure 2A) and the weak immunofluorescence signal, we could not make an unambiguous conclusion. With the increased accumulation of Ndt80 in zip1/zip1 NDT80/NDT80-bc cells, we were able to analyze nuclear localization of Ndt80 more efficiently and to compare localization of Ndt80 with that of Ndt80-bc in the same cells.
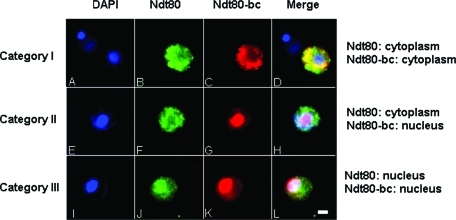
A ZIP1/ZIP1 NDT80/NDT80-bc strain and the zip1/zip1 NDT80/NDT80-bc strain used in the Western blot analysis were examined for subcellular localization of Ndt80 and Ndt80-bc before meiosis I. To avoid any bias due to the failure of staining and for the purpose of comparison between Ndt80 and Ndt80-bc, only cells with both signals were scored. Three categories of differential localization of Ndt80 and Ndt80-bc within a single cell were observed in the analysis. In category I, both the Ndt80 signal and Ndt80-bc signal were in the cytoplasm (Figure 3, A–D). In category II, the Ndt80 signal was in the cytoplasm, and the Ndt80-bc signal was concentrated in the nucleus (Figure 3, E–H). In category III, both the Ndt80 signal and Ndt80-bc signal were concentrated in the nucleus (Figure 3, I–L). Cells with nuclear Ndt80 signal and cytoplasmic Ndt80-bc signal had never been observed.
FIGURE 3:
Differential localizations of Ndt80 and Ndt80-bc. NDT80-HA/NDT80-bc-myc cells were stained with DAPI (A, E, and I), anti-HA antibodies (B, F, and J), and anti-myc antibodies (C, G, and K), to detect nuclei, Ndt80, and Ndt80-bc, respectively. Merged images are shown in (D), (H), and (L). Three categories of differential localization of Ndt80 and Ndt80-bc (A–D, E–H, and I-L) were as indicated and described in Results. Scale bar, 2 μm.
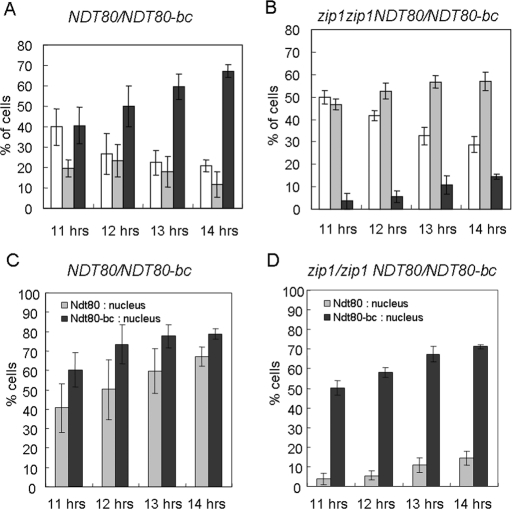
In the control ZIP1/ZIP1 NDT80/NDT80-bc strain, most cells examined had both Ndt80 and Ndt80-bc localized to the nucleus, and the proportion was increased with the progression of meiosis (Figure 4A). The proportion of cells containing cytoplasmic Ndt80 and Ndt80-bc gradually decreased as meiosis progressed (Figure 4A). The fraction of cells in which Ndt80 and Ndt80-bc had different localization patterns was small (Figure 4A). The sum totals of nuclear localization of Ndt80 and Ndt80-bc were also separately calculated and plotted for each time point. Both Ndt80 and Ndt80-bc were localized in the nucleus in most meiotic cells (Figure 4C). In contrast, in the zip1/zip1 NDT80/NDT80-bc strain, most cells had Ndt80-bc localized to the nucleus, but Ndt80 remained in cytoplasm (Figure 4B). The sum total of Ndt80 nuclear localization was significantly less than that of the Ndt80-bc in the zip1/zip1 NDT80/NDT80-bc cells (Figure 4D). These results indicated that the nuclear localization of Ndt80, but not Ndt80-bc, was severely affected by the zip1 mutation. Nuclear localization of Ndt80-bc was slightly delayed in zip1/zip1 NDT80/NDT80-bc cells, compared to that in ZIP1/ZIP1 NDT80/NDT80-bc or in zip1/zip1 NDT80-bc/NDT80-bc cells (Figure 4, C and D; Supplemental Figure S5).
