Abstract
Unstructured proteins, RNA or DNA components provide functionally important flexibility that is key to many macromolecular assemblies throughout cell biology. As objective, quantitative experimental measures of flexibility and disorder in solution are limited, small angle scattering (SAS), and in particular small angle X-ray scattering (SAXS), provides a critical technology to assess macromolecular flexibility as well as shape and assembly. Here, we consider the Porod-Debye law as a powerful tool for detecting biopolymer flexibility in SAS experiments. We show that the Porod-Debye region fundamentally describes the nature of the scattering intensity decay, which captures information needed for distinguishing between folded and flexible particles. Particularly for comparative SAS experiments, application of the law, as described here, can distinguish between discrete conformational changes and localized flexibility relevant to molecular recognition and interaction networks. This approach aids insightful analyses of fully and partly flexible macromolecules that is more robust and conclusive than traditional Kratky analyses. Furthermore, we demonstrate for prototypic SAXS data that the ability to calculate particle density by the Porod-Debye criteria, as shown here, provides an objective quality assurance parameter that may prove of general use for SAXS modeling and validation.
INTRODUCTION
Our view of the nature of bio-macromolecular function has been greatly enhanced by recent observations that many complex biological processes require structural flexibility within the biological machine. Flexibility contributes to the dynamics of the macromolecular particle over large (delocalized) and small (localized) scales that can be characterized by solution methods such as NMR and SAS1,2. Many cellulases, DNA repair proteins, transcription factors and extracellular matrix proteins contain long flexible linkers that tether a core domain to one or more catalytic or specificity domains3–5. This type of flexibility enhances the functional dynamics of the macromolecule thereby delocalizing the functional domain over a large volume. Particularly for DNA repair proteins (e.g., DNAPK, Nbs1, ligase III, RPA, Mre11 polynucleotide kinase, and XPF) flexible extensions allow the core DNA damage recognition domain to tightly bind a damaged DNA end while recruiting the requisite repair proteins to the site of DNA damage6–10. In contrast, localized flexibility can functionally provide a macromolecular domain with the ability to switch between distinct conformational states or destabilize in the absence of a signaling molecule, as seen with many ATPases11–14.
Objective evaluation of macromolecular flexibility is of great scientific and practical significance for biology and medicine. We know that the binding of the correct metal ion in many proteins and RNA is key for domain folding and that most metalloproteins and structured RNAs remain uncharacterized, so an efficient means to examine solution structures of biopolymers in the presence of metals is of great value15,16. Aberrant flexibility from either metal ion absence or mutation can cause disease by reducing specificity of fold and assembly, as seen for reactive oxygen control enzyme superoxide dismutase and DNA repair helicase XPD12,17,18. Furthermore, flexibility can be induced. The binding of the RNA chaperone DEAD-box protein Mss116 to the ai5γ group II intron RNA assists the RNA during folding by promoting dynamic sampling of folding states thereby avoiding a kinetic trap19.
Structural information in cell biology includes shapes, flexibility and assemblies with many biopolymers mimicking the shape of an unrelated biopolymer. Such shape mimicry occurs among protein modification domains, such as SUMO and even across biopolymer classes, such as protein mimicry of DNA or RNA20–22. The efficiency and robustness of SAS is creating a renaissance in structural biology by providing three-dimensional models based on the X-ray and neutron scattering data from macromolecules in solution1,23. These models are shapes that can be quickly compared with known shapes to gain insights, such as mimicry. Furthermore, advanced SAXS synchrotron technologies now provide the throughput for systematic examination of macromolecular interactomes in pathways and networks24. With the development of SAXS technologies combined with other results for defining accurate macromolecular structures, conformations and assemblies in solution, SAS is poised to address many important challenges for biological systems25. As a solution technique, a major potential advantage of SAS is the ability to assess flexibility and to even characterize unstructured to structured transitions for biopolymers under near physiological conditions in solution. Detecting conformational switching, destabilization or long-range delocalized flexibility in solution can be made using small-angle X-ray scattering (SAXS), typically through qualitative assessments of the scattering data in a Kratky plot26. Here, we analyze the behavior of flexible particles in light of what is known as the Porod-Debye law and show that application of the law provides a new and reliable means for identifying flexibility in small angle scattering (SAS) experiments.
