This article identifies novel stress-protective proteins that belong to the family of intrinsically unstructured proteins, DprA and DprB, which are associated with the cytosol and the peroxisomes in Aspergillus fumigatus.
Abstract
During a search for genes controlling conidial dormancy in Aspergillus fumigatus, two dehydrin-like genes, DprA and DprB, were identified. The deduced proteins had repeated stretches of 23 amino acids that contained a conserved dehydrin-like protein (DPR) motif. Disrupted DprAΔ mutants were hypersensitive to oxidative stress and to phagocytic killing, whereas DprBΔ mutants were impaired in osmotic and pH stress responses. However, no effect was observed on their pathogenicity in our experimental models of invasive aspergillosis. Molecular dissection of the signaling pathways acting upstream showed that expression of DprA was dependent on the stress-activated kinase SakA and the cyclic AMP-protein kinase A (cAMP-PKA) pathways, which activate the bZIP transcription factor AtfA, while expression of DprB was dependent on the SakA mitogen-activated protein kinase (MAPK) pathway, and the zinc finger transcription factor PacC. Fluorescent protein fusions showed that both proteins were associated with peroxisomes and the cytosol. Accordingly, DprA and DprB were important for peroxisome function. Our findings reveal a novel family of stress-protective proteins in A. fumigatus and, potentially, in filamentous ascomycetes.
INTRODUCTION
The human pathogen Aspergillus fumigatus has the remarkable ability to survive extended periods of adverse environmental conditions. This filamentous fungus produces large amounts of dormant asexual spores (conidia). Dormant conidia exhibit resistance to a variety of environmental stresses, including desiccation, extreme temperatures, and osmotic or oxidative stress. When favorable conditions occur, these metabolically inactive cell types will reactivate the cell biochemical and genetic machineries, and alleviate resistance-associated functions. It is expected that the expression of genes involved in the establishment of conidial dormancy would be down-regulated during germination. Indeed, transcriptomics studies initiated at a time the genome was not yet sequenced indicated that around one-fourth of the A. fumigatus genes had transcripts in the dormant conidia, and that the expression of 22% of these genes was down-regulated during germination (Lamarre et al., 2008), leading to the hypothesis that at least some of these genes would be essential for the establishment and control of conidial dormancy. Using this strategy led us to identify Afu4g00860, a gene constitutively transcribed in dormant conidia, the expression of which was down-regulated within 30 min in yeast extract peptone-dextrose (YPD; Lamarre et al., 2008). This gene codes for a protein of unknown function harboring five repeated domains and was called DprA based on the amino acid repeats. Another gene, encoding nine such domains, was also found in the genome of A. fumigatus (DprB: Afu6g12180). This study reports on the functional analysis of DprA and DprB. Phenotype analysis of disruption mutants indicated the involvement of DprA and DprB in specific stress responses. The use of signal transduction mutants in monitoring Dpr expression levels suggested convergence of several pathways on the regulation of Dpr genes. Subcellular localization revealed that both proteins were associated with the cytosol and peroxisomes. This study uncovers novel proteins involved in stress protection in A. fumigatus.
RESULTS
DprA (Afu4g00860) and DprB (Afu6g12180) encode fungal dehydrin-like proteins
The gene Afu4g00860 was retrieved from an expression profiling study conducted in A. fumigatus aiming at the identification of germination-regulated genes (Lamarre et al., 2008). The deduced sequence corresponded to a protein containing 246 amino acids. Scanning of the sequence by prediction software neither revealed known localization signatures nor gave indications of protein function. However, blast results gave a hit with a 435-residue protein from A. fumigatus, encoded by Afu6g12180. Remarkably, blast results aligned only on small portions of the proteins, with very poor homology in between. The homologous sequences were repeated five and nine times, respectively, and corresponded to the signature pattern of fungal dehydrins (Abba et al., 2006). The repeated domain consisted of a stretch of 23 amino acids, containing a conserved dehydrin-like protein (DPR) motif (Figure 1). For this reason, the genes were called Dpr. Dehydrins were described in plants, where they are involved in the protection against dehydration-related stresses (Rorat, 2006). However, they have not been studied in fungi. In silico analysis confirmed that DprA and DprB were dehydrin-like proteins, by virtue of their physicochemical properties (Wise, 2003; Abba et al., 2006; Supplemental Figure S1). The presence of hydrophobic residues, predicted phosphorylation sites, and proline residues within the DPR domains (Figures 1 and S1B) suggest that DPR domains could form a hydrophobic core, within which protein–protein interactions would take place.
FIGURE 1:
Alignment of the DPR domains from DprA and DprB. Conserved amino acids are boxed in black (identical) or gray (similar). D1A-D5A designate the five domains from DprA and D1B-D9B refer to the nine domains from DprB, numbered from N- to C-terminal end. Numbers indicate the amino acid positions. The DPR motif is indicated with a red border. Asterisks designate a predicted phosphorylation site conserved in all DPR domains.
DprA and DprB are down-regulated upon conidial germination
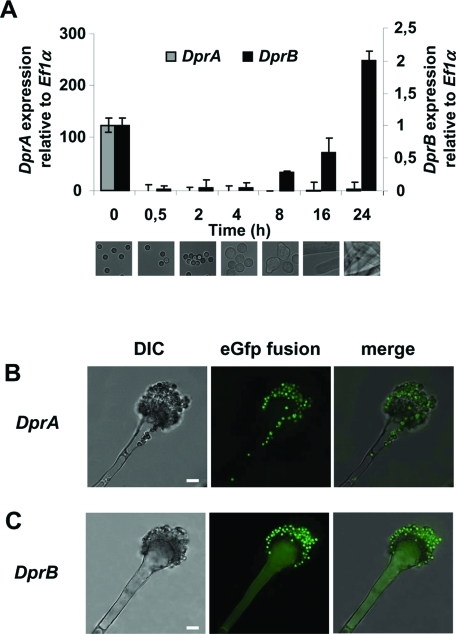
Expression of DprA and DprB was assessed during conidial germination (Figure 2A). In dormant conidia (time 0), DprA transcripts were ∼120 times more abundant than DprB. Both transcript types underwent significant down-regulation with a 1000- and a 350-fold decrease in expression level from 0 to 30 min, respectively. Very weak expression of DprA was detected beyond 30 min, at least up to 24 h. In contrast, DprB transcripts became abundant again after 8 h and reached a twofold increase at 24 h.
FIGURE 2:
Expression of DprA and DprB. (A) Evaluation of the expression levels of DprA and DprB by real-time PCR. RNA was extracted from dormant conidia of the AkuB strain (0 h) and conidia that were incubated for 0.5, 2, 4, 8, 16, or 24 h in liquid YPD medium at 37°C, 150 rpm. An arbitrary value of 1.0 was attributed to the expression level of DprB at time 0. Data are from three independent experiments ± SE. Developmental stages corresponding to the selected time points are shown. (B) Assessment of DprA expression by eGfp fusion in hyphae undergoing conidiation (7 d at 25°C). Note that DprA-eGfp is present only in the conidia. (C) Same as (B), with DprB expression. Scale bars: 5 μm.
To monitor the expression during development, the coding sequences of DprA and DprB were fused to eGfp under the control of their own promoters. DprA-eGfp fluorescence was detected only in dormant conidia (Figure 2B). In agreement with the real-time PCR data, DprB-eGfp fluorescence was observed in dormant conidia. However, in contrast to DprA-eGfp, DprB-eGfp was also observed in the conidiophores (Figure 2C) and in hyphae (unpublished data), indicating DprB was associated with late stages of development, unlike DprA.