FIGURE 4:
Quantification of subcellular localizations of Ndt80 and Ndt80-bc. The subcellular localizations of Ndt80 and Ndt80-bc in NDT80-HA/NDT80-bc-myc cells (A) and zip1/zip1 NDT80-HA/NDT80-bc-myc cells (B) were examined by anti-HA and anti-myc antibodies at indicated time points after inoculation into sporulation medium. Only mononucleated cells with both HA and myc signals were scored and classified into three categories as shown in Figure 3: I. both Ndt80 and Ndt80-bc are in the cytoplasm (white bars). II. Ndt80 is in cytoplasm, but Ndt80-bc is in the nucleus (gray bars). III. both Ndt80 and Ndt80-bc are in the nucleus (black bars). The data shown in (A) and (B) are reorganized to present the individual fractions of nuclear localization for Ndt80 (gray bars) and Ndt80-bc (black bars) in NDT80-HA/NDT80-bc-myc (C) and in zip1/zip1 NDT80-HA/NDT80-bc-myc (D) cells. Averages of five repeats for each strain at each time point are presented. A total of at least 249 cells were scored at each time point for each strain.
To determine whether the nuclear localization defect of Ndt80 is specific to the zip1 mutation or it represents a more general regulatory mechanism by the pachytene checkpoint, nuclear localization of Ndt80 was examined in zip1/zip1 pch2/pch2 NDT80/NDT80-bc and hop2/hop2 NDT80/NDT80-bc cells. The pch2 mutation restored the nuclear localization of Ndt80 in zip1 cells to the level of the NDT80/NDT80-bc cells, and the hop2 mutation inhibited Ndt80 nuclear localization, similar to the zip1 mutation (Supplemental Figure S6). These results supported that nuclear localization of Ndt80 is controlled by the pachytene checkpoint.
DISCUSSION
Ndt80 protein is a target of the pachytene checkpoint
Our results show that the NDT80-bc mutation bypasses the checkpoint-mediated arrest of the zip1, dmc1, and hop2 mutants. Because the promoter or any known regulatory region for NDT80 transcription is not altered in the NDT80-bc allele, the mechanism of suppression is unlikely due to a failure of transcriptional repression by the pachytene checkpoint. The differential localization of Ndt80 and Ndt80-bc in the zip1 cells further indicates that the suppression is due to the insensitivity of Ndt80-bc protein itself to the control of the pachytene checkpoint. These observations point to a mechanism for the posttranslational control of Ndt80 activity by the pachytene checkpoint.