RESULTS
Bio-SAXS FUNDAMENTALS
In most biological SAXS studies, the observed particle scattering is assumed to occur from a discrete electron density contrast between the solvent and particle26. This assumption holds true for folded biopolymers such as proteins, RNA or mixed nucleic acid-protein complexes where, at low resolution, the average electron density of the particle is greater than that of the solvent. If we consider that the scattering angle, q (Å−1), defines a resolution ruler (d = 2π / q), then at sufficiently low resolution (as q → 0) the scattering will be described by an average effective electron density contrast (Δρ) between the particle and solvent that becomes increasingly difficult to discern at higher resolution similar to a close visual inspection of an impressionism painting (Figure 1). The existence of this contrast typically allows for the determination of I(0), the scattering intensity at q=0, and the particle’s radius-of-gyration, Rg, using the Guinier approximation27 (Figure 2A). I(0) relates directly to the particle’s volume (V) and Δρ scaled by concentration, c. This relationship between I(0) and V serves as the basis for determining the mass of the scattering particle against a set of known standards28,29. The assumption of a discrete scattering contrast can be challenged by flexible systems, such that in the extreme case for a fully unfolded biopolymer, the scattering contrast is continuous making it difficult to clearly distinguish between the particle and solvent at higher resolution.
FIGURE 1.

At low resolution (standing far from picture), differing contrasts contribute to the identification of objects within the picture. The hat, dress and grassy surroundings are clear features of the paintings with differing contrasts. Under close inspection (high resolution), only high contrast differences, such as the blue dress, allow for individual features to be distinguishable whereas differences with low contrast (boxed region) become difficult to discern. The difficulty of identifying the hat against the grassy background is analogous to the electron density contrast becoming continuous between the particle and solvent for flexible systems. This image was adapted from Matin a Eragny by Camille Pissarro and kindly prepared by Mike Pique at the Scripps Research Institute.
FIGURE 2.

Canonical SAXS scattering equations. A) Guinier approximation describing the linear relationship between the observed scattering intensities, I(q), and scattering angle, q. The approximation determines the radius-of-gyration, Rg, and scattering at q=0, I(0). I(0) is directly related to the particle’s volume, V, and electron density contrast, Δρ. B) Kratky plot of scattering data illustrating changes in the behavior of the curve for folded (sphere), partially folded (sphere-random coil) and completely unfolded particles (random coil). For a folded particle, the integrated area under the curve determines the Porod invariant, Q, and is scaled by concentration, c. C) SAXS derived structural parameters. The ratio of I(0) to Q determines the volume of the scattering particle sometimes known as the Porod volume, VP. Application of the Porod-Debye law determines the surface area, S, of the scattering particle that is scaled by concentration and Δρ that can be normalized using Q.
In the discrete contrast case (folded), G. Porod noted that the scattering data transformed as q2 I(q) vs. q, should capture a defined area26 (Figure 2B), known as the Porod invariant, Q (Figure 2B). Q is a direct measurement of Δρ that is scaled by the particle’s V and c. The determination of Q can, in theory, be used to determine several structural parameters such as the particle’s V, surface-to-volume ratio and correlation length, lc (Figure 2C)26,30.
For flexible particles, the ability to derive the structural values from SAS data becomes increasingly difficult depending on the degree to which the particle behaves as a random coil. If the particle is composed of a flexible polymeric extension with a single well-folded domain, then at sufficient q, the scattering should be due to two different electron density contrasts. This behavior can be observed in a Kratky plot where the transformed curve will be described by an initial parabolic peak followed by an elevated baseline at high q (Figure 2B). Further elevation of the baseline towards a hyperbolic-like curve will be observed for particles behaving as a random coil or Gaussian chain in solution26 (Figure 2B). These visual features in a Kratky plot have provided a means to qualitatively assess the degree of flexibility within the scattering particle. Furthermore, a dimensionless Kratky plot has been proposed as an alternative method for comparing macromolecules of differing masses and conformational states31 (Figure 3). This method normalizes the scattering data to I(0) and skews the scattering angle by Rg (Figure 3C), such that the resulting SAS data in a Kratky plot is normalized for c, V and Rg thereby allowing for a comparison of scattering profiles on a common scale. The dimensionless Kratky plot will be sensitive to the accuracy in the determination of the Guinier region (Figure 3D). These observations highlight the limitations of the Kratky plot as a tool for assessing flexibility requiring SAS data to be collected to sufficiently high q from particles that are sufficiently flexible to diverge the baseline.
FIGURE 3.
Dimensionless Kratky analysis. A) SAXS data of a 70 kDa protein collected at two concentrations (open gray circles represent a 70% dilution of the sample in red). B) Kratky plot of the data in A where the data in gray was scaled by 1.4× dilution factor. C) Transformation equations for the dimensionless Kratky plot using Guinier parameters: Rg and I(0). D) Dimensionless Kratky transformation using the experimental Guinier parameters for each respective curve in A.