DprA is involved in the oxidative stress response of conidia
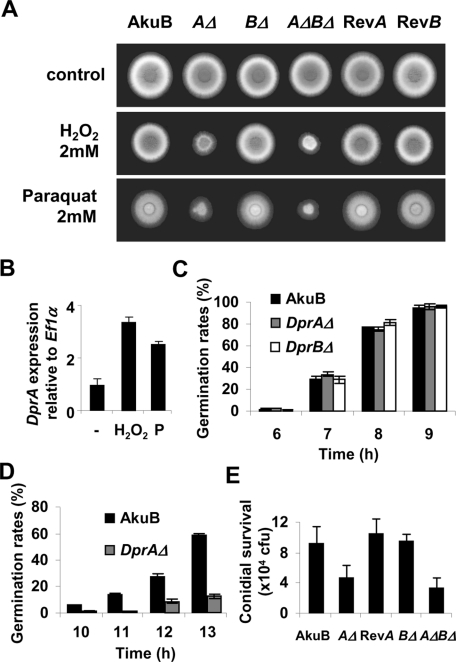
Growth of DprAΔ (but not DprBΔ) mutants was inhibited by oxidative stress generated by hydrogen peroxide or paraquat at concentrations > 2 mM (Figure 3A). Consistent with this, DprA expression was up-regulated upon treatment with 2 mM H2O2 or 2 mM paraquat (Figure 3B). In the absence of stress, no difference was observed between the germination curves of the mutant and control strains (Figure 3C). However, when 2 mM H2O2 was added to the medium, germination of the DprAΔ (and DprAΔ DprBΔ) mutants was impaired (Figure 3D). After 13 h, germination had reached its maximum stage. At that time point, only 30% of the mutant conidia had undergone swelling, compared to 70% for the control strains. This defect in swelling, and subsequently in germination, could be explained by the higher susceptibility of DprAΔ conidia to the fungicidal effect of H2O2. To check the behavior of conidia challenged with host-derived reactive oxidant species, conidial survival was assessed after 36 h in the lungs of immunocompetent mice. Accordingly, the conidia of the DprAΔ and DprAΔ DprBΔ mutants were hypersensitive to killing by the lung phagocytes (Figure 3E). However, no difference was observed in the virulence of the strains in two experimental models of invasive aspergillosis (Figure S2).
FIGURE 3:
Oxidative stress-induced phenotype of DprAΔ mutants. (A) Growth of DprΔ mutants (AΔ, BΔ and AΔBΔ) in the presence or absence of oxidative stress induced by H2O2 or paraquat, compared with the control strains (AkuB, or the revertant strains RevA and RevB). Conidia (105) were spotted on Sabouraud medium supplemented with 2 mM H2O2 or 2 mM paraquat (P). The plates were incubated at 37°C for 30 h. (B) Expression levels of DprA under oxidative stress. Overnight cultures of the AkuB strain were treated for 1 h with 2 mM H2O2 or 2 mM paraquat at 37°C 150 rpm, prior to RNA extraction. An arbitrary value of 1.0 was attributed to the expression level corresponding to the control without stress. Data are from three independent experiments ± SE. (C) Germination rates of the control (AkuB) and the DprΔ mutant conidia on Sabouraud medium at 37°C. Conidia undergoing germination were scored on a total of 100 conidia at 1 h intervals, in three independent experiments ± SE. (D) Same as (C), but in the presence of 2 mM H2O2. (E) Conidial killing in immunocompetent mice 36 h after intranasal infection. Conidial survival was evaluated by plating serial dilutions of bronchoalveolar lavages. Data are the mean value obtained with 3 mice ± SE.
DprB is involved in the osmotic stress response
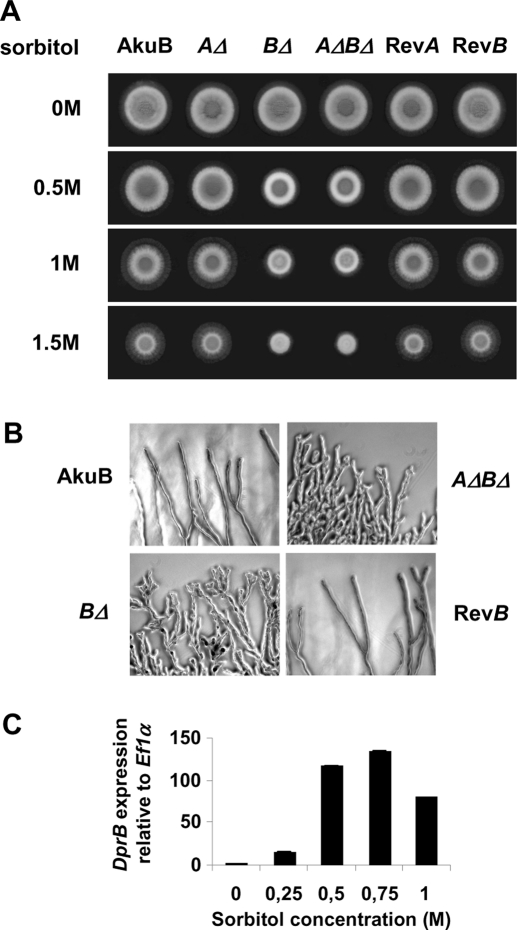
When grown in the presence of sorbitol, DprBΔ (in contrast to DprAΔ mutants), displayed an abnormal colony morphology, which could be observed as from 0.5 M sorbitol (Figure 4A). DprBΔ colonies were restricted to forming a “central zone,” in which conidiation would take place, with no “peripheral zone,” in which the colony would normally expand by apical extension of the hyphae. With 1.5 M sorbitol, the DprBΔ colonies had one-half the diameter of the control strain colonies. Microscopic observation of the colony edges showed extensive branching (Figure 4B). The results were similar irrespective of the osmoticum (sorbitol, glycerol, mannitol, NaCl, KCl) or medium (Sabouraud, YPD, minimal medium, malt extract; unpublished data) used. Addition of 1.5 M sorbitol to the medium did not affect the germ tube emergence nor the germination kinetics of the mutant conidia compared with the control strains (unpublished data), indicating that osmostress did not affect the DprBΔ mutants at the conidial stage, but rather at later stages of development. When the wild-type strain was subjected to sorbitol concentrations ranging from 0 to 1 M, DprB transcript levels were up-regulated (Figure 4C). Regulation was dose-dependent, with an expression peak with 0.75 M sorbitol.
FIGURE 4:
Osmotic stress-induced phenotype of DprBΔ mutants. (A) Growth of DprΔ mutants (AΔ, BΔ, and AΔBΔ) in the presence or absence of osmotic stress induced by sorbitol, compared with the control strains (AkuB, or the revertant strains RevA and RevB). Conidia (105) were spotted on Sabouraud medium supplemented with 0, 0.5, 1.0, or 1.5 M sorbitol. The plates were incubated at 37°C for 30 h. (B) Microscopic observation of the colonies grown on solid Sabouraud supplemented with 0.5 M sorbitol. Note the hyperbranched mycelium of the DprBΔ and DprAΔ DprBΔ mutants, compared to the parental and revertant strains. (C) Expression levels of DprB under osmotic stress assessed by real-time PCR. Extracts are from overnight liquid cultures of the wild-type strain subjected to a range of sorbitol concentrations (0, 0.25, 0.5, 0.75, 1 M) for 30 min at 37°C. An arbitrary value of 1.0 was attributed to the expression level without addition of sorbitol. Data are from three independent experiments ± SE.