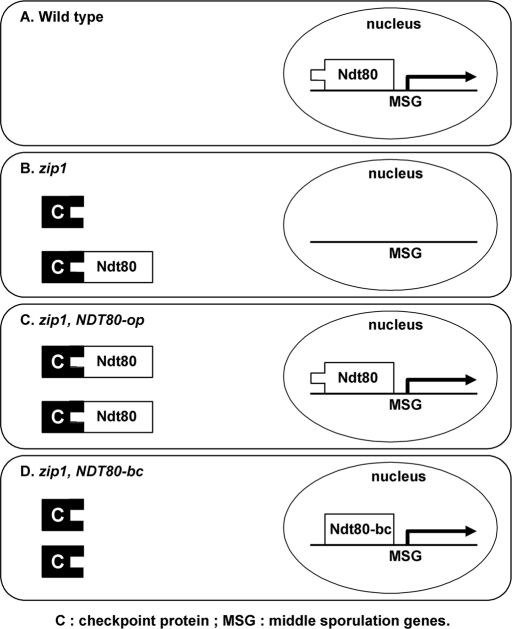
The DNA-binding domain of Ndt80 is defined in the N terminus (residues1∼340; Lamoureux et al., 2002; Montano et al., 2002), and the transcriptional activation domain is located at the C terminus (residues 426∼627; Sopko et al., 2002). The 57 amino acids (residues 346∼402) deleted in the Ndt80-bc is neither in the DNA-binding domain nor in the activation domain. The Ndt80-bc protein functions as well as Ndt80, except that it is no longer regulated by the pachytene checkpoint. The most economical explanation for the loss of regulation is that the deleted region overlaps the target domain for checkpoint control. We propose that, when the pachytene checkpoint is triggered, an unidentified checkpoint protein physically interacts with Ndt80 at this target domain to inhibit its function (Figure 5, A and B). Without this target domain, Ndt80-bc is functional in zip1, dmc1, and hop2 cells (Figure 5D). The observation that the NDT80-bc allele is dominant to the wild-type NDT80 is consistent with the proposed model. Furthermore, it has been shown that overproduction of Ndt80 suppresses checkpoint-arrested mutants (Tung et al., 2000). Our interpretation is that overproduction of Ndt80 would titrate out the unidentified inhibitor, thus allowing some of the Ndt80 to perform its function (Figure 5C).
FIGURE 5:
Model of posttranslational regulation of Ndt80 activity. (A) In wild-type cells, Ndt80 protein is able to activate the expression of middle sporulation genes (MSGs) after meiotic recombination has been completed. (B) In zip1 cells, Ndt80 is directly targeted by the pachytene checkpoint and unable to activate transcription of MSG. (C) When Ndt80 is overproduced in zip1 cells, some Ndt80 might escape the control of checkpoint protein and induce the expression of MSG. (D) In zip1 NDT80-bc cells, the checkpoint cannot target the Ndt80-bc, and the transcription of MSG is induced.
Nuclear localization of Ndt80 is controlled by the pachytene checkpoint
The observation that the progression of meiosis is delayed in the zip1/zip1 NDT80/NDT80-bc cells, but not in the ZIP1/ZIP1 NDT80/NDT80-bc or zip1/zip1 ndt80Δ/NDT80-bc cells, indicates that the delay is both checkpoint- and Ndt80-dependent. This result suggests that the checkpoint-inhibited Ndt80 and checkpoint-insensitive Ndt80-bc coexist in the zip1/zip1 NDT80/NDT80-bc cells for a certain period of time before nuclear divisions. Our results show that although expression patterns are similar for Ndt80 and Ndt80-bc in the zip1/zip1 NDT80/NDT80-bc cells, their subcellular localizations are different. In the zip1/zip1 NDT80/NDT80-bc cells, the nuclear localization of Ndt80, but not Ndt80-bc, is blocked. We propose that nuclear import of Ndt80 is regulated by the pachytene checkpoint. In those recombination-defective cells, the Ndt80 protein is retained in the cytoplasm by an unidentified checkpoint protein, preventing Ndt80 from activating transcription of its target genes, and cells arrest at the pachytene stage (Figure 5). To identify this checkpoint, anchor protein(s) will be the most important task for further understanding the pachytene-checkpoint machinery.