THE POROD-DEBYE FOURTH POWER LAW
The Porod-Debye law describes a fourth power law approximation to the relationship between q and the observed intensities, I(q)32,33. Similar to the Guinier approximation, the Porod-Debye law is an approximation that occurs within a limited range of scattering angles and suggests that the scattering of a folded particle should decay as q−4 scaled by the particle’s surface area (S), c and Δρ (Figure 2C). Thus, transformation of the scattering data as q4 I(q) vs. q or q4 I(q) vs. q4 (Porod-Debye plot) should display a curve asymptotically approaching a constant value as q approaches infinity. The Porod-Debye law can be used to extract limited information from the SAXS curve, namely S, and dividing by the Porod invariant determines the surface-to-volume ratio of the particle (Figure 2C). In practice, the asymptotic relationship is rarely observed, as data at higher q contains significant shape information thereby destroying the asymptotic behavior predicted by the Porod-Debye law34. In addition, experimental buffer subtracted SAXS data contains contributions from the internal fluctuations of both the particle and its excluded volume26. Though insignificant at low angles, these contributions over-estimate Q such that the determination of V, lc, or S/V ratio will be underestimated. Based on the difficulty of applying the Porod-Debye law and the evaluation of Q, it is not surprising that the aforementioned parameters are rarely reported from a SAXS study of biological particles!
If we consider that the Porod-Debye law predicts a behavior within a limited range of the SAXS data (akin to the linear relationship predicted by Guinier (q·Rg < 1.3) for the determination of Rg), then application of the law predicts a plateau within the low resolution region of the SAXS data when transformed by q4. The presence of the Porod-Debye plateau validates the assumptions that the scattering particle exists with a sharp homogenous electron density contrast between the particle and solvent. Limiting the determination of Q to the Porod-Debye region of the data (e.g. using the program PRIMUS35) can now provide a reliable determination of the particle’s macromolecular volume.