The DprBΔ mutant displays a pH-dependent phenotype that is PacC-related
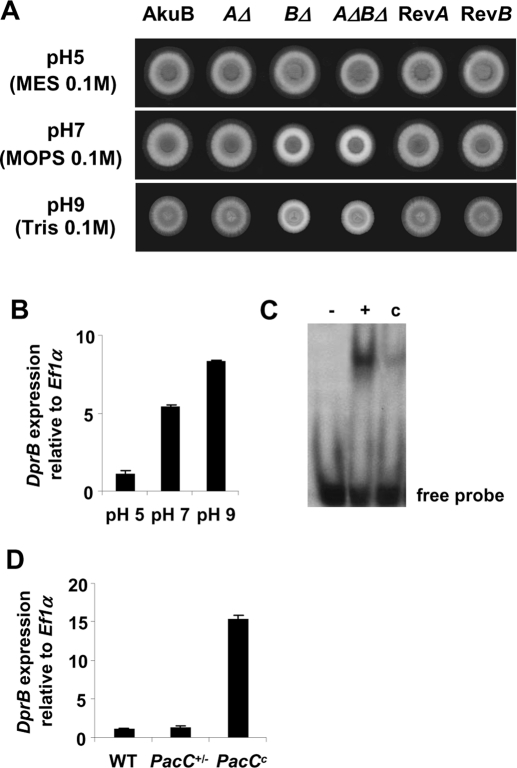
Growth of the DprBΔ (but not of the DprAΔ) mutant was impaired at pH 7 and pH 9, but not at pH 5 (Figure 5A). At pH 9, after 72 h of growth, the DprBΔ colonies had one-half the diameter of the control strain colonies (unpublished data). Real-time PCR indicated DprB was expressed preferentially at neutral or alkaline pH (Figure 5B), consistent with the phenotype of the DprBΔ mutant. As seen in the assays under osmostress, germination of the conidia was not affected by pH stress (unpublished data). The PacC transcription factor controls pH-regulated genes in Aspergillus spp. (Tilburn et al., 1995). A putative binding site of PacC (GCCAGG) was detected in the promoter of DprB at position −264. The binding of recombinant PacC to the DNA sequence was confirmed (Figures 5C and S3). In Aspergillus spp., PacC acts as both an activator of alkaline-expressed genes and a repressor of acid-expressed genes. Loss-of-function mutations of PacC (PacC+/−) cause an acidity-mimicking phenotype and result in an increased expression of acid-expressed genes, and a reduced expression of alkaline-expressed genes. Gain-of-function, alkalinity-mimicking mutations of PacC (PacCc) result in a phenotype opposite that of acidity-mimicking mutations. DprB expression was checked in the alkalinity-mimicking and in the acidity-mimicking strains (Amich et al., 2009). In agreement with a positive regulation of DprB by PacC, the expression of DprB in the acidity-mimicking strain was similar to that in the wild-type, whereas it was overexpressed in the alkalinity-mimicking strain (Figure 5D).
FIGURE 5:
pH stress-induced phenotype of DprBΔ mutants. (A) Growth of DprΔ mutants (AΔ, BΔ, and AΔBΔ) under pH stress, compared with the control strains (AkuB, or the revertant strains RevA and RevB). Conidia (105) were spotted on Sabouraud medium supplemented with 0.1 M MES, MOPS, or Tris and adjusted to pH 5, 7, or 9. The plates were incubated at 37°C for 30 h. (B) DprB expression according to extracellular pH. Real-time PCR was performed on extracts from overnight cultures shifted for 1 h to medium at pH 5, 7, or 9 at 37°C, 150 rpm. An arbitrary value of 1.0 was attributed to the expression level corresponding to pH 5. Data are from three independent experiments ± SE. (C) Gel-mobility shift assay using the GST::PacC (30–195*) protein produced in Escherichia coli and a 37 bp fragment of the DprB promoter. The 32P-labeled DNA fragment (40 fmol) was incubated in the absence (−) or presence (+) of the purified GST::PacC protein (40 fmol). Unlabeled probe (4 pmol) was used as competitor (c) to check the specificity of the binding. (D) DprB expression in PacC mutants. Real-time PCR was performed on extracts from overnight cultures at pH 7 of the reference strain AkuB, the acid-mimicking strain PacC+/−, and the basic-mimicking strain PacCc. An arbitrary value of 1.0 was attributed to the expression level obtained with the wild-type strain (WT). Data are from three independent experiments ± SE.
DprA and DprB act downstream of the SakA MAPK
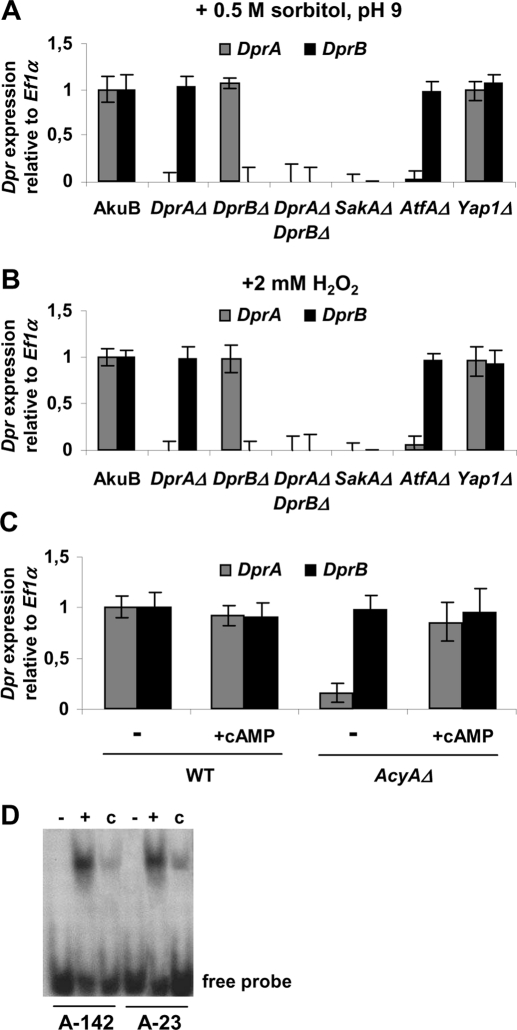
To gain insight into the signaling cascades involved in the recruitment of Dpr proteins, the expression of Dpr genes was assessed in signal transduction mutants from the mitogen-activated protein kinase (MAPK) pathways (SakAΔ, MpkAΔ, MpkBΔ, and MpkCΔ; Du et al., 2006; Reyes et al., 2006; Valiante et al., 2008), the cyclic AMP (cAMP) signaling pathway (AcyAΔ, PkaC1Δ, and PkaRΔ; Liebmann et al., 2003, 2004; Grosse et al., 2008), and the calcium/calcineurin transduction pathway (calAΔ; da Silva Ferreira et al., 2007). In contrast with other signaling mutants, DprA and DprB transcripts were not detected in the SakAΔ mutant (Figure 6, A and B), indicating DprA and DprB acted downstream of the stress-activated kinase (SAK) SakA MAPK cascade. In A. fumigatus, the SakA signaling pathway regulates the response to hyperosmotic and oxidative stress (Du et al., 2006; Reyes et al., 2006). An in silico comparative analysis was undertaken to identify targets of SakA that could link the MAPK to Dpr genes. In Saccharomyces cerevisiae, the SakA homologue Hog1 interacts with 4 transcription factors, Msn2, Msn4, Sko1, and Hot1 (Hohmann et al., 2007). However, A. fumigatus lacked clear orthologues of these proteins (Bahn, 2008), indicating that other sets of transcription factors achieved stress-response regulation downstream of SakA. In Schizosaccharomyces pombe, two bZIP-type transcription factors, Atf1 and Pap1, intervene downstream of the SakA-related MAPK cascade in response to environmental stress signals (Toda et al., 1991; Takeda et al., 1995; Shiozaki and Russell, 1996; Gaits et al., 1998; Toone et al., 1998). Aspergillus spp. possess an Atf1 and a Pap1 homologue, and SakA was shown to interact with AtfA in A. nidulans (Lara-Rojas et al., 2011). Mutant strains for AtfA (Afu3g11330) and Yap1 (Afu6g09930) were constructed, and real-time PCR analysis showed that DprA expression was impaired in the AtfAΔ but not in the Yap1Δ mutant, indicating that DprA acted downstream of AtfA (Figure 6, A and B).