There are other examples in which the nuclear localization of a transcription factor is controlled by cytoplasmic sequestration, such as p53 and nuclear factor-κB (NF-κB) (Hunt, 1989; Calkhoven and Ab, 1996; Vandromme et al., 1996; Hood and Silver, 1999; Komeili and O'Shea, 2000; Liang and Clarke, 2001; Li and Verma, 2002). The localization of the p53 tumor suppressor is tightly regulated through multiple mechanisms, including the nuclear import and export signals, oligomerization, Mdm2-mediated export, and phosphorylation (Komeili and O'Shea, 2000; Liang and Clarke, 2001). In addition, it was suggested that a hsp70 family member, mot-2, could interact with p53 and inhibit its nuclear import (Wadhwa et al., 1998). The NF-κB of the Rel-related transcription factor is a well-studied example of cytoplasmic sequestration (Calkhoven and Ab, 1996; Vandromme et al., 1996; Hood and Silver, 1999; Li and Verma, 2002). NF-κB is constitutively expressed, but in uninduced cells it is sequestered in the cytoplasm with the retention protein IκB (Baeuerle and Baltimore, 1988). Cytokine stimulation leads to phosphorylation and degradation of IκB, and then the NF-κB is imported into the nucleus (Beg et al., 1993).
The structure of the Ndt80 DNA-binding domain reveals that Ndt80 is a member of the immunoglobulin (Ig)-fold family of transcription factors (Lamoureux et al., 2002; Fingerman et al., 2004). It is interesting that both p53 and NF-κB belong to the Ig-fold family (Rudolph and Gergen, 2001).
Competition between Ndt80 and Ndt80-bc in zip1 cells
Because Ndt80 functions as a monomer in activating transcription (Lamoureux et al., 2002; Montano et al., 2002), the delay in meiotic progression of the zip1/zip1 NDT80/NDT80-bc cells is unlikely due to the formation of nonfunctional heterodimers. Alternatively, the Ndt80 might interfere with Ndt80-bc by competing for a processing step that is necessary for Ndt80 or Ndt80-bc to function. The difference in nuclear localization between Ndt80 and Ndt80-bc indicates that the competition step is probably at or before nuclear import. Consistent with this idea, nuclear localization of Ndt80-bc is retarded in zip1/zip1 NDT80/NDT80-bc compared to ZIP1/ZIP1 NDT80/NDT80-bc or zip1/zip1 NDT80-bc/NDT80-bc. The nuclear localization of Ndt80-bc appears to be affected in the presence of Ndt80. One possibility is that both the Ndt80 and Ndt80-bc can bind to the nuclear import receptor; however, the unproductive binding of Ndt80 occupies the nuclear import machinery, thus impairing transport of Ndt80-bc. Alternatively, the competition might occur before the binding to the nuclear import machinery.
Role of phosphorylation in the regulation of Ndt80 activity
There are multiple steps in regulation of transcription factors (Calkhoven and Ab, 1996). Multiple regulation mechanisms could also be applied to a single protein, for example, the transcription factor Pho4. Similar to Ndt80, Pho4 contains multiple phosphorylation sites. These multiple phosphorylation sites play distinct and separable roles in regulating Pho4 activity (Komeili and O'Shea, 1999). Phosphorylation of Ndt80 has been correlated to its activation (Tung et al., 2000; Sopko et al., 2002; Shubassi et al., 2003), but the role of phosphorylation has not been identified. We found that both the Ndt80 and Ndt80-bc are phosphorylated in the wild type and the zip1/zip1 NDT80/NDT80-bc cells. We have noticed that the amounts of unphosphorylated and phosphorylated Ndt80 appear to be equivalent in these cells. Interestingly, in the case of Ndt80-bc, the amounts of phosphorylated forms are significantly smaller than that of the unphosphorylated form. The reason for the difference in the extent of phosphorylation between Ndt80 and Ndt80-bc is not clear.
Although our results indicate that the Ndt80-bc protein bypasses the nuclear import block of the pachytene checkpoint, we have not ruled out the possibility that there are other checkpoint mechanisms for Ndt80 regulation in addition to nuclear import. For example, the binding of Ndt80 to MSEs and its interaction with other transcription factors could also be regulated. It would be unusual if all these regulatory mechanisms target the same region that is deleted in Ndt80-bc. A more detailed study in the checkpoint target domain of Ndt80 should clarify these possibilities.