FIGURE 6:
Signal transduction pathways involved in the expression of Dpr genes. (A) Expression of Dpr genes in A. fumigatus mutants under 0.5 M sorbitol, pH 9. Transcript levels of DprA and DprB were estimated by real-time PCR in the reference strain AkuB; the Dpr mutants DprAΔ, DprBΔ, DprAΔ DprBΔ; the MAPK mutant SakAΔ; and the transcription factor mutants AtfAΔ and Yap1Δ. An arbitrary value of 1.0 was attributed to the expression levels in the AkuB strain. (B) Same as (A), under 2 mM H2O2. (C) Expression of Dpr genes in the adenylyl cyclase mutant AcyAΔ, and the effects of adding 50 mM exogenous cAMP as determined by real-time PCR. An arbitrary value of 1.0 was attributed to the expression levels in the AkuB strain without treatment with cAMP. (D) Gel-mobility shift assay using the AtfA::6His recombinant protein and 37–base pair fragments from the DprA promoter (A-142 or A-23). The 32P DNA fragments (40 fmol) were incubated in the absence (−) or presence (+) of the recombinant AtfA::6His protein (40 fmol). Unlabeled probe (4 pmol) was used as competitor (c) to check the binding specificity.
The expression of DprA was also affected in the adenylyl cyclase mutant AcyAΔ (Figure 6C). When the AcyAΔ strain was grown on medium supplemented with 25 mM cAMP, the wild-type expression level of DprA was restored, indicating regulation by the cAMP-related pathway. Consistent with this finding, putative cAMP-responsive element (CRE) sequences were found in the promoter of DprA at positions −142 (TGACGTAA) and −23 (GAACGTCA), to which a recombinant AtfA protein was able to bind (Figures 6D and S3).
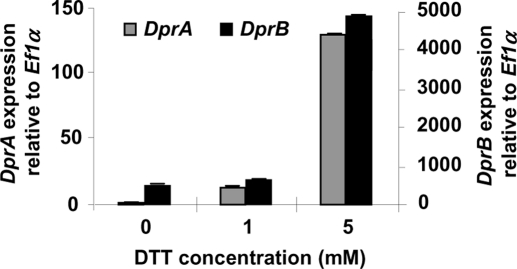
DprA and DprB are induced by DTT
A major class of stress-protective molecules is represented by molecular chaperones. These molecules are essential for cells to prevent the aggregation of partially unfolded proteins. This requirement is increased when cells experience protein unfolding stresses. Upon treatment with dithiothreitol (DTT), an inducer of the unfolded-protein response, significant up-regulation of DprA and DprB expression was observed (Figure 7), suggesting a potent role of the corresponding proteins as molecular chaperones.
FIGURE 7:
Expression of DprA and DprB upon treatment with DTT. Real-time PCR was performed on RNA extracted from overnight cultures of the AkuB strain treated for 1 h with 0, 1, or 5 mM DTT at 37°C, 150 rpm. An arbitrary value of 1.0 was attributed to the expression level of DprA in the absence of DTT. Data are from three independent experiments ± SE.
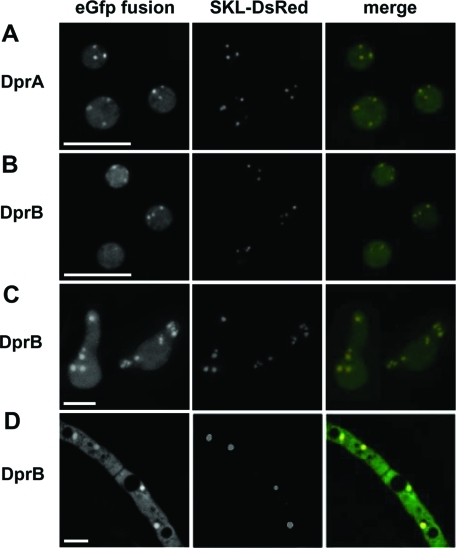
DprA and DprB fused to eGfp are associated with the cytosol and the peroxisomes
To check their subcellular localization, DprA and DprB were fused at their carboxy-terminal end to eGfp, under the control of their native promoters, in the respective mutant strains. The functionality of the constructs was checked by the restoration of the wild-type phenotype (unpublished data). Both DprA-eGfp and DprB-eGfp fusion proteins accumulated in the cytoplasm and in punctuate organelles. The labeled organelles did not stain with the membrane- and endocytosis-selective dye FM4–64 (Supplemental Movie S1; Fischer-Parton et al., 2000). To test the hypothesis that the organelles might be peroxisomes, the DprA- and DprB-eGfp strains were transformed with a plasmid bearing a DsRed-serine-lysine-leucine (DsRed-SKL) fusion typical of the type 1 peroxisomal targeting sequence PTS1 (Ruprich-Robert et al., 2002; Elleuche and Pöggeler, 2008). Colocalization of eGfp and DsRed showed that DprA and DprB were associated with peroxisomes (Figure 8 and Movie S2).
FIGURE 8:
Subcellular localization of DprA and DprB using eGfp fusions. DprA and DprB were fused at their 3′ end to the coding sequence of eGfp. Expression was driven by the gene's own promoter in the corresponding DprAΔ and DprBΔ strains. Localization of DprA in dormant conidia (A), and localization of DprB in dormant conidia (B), in germinating conidia (C), and in hyphae (D). Scale bars: 5 μm
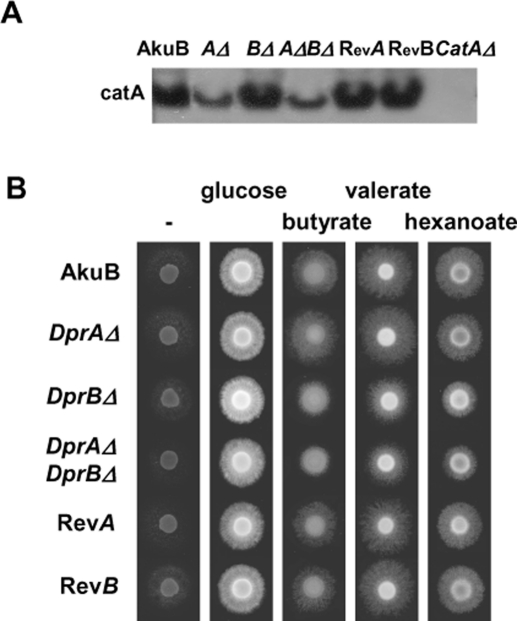
DprΔ mutants have altered catalase and β-oxidation activities
Peroxisomes contain a large battery of enzymes that are important notably for oxygen species detoxification and β-oxidation of fatty acids. Catalases have a protective role against H2O2 and have been shown to be localized in peroxisomes (Schrader and Fahimi, 2006). A. fumigatus possesses three catalase activities: CatA, which is produced exclusively in conidia, and Cat1 and Cat2, which are produced in the mycelium (Paris et al., 2003). In an attempt to explain the higher sensitivity of DprAΔ conidia to H2O2, catalase activity was assessed in conidial protein extracts. As shown by in-gel detection, activity due to the conidial catalase CatA was reduced in the DprAΔ mutant (Figure 9A). This was not the case for the mycelial catalases Cat1 and Cat2 when mycelial extracts were assessed (unpublished data).
FIGURE 9:
Effect of DprΔ mutation on peroxisomal functions. (A) In-gel detection of catalase activity. Each line was loaded with 30 μg total protein. (B) Growth on different carbon sources. The following carbon sources were added to modified minimal medium: 1% glucose, 10 mM butyrate, 10 mM valerate, 5 mM hexanoate. Growth was for 3 d at 37°C.
Growth of the DprBΔ mutant was impaired when short-chain fatty acids (butyrate [C4], valerate [C5], hexanoate [C6]) were used as sole carbon sources (Figure 9B), but not with longer-chain fatty acids such as oleic acid (C18), or Tween 20, whose major component is lauric acid (C12; unpublished data). In Aspergillus spp., β-oxidation of short-chain fatty acids with odd-numbered carbons (notably valerate) requires the peroxisomal β-oxidation pathway (Hynes et al., 2008). The two results are in agreement with a peroxisomal association of Dpr proteins.
DISCUSSION
Dpr genes encode specific stress-response proteins
While the DprAΔ mutant was impaired in growth on media supplemented with H2O2 or paraquat, the DprBΔ mutant growth was affected under hyperosmotic conditions or when pH was maintained at 7 or 9. Other stress conditions, e.g., extreme temperatures (storage at −20°C or 4°C, culture at 50°C), drug treatments (with caspofungin, Congo red, calcofluor white, or SDS), or high concentrations of metal ions (Fe2+, Cd2+, Co2+, Zn2+, or Ni2+) had no effect on the development of the mutants (unpublished data), indicating DprA and DprB were involved specifically but differently in oxidative, osmotic, and pH stress.
The protection effect exerted by Dpr proteins against stress was confirmed by overexpression studies of DprA and DprB that resulted in enhanced resistance to oxidative stress, and osmotic and pH stress, respectively (Figure S4). This is in good accordance with studies related to the overexpression of dehydrins and molecular chaperones, in either homologous or heterologous systems (Swire-Clark and Marcotte, 1999; Brini et al., 2007; Montero-Barrientos et al., 2008), although the stresses involved were different.
An excellent correlation was obtained between expression levels under stress and mutant phenotypes, suggesting that Dpr regulation was predominantly transcriptional. Gene expression was modulated transiently within the time frames described previously in filamentous fungi for the regulation of oxidative stress (30–60 min; Li et al., 2008), osmotic stress (30–90 min; Han and Prade, 2002), and the pH response (30–120 min; Galindo et al., 2007), indicating a genetic program involving DprA and DprB was rapidly switched on to reestablish homeostasis. However, posttranslational regulation cannot be excluded, since putative phosphorylation sites were detected (Figures 1 and S1).
Signal transduction pathways involved in Dpr-related stress response
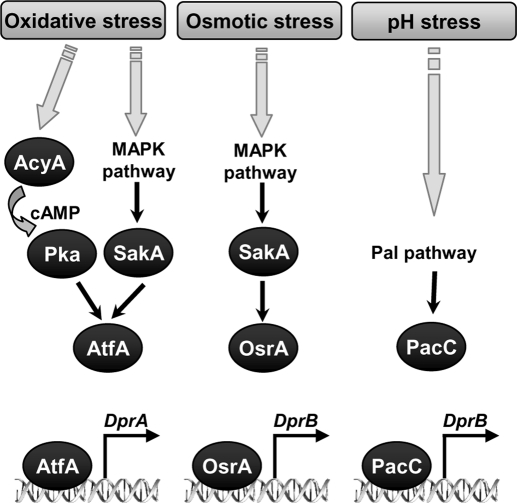
In this study, we showed that several pathways were involved in the regulation of Dpr genes. A hypothetical model is proposed in Figure 10. Common to Dpr gene regulation, the SakA pathway was known to control stress responses, either oxidative- (Nguyen et al., 2000; Du et al., 2006; Hagiwara et al., 2009; Zhang et al., 2009; Balazs et al., 2010) or osmotic-related (Han and Prade, 2002; Liu et al., 2008; Zhang et al., 2009). Together with the cAMP-related pathway, SakA contributes to the oxidative stress response mediated by DprA. The AtfA transcription factor, known to intervene downstream of the cAMP-related pathway (Rehfuss et al., 1991; Neely and Hoffman, 2000), is activated by SakA, most probably upon phosphorylation (Lara-Rojas et al., 2011). The acquisition of DprA-related oxidative stress tolerance in the conidia is consistent with the fact that Pka- and AtfA-mediated oxidative stress tolerance in Aspergillus spp. is acquired specifically in conidia (Kawasaki et al., 2002; Zhao et al., 2006; Hagiwara et al., 2008). Upon osmotic stress, transcription of DprB was dependent on the SakA-related pathway and on a hypothetical osmotic stress regulator, OsrA. The SakAΔ strain was unaffected by pH stress, whereas the PacC mutants were unaffected by osmostress (unpublished data), suggesting the two pathways converged independently on DprB.
FIGURE 10:
Proposed model for the regulation of Dpr genes. In response to oxidative stress, cAMP is synthesized by the adenylyl cyclase AcyA. Due to high levels of cAMP, protein kinase A (PKA) is activated. In parallel, oxidative stress also activates signal transduction via the SakA-related MAPK pathway. Both pathways converge on the bZIP transcription factor AtfA. Once activated, AtfA binds to the promoter of its target gene DprA to positively regulate its transcription. In response to osmotic stress, the signal is transduced via the SakA-related MAPK pathway to activate a hypothetical osmotic stress regulator (OsrA), leading to the transcription of DprB. When subjected to pH-related stress, the signal is transduced via the extracellular pH response pathway (Pal) pathway mediated by the zinc finger transcription factor PacC. PacC binds to the promoter of DprB to positively regulate its transcription.
The involvement of SakA in two distinct responses raises the question of signaling specificities. In S. cerevisiae, the Fus3- and the Kss1-related MAPK pathways converge on the Ste12 transcription factor to regulate mating or filamentation, respectively, in response to different stimuli. Signaling specificity is achieved by selective ubiquitination and sumoylation of Ste12 and its cofactor Tec1, leading to either degradation or protection of the respective transcription factors (Bruckner et al., 2004; Chou et al., 2008). Cross-activation of AtfA and OsrA in A. fumigatus could be prevented by a similar interplay between ubiquitination and sumoylation. This hypothesis is supported by the presence of two sumoylatable sites on AtfA (positions 399 and 474). Alternatively, dual phosphorylation of SakA by specific phosphatases could lead to the activation of different downstream effectors or OsrA could interact with SakA in the absence of phosphorylation, as shown for Hot1 (Saito and Tatebayashi, 2004).
Possible cellular function of Dpr proteins
Widely distributed in eukaryotes, intrinsically unstructured proteins (IUPs), which lack defined folding and three-dimensional structures, have high intramolecular flexibility (Tompa, 2002; Dyson and Wright, 2005). Dehydrins are IUPs that typically accumulate in plants in response to environmental stimuli with a dehydrative component (notably drought), salinity, and seed maturation (Battaglia et al., 2008). They possess one or several K-segments (EKKGIMDKIKEKLPG) that are presumed to interact with key cellular regulators prone to misfolding, which allows their stabilization in a chaperone-like manner (Close, 1996; Kovacs et al., 2008), thus precluding aggregation of proteins fatally damaged by stress. Tompa and Kovacs (2010) proposed that in virtue of their structural plasticity, IUPs may serve as potent chaperones. The possible role of Dpr proteins as molecular chaperones is supported by several lines of evidence: 1) Dpr genes are induced under stress and Dpr proteins subsequently exert a protective role against stress. 2) DprA and DprB expression is up-regulated upon addition of DTT, an inducer of protein misfolding. 3) The flexible structure of Dpr proteins, proline residues, and phosphorylation sites within the DPR domains are compatible with the physicochemical features expected in chaperones (Tompa and Kovacs, 2010). 4) Their association with the cytoplasm and the peroxisomes is consistent with the shuttling of peroxisomal precursors from the cytoplasm, where they are synthesized, to the peroxisomes, where they are addressed (Götte et al., 1998; Hettema et al., 1998).