MATERIALS AND METHODS
Yeast strains and plasmids
Most experiments were carried out in isogenic derivatives of BR2495 (Rockmill and Roeder 1990), which has the following genotype:
MATa/MATα leu2–27/leu2–3,112 his4–280/his4–260 arg4–8/ARG4 thr1–1/thr1–4 ade2–1/ade2–1 ura3–1/ura3–1 trp1–1/trp1–289 cyh10/CYH10
The SK1 strains used for tetrad analysis are isogenic derivatives of MY261 (Sym and Roeder, 1994), which has the following genotype:
MATa/MATα CENIII::URA3/CENIII::TRP1 leu2::hisG/leu2::hisG HIS4/his4-B-LEU2 lys2/lys2 ho::LYS2/ho::lys2 trp1-H3/trp1-H3 ura3/ura3
Yeast manipulations were performed and media were prepared using standard procedures (Sherman et al., 1986). Cells were grown and induced for meiosis at 30°C. All the strains were constructed by substitutive transformations (Rothstein, 1991). Yeast transformations were carried out by using the lithium acetate method (Ito et al., 1983).
Plasmids for disruption of ZIP1 (Sym et al., 1993; Sym and Roeder, 1994, 1995), PCH2 (San-Segundo and Roeder, 1999), DMC1 (Bishop et al., 1992), and HOP2 (Leu et al., 1998) have been described.
The NDT80-Δ346–402-HA (NDT80-bc-HA) deletion was first derived from the plasmid TP124, which contains the NDT80-HA allele (Tung et al., 2000) on the YEp351 vector (Hill et al., 1986). The deletion was constructed by a three-fragment ligation strategy as follows: For fragment 1, a 1254 bp fragment containing the region from position –199 to +1055 of the NDT80 open reading frame (ORF) was obtained by PCR using primers NDT80-XhoI (5′-GATTCTCAAATATCTCGAGGCCTGT-3′) and NDT80–346D (5′-GTGCTGTTTTGTGAAGCTTTGACACTCGACGGTGTTC-3′). The NDT80–346D primer introduced a new HindIII site in the PCR product at position +1037 of the NDT80 ORF. The PCR product was cut with XhoI and HindIII to release a 1223 bp XhoI-HindIII fragment containing the coding sequence for the N-terminal 345 amino acid of Ndt80. For fragment 2, a 967 bp HindIII-SacI fragment encoding the C-terminal portion of Ndt80-HA from residue 403 was released by cutting the YEp351-NDT80-HA plasmid with HindIII and SacI. For fragment 3, a 6.4 kb XhoI-SacI fragment containing the upstream region and the YEp351 portion was obtained by cutting the YEp351-NDT80-HA plasmid with XhoI and SacI. These three fragments were ligated together to get the plasmid T324 which contains the NDT80-bc-HA allele on the YEp351 vector. A 3.0 kb SalI-SacI fragment containing the NDT80-bc-HA mutation was subcloned into the pRS306 vector (Sikorski and Hieter, 1989) to get the plasmid T330. T330 was used to replace the chromosomal copy of NDT80 with NDT80-bc by two-step transplacement. T330 was targeted to chromosome by cutting with SpeI for the purpose of substitution.
To construct the NDT80-bc-myc allele, the HA coding sequence on T324 was removed by cutting with NotI, and two copies of a 120 bp NotI-NotI fragment encoding three copies of the c-myc epitope was inserted into the NotI site to create plasmid T455. A 0.9 kb HindIII-EcoRI fragment including the epitope-coding sequence from T455 was inserted into the HindIII-EcoRI sites of the plasmid TP121 (Tung et al., 2000) to get plasmid T456. T456 was used to replace the chromosomal copy of NDT80-bc with NDT80-bc-myc, targeting for integration by cutting with BglII.
Cytology
At the indicated time points, meiotic cells were collected and fixed in 3.7% formaldehyde at room temperature for 20 min and washed three times with 1.2 M sorbitol, 50 mM potassium phosphate buffer, pH 7.0 (solution A). Fixed cells were incubated in solution A containing 0.1% β-mercaptoethanol, 0.02% glusulase, and 10 mg/ml Zymolyase 100T at 37°C for 60 min to prepare spheroplasts, which were washed twice with solution A and settled onto poly-l-lysine–coated slides. They were washed with phosphate-buffered saline (PBS)+1% bovine serum albumin (BSA) three times. For the detection of Ndt80-bc-myc and Ndt80-HA proteins, cells were incubated with mouse anti-myc (9E10; Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-HA (16B12; Covance, Berkeley, CA) antibodies at 1:150 (vol/vol) dilution at 4°C overnight. After incubation of the primary antibodies, cells were washed once with PBS and three times with PBS+1% BSA. Secondary antibody incubations were performed at room temperature for 2 h using Texas Red–conjugated donkey anti–mouse and fluorescein isothiocyanate–conjugated goat anti–rabbit antibodies (Jackson ImmunoResearch, West Grove, PA) at a 1:200 (vol/vol) dilution. After the incubation with the secondary antibodies, cells were washed once with PBS, twice with PBS+1% BSA, and then once with PBS. Samples were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) containing 1 μg/ml 4’,6-diamidino-2-phenylindole (DAPI). An Olympus AX70 fluorescence microscope (Olympus, Tokyo, Japan) and an Uplan Apo100× objective (Olympus) were used to observe antibody-stained preparations. The images were captured using a PXL CCD camera (Photometrics, Tucson, AZ) and processed with the V for Windows software (Digital Optics, Auckland, New Zealand). For quantification of subcellular localization of Ndt80-HA and Ndt80-bc-myc, only mononucleated cells with both signals of Ndt80-HA and Ndt80-bc-myc were counted.
Western blot analysis
Meiotic cell protein extraction, electrophoresis, and blotting procedures were performed as described previously (Tung et al., 2000). Proteins were probed with either 1) mouse anti-HA antibody 16B12 (Covance) at a 1:1000 (vol/vol) dilution to detect Ndt80-HA or 2) mouse anti-myc antibody 9E10 (Santa Cruz) at a 1:1000 (vol/vol) dilution to detect Ndt80-bc-myc. Rat anti-tubulin antibody YOL1/34 (Harlan Laboratories, Hillcrest, UK) was used to detect tubulin as a loading control. Primary antibodies were detected by using goat anti–mouse or donkey anti–rat alkaline phosphatase–conjugated antibodies (Jackson ImmunoResearch) at a 1:2000 (vol/vol) dilution followed by 5-bromo-4-chloro-3’-indolyphosphate and nitro-blue tetrazolium colorimetric detection. The kinetics of Ndt80 and Ndt80-bc accumulation during meiosis were analyzed with Image-Pro Plus software (Media Cybernetics, Silver Spring, MD).
Supplementary Material
Acknowledgments
We are grateful to Beth Rockmill and Jun-Yi Leu for helpful comments on the manuscript. This work was supported by National Science Council grants NSC92–2311-B002–102 and NSC93–2311-B002–028, Taiwan, Republic of China.
Abbreviations used:
- BSA
bovine serum albumin
- DAPI
4’,6-diamidino-2-phenylindole
- HA
hemagglutinin
- Ig
immunoglobulin
- MSE
middle sporulation element
- MSG
middle sporulation gene
- NF-κB
nuclear factor-κB
- ORF
open reading frame
- PBS
phosphate-buffered saline
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-1011) on April 6, 2011.