The absence of peroxisome-targeting signal (PTS) is in favor of a peripheral association of Dpr proteins with the peroxisomes. Other proteins, notably peroxins, which are known to be associated both with the cytosol and the peroxisomes, participate in peroxisomal protein import and do not exhibit any obvious PTS (Pex1 [ Tamura et al., 2006], Pex5 [ van der Klei et al., 1995], Pex7 [ Marzioch et al., 1994], Pex19 [ Sacksteder et al., 2000], Pex18/21 [ Purdue et al., 1998]). Such proteins are either “piggybacked” by partners that have a PTS (Glover et al., 1994; Lee et al., 1997; Klein et al., 2002; Islinger et al., 2009) or are cytosolic proteins associated with the outer sides of peroxisomes (Götte et al., 1998 ; Hettema et al., 1998; Purdue et al., 1998).
Molecular chaperones associated with proteins being imported to the peroxisomes belong predominantly to the heat shock protein Hsp70/Hsp40 family, members of which were first identified due to their specific induction during the cellular response to stress conditions. Requirement of members of the chaperone Hsp70/Hsp40 family for peroxisomal matrix protein import was demonstrated (Walton et al., 1994; Legakis and Terlecky, 2001). Other molecular chaperones involved in peroxisomal protein import include the peroxin Pex19 (Jones et al., 2004; Vizeacoumar et al., 2006) or the ankyrin repeat protein Ankr2A (Shen et al., 2010).
In conclusion, our study uncovered novel proteins involved in the protection against stress. DprA and DprB likely act in the cell as molecular chaperones. Their identification from a conidial dormancy transcript profiling study points out the necessity for the fungus to exhibit a high level of stress resistance during the dormancy period.
MATERIALS AND METHODS
Strains and growth conditions
The A. fumigatus strains used in this study are listed in Supplemental Table S1. The strain AkuB (da Silva Ferreira et al., 2006) was used as the recipient strain for genetic transformation, unless otherwise specified. All strains were maintained on 2% malt agar slants at 25°C. Conidia were recovered following a 7-d culture at 25°C, in sterile 0.05% Tween 20, and filtered through a 40 μm pore size filter (BD Falcon, le Pont de Claix, France). Recombinant proteins were produced in Escherichia coli BL21 (DE3) or Pichia pastoris GS115.
In silico analysis
Blast analyses were submitted to the National Center for Biotechnology Information facility (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Estimation of intrinsic protein disorder was done with PrDOS (http://prdos.hgc.jp/cgi-bin/top.cgi; Ishida and Kinoshita, 2007). Prediction of phosphorylation sites was done with NetPhos software (http://www.cbs.dtu.dk/services/NetPhos/). The hydrophobic cluster analysis (Gaboriaud et al., 1987) plots were generated by the Mobyle Project's HCA server (http://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py?form=HCA). Prediction of sumoylation sites was performed using SUMOplot (http://www.abgent.com/tools/sumoplot).
Construction of A. fumigatus deletion, overexpression, and fusion strains
The cassettes used for the genetic transformation of A. fumigatus were generated by fusion PCR, or inverse PCR combined with conventional cloning, as described previously (Karababa et al., 2006; Lamarre et al., 2007). The primers are listed in Tables S2 and S3 and the constructs are schematized in Figures S5–S7. The deletion constructs consisted of a selection marker cassette encompassed by ∼1 kb flanking regions of the target gene. Complementation cassettes were made out of 1 kb of the 5′ flanking region, followed by the wild-type coding sequence and the selection marker cassette. The resistance genes to hygromycin (Hph, amplified from pAN7.1; Punt and van den Hondel, 1992) or phleomycin (Ble, amplified from pAN8.1; Punt and van den Hondel, 1992) were used to construct the cassettes. The double mutant was constructed by deletion of DprA in the DprBΔ mutant. For the overexpression of DprA and DprB, the genomic sequences of DprA and DprB encompassed with 1 kb upstream and downstream regions were amplified and cloned into the pPTRII vector (Takara, Saint-Germain-en-Laye, France) in the HindIII and SmaI restriction sites, respectively. For the subcellular localization of DprA and DprB, eGfp (Thastrup et al., 1999) was fused to the carboxy-terminal ends and the expression was driven by 1 kb of the gene native promoters. The mutant strains DprAΔ and DprBΔ were cotransformed with the eGfp fusion cassettes together with pAN8.1 or the pAN7.1 linearized with NdeI or HindIII, respectively. For colocalization experiments with the SKL-DsRed construct, the A. fumigatus strain CBS144–89 was used as the recipient strain for the eGfp fusions. The strains obtained were cotransformed with the plasmid bearing the linearized SKL-DsRed construct (Elleuche and Pöggeler, 2008), together with linearized pAN7.1 or pAN8.1, respectively. Transformation of A. fumigatus was carried out by conidial electroporation, as previously described (Lambou et al., 2010). The transformants were verified by Southern blot analysis and real-time PCR.
Phenotype analyses
Phenotypic tests were carried out mostly in Sabouraud medium (2% glucose, 1% mycopeptone; Oxoid, Dardilly, France). When mentioned, YPD, 2% malt extract, or minimal medium (Cove, 1966) were used as alternatives. For pH response assays, Sabouraud medium was amended with 0.1 M Tris, MOPS, or HEPES, according to the desired buffer range. For carbon source assays, 10 mM ammonium chloride, instead of ammonium tartrate, was added to minimal medium. In-gel catalase activity and germination rates were determined as described previously (Paris et al., 2003; Du et al., 2006).
Production of recombinant proteins and gel-mobility shift assays
The plasmid pGEX-PacC (Tilburn et al., 1995) was used to transform E. coli. The fusion protein was produced and purified on glutathione agarose (Sigma, Saint Quentin Fallavier, France) according to the manufacturer's instructions. The coding sequence of AtfA was amplified from A. fumigatus AkuB cDNA with the primers listed in Table S4, and cloned into the pKJ113 vector (Borg-von Zepelin et al., 1998) between the XhoI and NotI restriction sites. Following transformation of P. pastoris, the recombinant 6×His-tagged protein was produced and purified on Probond resin (Invitrogen, Cergy Pontoise, France) as suggested by the manufacturer. The DNA sequences used as probes are listed in Table S4. Probe labeling and gel-mobility shift experiments were carried out as described by Herbert et al. (2002).
Real-time PCR
Total RNA (5 μg) was reverse-transcribed using the Superscript II Reverse Transcriptase kit (Invitrogen, Cergy Pontoise, France). Quantitative PCR assays were performed using 96-well optical plates (Thermo Scientific, Courtaboeuf, France) in an iCycler iQ (170–8740; Bio-Rad, Marnes-la-Coquette, France) according to Bio-Rad's manufacturer instructions. Each run was assayed in triplicate in a final volume of 20 μL containing the DNA template at an appropriate dilution, 1× ABsolute SYBR green Fluorescein (Thermo Scientific), and 100 mM each primer (Beacon Designer 4.0 software; Premier International Software, Palo Alto, CA; Table S5). The cycling program was 95°C for 15 min, 40 cycles of 95°C for 30 s, and 55°C for 30 s. Amplification of one single specific target DNA was checked with a melting curve analysis at the end of the PCR (+0.5°C ramping for 10 s, from 55°C to 95°C). The generated data were then analyzed with Optical Systems Software 3.1 (Bio-Rad). The expression ratios were normalized to Ef1α expression, and calculated according to the ΔΔCt method (Livak and Schmittgen, 2001). To verify the absence of genomic DNA contamination, negative control templates in which reverse transcriptase was omitted were used for each gene set.
Live-cell imaging
For all microscopy experiments, A. fumigatus was cultured in Lab-Tek 8-well chamber-slides (Nalge Nunc International, Roskilde, France) containing Vogel's minimal medium N (Vogel, 1964). Wide-field epifluorescence imaging was performed on an inverted Nikon TE2000 microscope equipped with a Hamamatsu Orca ER CCD camera and a pE-2 LED excitation system (CoolLED) as the epifluorescence light source. Exposure times ranged from 200 to 400 ms; z-stacks of optical sections at 0.5 μm steps. Images were processed through 10 iterative deconvolutions using AutoQuant X software (Media Cybernetics, Bethesda, MD). For time-course experiments, images were acquired every 30 s for 10 min. Movies were generated and manipulated using ImageJ software (http://rsbweb.nih.gov/ij/). For membrane staining, germlings were incubated in 10 μM FM4–64 (Molecular Probes, Cergy Pontoise, France) for 30 min at 30°C.
In vivo experiments
Conidial survival assays in mouse lungs were carried out with inoculation of 5 × 108 conidia as described by Lambou et al. (2010). The virulence of the strains was tested in two models of invasive aspergillosis: cyclophosphamide and cortisone acetate-treated mice (Smith et al., 1994) and Galleria mellonella waxworms (Renwick et al., 2006). The virulence in immuno-suppressed mice was as described by Mouyna et al. (2010), except that the inoculation was with 1.5 × 105 conidia, and the cyclophosphamide dosage was 150 mg/kg. For the G. mellonella experiments, waxmoth (about 0.3–0.4 g in body weight) in the final larval stage (RJ Mous Livebait WOF, Netherlands) was used (10 per strain). 1.5 × 104 spores in phosphate-buffered saline (PBS) per larvae were used for injection into the hemocoel of 10 larvae per strain. The waxmoth larvae were then incubated at 37ºC for up to 10 d, and survival was recorded daily.
Supplementary Material
Acknowledgments
This work was supported by postdoctoral fellowships to J.W.S.H. from Region Ile de France and Fondation pour la Recherche Médicale. We are grateful to Miguel Peñalva, Judith Benhsen, and Stefanie Pöggeler for providing pGEX-PacC, eGFP, and pSKL-DsRed plasmids; to Jorge Amich, Vito Valiante, and Gregory May for providing PacC and signal transduction mutants; and to Greg Jedd for helpful advice. We acknowledge ESF Fuminomics for a short-visit grant to Nick Read's laboratory for the microscopic analyses.
Abbreviations used:
- cAMP
cyclic adenosine monophosphate
- DPR
dehydrin-like protein
- DTT
dithiothreitol
- IUP
intrinsically unstructured protein
- MAPK
mitogen-activated protein kinase
- PKA
protein kinase A
- PTS
peroxisome targeting signal
- YPD
yeast extract peptone-dextrose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0914) on April 13, 2011.
REFERENCES
- Abba S, Ghignone S, Bonfante P. A dehydration-inducible gene in the truffle Tuber borchii identifies a novel group of dehydrins. BMC Genomics. 2006;7:39–53. doi: 10.1186/1471-2164-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amich J, Leal F, Calera JA. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int Microbiol. 2009;12:39–47. [PubMed] [Google Scholar]
- Bahn YS. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell. 2008;7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs A, et al. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol Genet Genomics. 2010;283:289–303. doi: 10.1007/s00438-010-0513-z. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg-von Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol. 1998;28:543–554. doi: 10.1046/j.1365-2958.1998.00815.x. [DOI] [PubMed] [Google Scholar]
- Brini F, Hanin M, Lumbreras V, Amara I, Khoudi H, Hassairi A, Pages M, Masmoudi K. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep. 2007;26:2017–2026. doi: 10.1007/s00299-007-0412-x. [DOI] [PubMed] [Google Scholar]
- Bruckner S, Kohler T, Braus GH, Heise B, Bolte M, Mosch HU. Differential regulation of Tec1 by Fus3 and Kss1 confers signaling specificity in yeast development. Curr Genet. 2004;46:331–342. doi: 10.1007/s00294-004-0545-1. [DOI] [PubMed] [Google Scholar]
- Chou S, Zhao S, Song Y, Liu H, Nie Q. Fus3-triggered Tec1 degradation modulates mating transcriptional output during the pheromone response. Mol Syst Biol. 2008;4:212–218. doi: 10.1038/msb.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant. 1996;97:795–803. [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira ME, Heinekamp T, Hartl A, Brakhage AA, Semighini CP, Harris SD, Savoldi M, de Gouvea PF, de Souza Goldman MH, Goldman GH. Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet Biol. 2007;44:219–230. doi: 10.1016/j.fgb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Sarfati J, Latge JP, Calderone R. The role of the SakA (Hog1) and TcsB (Sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol. 2006;44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Elleuche S, Pöggeler S. Visualization of peroxisomes via SKL-tagged DsRed protein in Sordaria macrospora. Fungal Genet Rep. 2008;55:9–12. [Google Scholar]
- Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinson HA, Read ND. Confocal microscopy of FM4–64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J Microsc. 2000;198:246–259. doi: 10.1046/j.1365-2818.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gaits F, Degols G, Shiozaki K, Russell P. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 1998;12:1464–1473. doi: 10.1101/gad.12.10.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo A, Hervas-Aguilar A, Rodriguez-Galan O, Vincent O, Arst HN, Jr, Tilburn J, Penalva MA. PalC, one of two Bro1 domain proteins in the fungal pH signalling pathway, localizes to cortical structures and binds Vps32. Traffic. 2007;8:1346–1364. doi: 10.1111/j.1600-0854.2007.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Andrews DW, Rachubinski RA. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc Natl Acad Sci USA. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau WH, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse C, Heinekamp T, Kniemeyer O, Gehrke A, Brakhage AA. Protein kinase A regulates growth, sporulation, and pigment formation in Aspergillus fumigatus. Appl Environ Microbiol. 2008;74:4923–4933. doi: 10.1128/AEM.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Asano Y, Marui J, Yoshimi A, Mizuno T, Abe K. Transcriptional profiling for Aspergillus nidulans HogA MAPK signaling pathway in response to fludioxonil and osmotic stress. Fungal Genet Biol. 2009;46:868–878. doi: 10.1016/j.fgb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Hagiwara D, Asano Y, Yamashino T, Mizuno T. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci Biotechnol Biochem. 2008;72:2756–2760. doi: 10.1271/bbb.80001. [DOI] [PubMed] [Google Scholar]
- Han KH, Prade RA. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol Microbiol. 2002;43:1065–1078. doi: 10.1046/j.1365-2958.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- Herbert C, Jacquet C, Borel C, Esquerré-Tugayé MT, Dumas B. A cis-acting sequence homologous to the yeast filamentation and invasion response element regulates expression of a pectinase gene from the bean pathogen Colletotrichum lindemuthianum. J Biol Chem. 2002;277:29125–29131. doi: 10.1074/jbc.M201489200. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Ruigrok CC, Koerkamp MG, Van Den Berg M, Tabak HF, Distel B, Braakman I. The cytosolic DnaJ-like protein djp1p is involved specifically in peroxisomal protein import. J Cell Biol. 1998;142:421–434. doi: 10.1083/jcb.142.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- Hynes MJ, Murray SL, Khew GS, Davis MA. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics. 2008;178:1355–1369. doi: 10.1534/genetics.107.085795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:460–464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islinger M, Li KW, Seitz J, Volkl A, Luers GH. Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes–evidence for a natural piggyback import mechanism in mammals. Traffic. 2009;10:1711–1721. doi: 10.1111/j.1600-0854.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol. 2004;164:57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki L, Sanchez O, Shiozaki K, Aguirre J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol Microbiol. 2002;45:1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x. [DOI] [PubMed] [Google Scholar]