REFERENCES
- Baeuerle P, Baltimore D. IκB: a specific inhibitor of the NFκB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Beg AA, Finco TS, Nantermet PV, Baldwin AS., Jr Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism for NF-κB activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Barot A, Kleckner N. Yeast Pch2 promotes dominal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc Natl Acad Sci USA. 2008;105:3327–3332. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Ab G. Multiple steps in the regulation of transcription-factor level and activity. Biochem J. 1996;317:329–342. doi: 10.1042/bj3170329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Fingerman IM, Sutphen K, Montano SP, Georgiadis MM, Vershon AK. Characterization of critical interactions between Ndt80 and MSE DNA defining a novel family of Ig-fold transcription factors. Nucleic Acids Res. 2004;32:2947–2956. doi: 10.1093/nar/gkh625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Amon A. Checking your breaks: Surveillance mechanisms of meiotic recombination. Curr Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hong EJE, Roeder GS. A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev. 2002;16:363–376. doi: 10.1101/gad.938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JK, Silver PA. In or out? regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- Hunt T. Cytoplasmic anchoring proteins and the control of nuclear localization. Cell. 1989;59:949–951. doi: 10.1016/0092-8674(89)90747-2. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Nuclear transport and transcription. Curr Opin Cell Biol. 2000;12:355–360. doi: 10.1016/s0955-0674(00)00100-9. [DOI] [PubMed] [Google Scholar]
- Lamoureux JS, Stuart D, Tsang R, Wu C, Glover JNM. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 2002;21:5721–5732. doi: 10.1093/emboj/cdf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J-Y, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- Leu J-Y, Roeder GS. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol Cell. 1999;4:805–814. doi: 10.1016/s1097-2765(00)80390-1. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-κB regulation in the immune system. Nature Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Liang S-H, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–2783. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- Lindgren A, Bungard D, Pierce M, Xie J, Vershon A, Winter E. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcription repressor. EMBO J. 2000;19:6489–6497. doi: 10.1093/emboj/19.23.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Montano SP, Coté ML, Fingerman I, Pierce M, Vershon AK, Georgiadis MM. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc Natl Acad Sci USA. 2002;99:14041–14046. doi: 10.1073/pnas.222312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Segall J. Regulation of the premiddle and middle phase of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol Cell Biol. 2002a;22:6417–6429. doi: 10.1128/MCB.22.18.6417-6429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Segall J. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol Cell Biol. 2002b;22:6430–6440. doi: 10.1128/MCB.22.18.6430-6440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol Cell Biol. 2003;23:4814–4825. doi: 10.1128/MCB.23.14.4814-4825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H, Roeder GS. Roles of two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rudolph MJ, Gergen JP. DNA-binding by Ig-fold proteins. Nature Struct Biol. 2001;8:384–386. doi: 10.1038/87531. [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY:: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Shubassi G, Luca N, Pak J, Segall J. Activity of phosphoforms and truncated version of Ndt80, a checkpoint-regulated sporulation-specific transcription factor of Saccharomyces cerevisiae. Mol Gen Genomics. 2003;270:324–336. doi: 10.1007/s00438-003-0922-3. [DOI] [PubMed] [Google Scholar]
- Shuster EO, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Raithatha S, Stuart D. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol Cell Biol. 2002;22:7024–7040. doi: 10.1128/MCB.22.20.7024-7040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Schwaca A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci USA. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Zip1-induced changes in synaptonemal complex structure and poly complex assembly. J Cell Biol. 1995;128:455–466. doi: 10.1083/jcb.128.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Tung K-S, Hong E-JE, Roeder GS. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc Natl Acad Sci USA. 2000;97:12187–12192. doi: 10.1073/pnas.220464597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K-S, Roeder GS. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 1998;149:817–832. doi: 10.1093/genetics/149.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandromme M, Gauthier-Rouvière C, Lamb N, Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Robert M, Yoshida A, Nomura H, Reddel RR, Mitsui Y, Kaul SC. Inactivation of tumor suppressor p53 by Mot-2, a hsp70 family member. J Biol Chem. 1998;273:29586–29591. doi: 10.1074/jbc.273.45.29586. [DOI] [PubMed] [Google Scholar]
- Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.