- Klein AT, Van Den Berg M, Bottger G, Tabak HF, Distel B. Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J Biol Chem. 2002;277:25011–25019. doi: 10.1074/jbc.M203254200. [DOI] [PubMed] [Google Scholar]
- Kovacs D, Kalmar E, Torok Z, Tompa P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre C, Ibrahim-Granet O, Du C, Calderone R, Latge JP. Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet Biol. 2007;44:682–690. doi: 10.1016/j.fgb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Lamarre C, Sokol S, Debeaupuis JP, Henry C, Lacroix C, Glaser P, Coppee JY, Francois JM, Latge JP. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics. 2008;9:417–431. doi: 10.1186/1471-2164-9-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambou K, Lamarre C, Beau R, Dufour N, Latge JP. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol Microbiol. 2010;75:910–923. doi: 10.1111/j.1365-2958.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- Lara-Rojas F, Sanchez O, Kawasaki L, Aguirre J. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol Microbiol. 2011;80:436–454. doi: 10.1111/j.1365-2958.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Mullen RT, Trelease RN. Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell. 1997;9:185–197. doi: 10.1105/tpc.9.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legakis JE, Terlecky SR. PTS2 protein import into mammalian peroxisomes. Traffic. 2001;2:252–260. doi: 10.1034/j.1600-0854.2001.90165.x. [DOI] [PubMed] [Google Scholar]
- Li Q, McNeil B, Harvey LM. Adaptive response to oxidative stress in the filamentous fungus Aspergillus niger B1-D. Free Radic Biol Med. 2008;44:394–402. doi: 10.1016/j.freeradbiomed.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Liebmann B, Gattung S, Jahn B, Brakhage AA. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol Genet Genomics. 2003;269:420–435. doi: 10.1007/s00438-003-0852-0. [DOI] [PubMed] [Google Scholar]
- Liebmann B, Muller M, Braun A, Brakhage AA. The cyclic AMP-dependent protein kinase A network regulates development and virulence in Aspergillus fumigatus. Infect Immun. 2004;72:5193–5203. doi: 10.1128/IAI.72.9.5193-5203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Leroux P, Fillinger S. The HOG1-like MAP kinase Sak1 of Botrytis cinerea is negatively regulated by the upstream histidine kinase Bos1 and is not involved in dicarboximide- and phenylpyrrole-resistance. Fungal Genet Biol. 2008;45:1062–1074. doi: 10.1016/j.fgb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta CT) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau WH. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13:4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Barrientos M, Hermosa R, Nicolas C, Cardoza RE, Gutierrez S, Monte E. Overexpression of a Trichoderma HSP70 gene increases fungal resistance to heat and other abiotic stresses. Fungal Genet Biol. 2008;45:1506–1513. doi: 10.1016/j.fgb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Mouyna I, et al. Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol Microbiol. 2010;76:1205–1221. doi: 10.1111/j.1365-2958.2010.07164.x. [DOI] [PubMed] [Google Scholar]
- Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol Cell Biol. 2000;20:6426–6434. doi: 10.1128/mcb.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AN, Lee A, Place W, Shiozaki K. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol Biol Cell. 2000;11:1169–1181. doi: 10.1091/mbc.11.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S, Wysong D, Debeaupuis JP, Shibuya K, Philippe B, Diamond RD, Latge JP. Catalases of Aspergillus fumigatus. Infect Immun. 2003;71:3551–3562. doi: 10.1128/IAI.71.6.3551-3562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt PJ, Van Den Hondel CA. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 1992;216:447–457. doi: 10.1016/0076-6879(92)16041-h. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfuss RP, Walton KM, Loriaux MM, Goodman RH. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem. 1991;266:18431–18434. [PubMed] [Google Scholar]
- Renwick J, Daly P, Reeves EP, Kavanagh K. Susceptibility of larvae of Galleria mellonella to infection by Aspergillus fumigatus is dependent upon stage of conidial germination. Mycopathologia. 2006;161:377–384. doi: 10.1007/s11046-006-0021-1. [DOI] [PubMed] [Google Scholar]
- Reyes G, Romans A, Nguyen CK, May GS. Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot Cell. 2006;5:1934–1940. doi: 10.1128/EC.00178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorat T. Plant dehydrins-tissue location, structure and function. Cell Mol Biol Lett. 2006;11:536–556. doi: 10.2478/s11658-006-0044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprich-Robert G, Berteaux-Lecellier V, Zickler D, Panvier-Adoutte A, Picard M. Identification of six loci in which mutations partially restore peroxisome biogenesis and/or alleviate the metabolic defect of pex2 mutants in Podospora. Genetics. 2002;161:1089–1099. doi: 10.1093/genetics/161.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem. 2004;136:267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Shen G, Kuppu S, Venkataramani S, Wang J, Yan J, Qiu X, Zhang H. ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell. 2010;22:811–831. doi: 10.1105/tpc.109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Smith JM, Tang CM, Van Noorden S, Holden DW. Virulence of Aspergillus fumigatus double mutants lacking restriction and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect Immun. 1994;62:5247–5254. doi: 10.1128/iai.62.12.5247-5254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swire-Clark GA, Marcotte WR., Jr The wheat LEA protein Em functions as an osmoprotective molecule in Saccharomyces cerevisiae. Plant Mol Biol. 1999;39:117–128. doi: 10.1023/a:1006106906345. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Yasutake S, Matsumoto N, Fujiki Y. Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J Biol Chem. 2006;281:27693–27704. doi: 10.1074/jbc.M605159200. [DOI] [PubMed] [Google Scholar]
- Thastrup O, Tullin S, Poulsen LK, Bjorn SP. Novel Variants of Green Fluorescent Protein. GFP, US patent, Denmark:: Novo, Nordisk A/S; 1999. [Google Scholar]
- Tilburn J, Sarkar S, Widdick DA, Espeso EA, Orejas M, Mungroo J, Penalva MA, Arst HN., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Tompa P, Kovacs D. Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol. 2010;88:167–174. doi: 10.1139/o09-163. [DOI] [PubMed] [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiante V, Heinekamp T, Jain R, Hartl A, Brakhage AA. The mitogen-activated protein kinase MpkA of Aspergillus fumigatus regulates cell wall signaling and oxidative stress response. Fungal Genet Biol. 2008;45:618–627. doi: 10.1016/j.fgb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Van Der Klei IJ, Hilbrands RE, Swaving GJ, Waterham HR, Vrieling EG, Titorenko VI, Cregg JM, Harder W, Veenhuis M. The Hansenula polymorpha PER3 gene is essential for the import of PTS1 proteins into the peroxisomal matrix. J Biol Chem. 1995;270:17229–17236. doi: 10.1074/jbc.270.29.17229. [DOI] [PubMed] [Google Scholar]
- Vizeacoumar FJ, Vreden WN, Aitchison JD, Rachubinski RA. Pex19p binds Pex30p and Pex32p at regions required for their peroxisomal localization but separate from their peroxisomal targeting signals. J Biol Chem. 2006;281:14805–14812. doi: 10.1074/jbc.M601808200. [DOI] [PubMed] [Google Scholar]
- Vogel HJ. Distribution of lysine pathway among fungi: evolutionary implications. Am Nat. 1964;98:435–446. [Google Scholar]
- Walton PA, Wendland M, Subramani S, Rachubinski RA, Welch WJ. Involvement of 70-kD heat-shock proteins in peroxisomal import. J Cell Biol. 1994;125:1037–1046. doi: 10.1083/jcb.125.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ. LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics. 2003;4:52–70. doi: 10.1186/1471-2105-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao J, Fang W, Zhang J, Luo Z, Zhang M, Fan Y, Pei Y. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl Environ Microbiol. 2009;75:3787–3795. doi: 10.1128/AEM.01913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Panepinto JC, Fortwendel JR, Fox L, Oliver BG, Askew DS, Rhodes JC. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect Immun. 2006;74:4865–4874. doi: 10.1128/IAI.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.