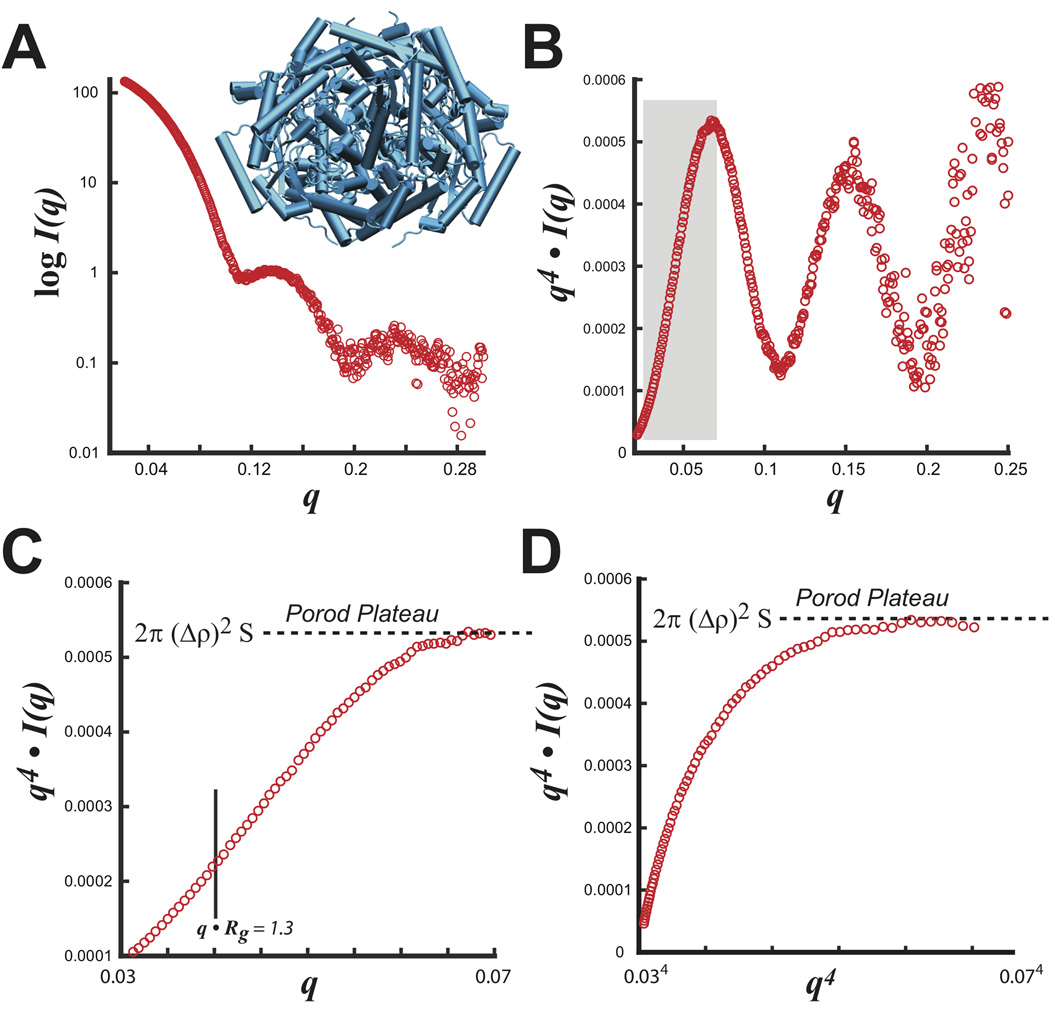
Experimentally, this behavior can be most readily demonstrated for glucose isomerase (GI), a nearly spherical tightly packed particle (Figure 4A). Transforming the entire SAXS dataset (0 < q < 0.32 Å−1) as q4 I(q) vs. q shows a curve with significant shape-dependent oscillatory features (Figure 4B). By noting that the Porod-Debye law resides within a limited region of the data, the plateau can be identified in a truncated dataset that removes most shape dependent scattering (Figure 4C, D). SAXS measurements of GI using 8 different concentrations determine an average V of 210,000 (±5,000) Å3, which imply a protein density (dprotein) of 1.36 (±0.04) gm·cm−3, in excellent agreement with the mean empirical density of proteins taken as 1.35 to 1.37 gm·cm−3 36,37. Applying the Porod-Debye law to the appropriate range of scattering data now provides additional opportunities for interpreting SAXS experiments.
FIGURE 4.
S. rubiginosus glucose isomerase. A) Experimental SAXS intensity plot with the tetrameric structure (blue). B) Porod plot of SAXS data transformed as q4 I(q) vs. q. Boxed region in gray describes the Porod-Debye region used in C and D. Examination of the boxed region in C) shows the Porod-Debye plateau that is proportional to 2π·(Δρ)2·S. D) Porod-Debye plot of SAXS data transformed as q4 I(q) vs. q4. Both plots in C and D aid in defining the Porod-Debye region for the determination of V using PRIMUS.
THE MACROMOLECULAR VOLUME IN SOLUTION
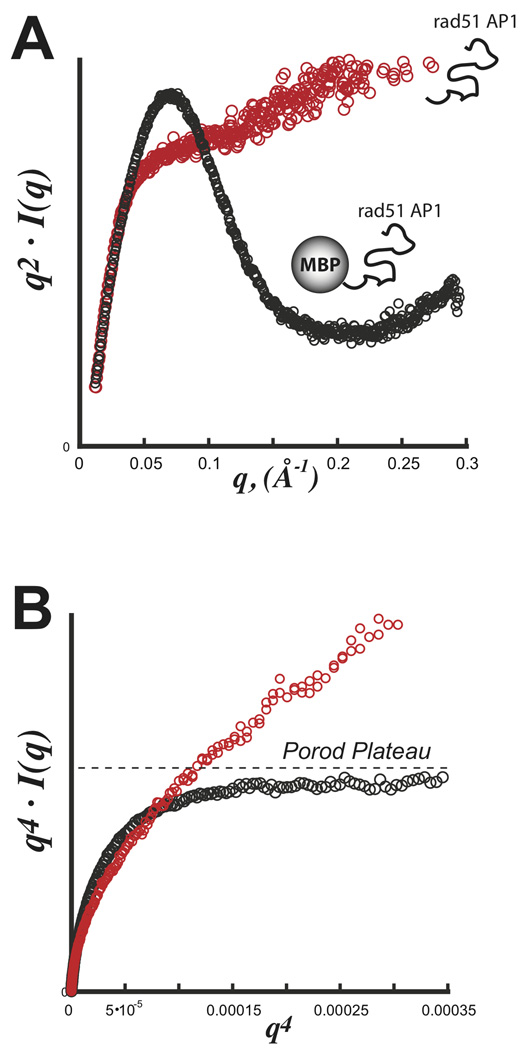
GI is a prototypical scattering particle that is ideal with a nearly spherical tightly packed core displaying little flexibility in solution. The macromolecular volume is simply the sum of the particle’s displaced volume and associated hydration shell. For flexible particles in solution, this concept of the macromolecular volume must be expanded to include the volumetric space occupied by the flexible domain. Consequently, the observed dprotein is expected to decrease reflecting the particle’s reduced packing density. This relationship is demonstrated using SAXS data from an intrinsically unfolded protein. Experiments on RAD51AP1 (36 kDa) confirm the protein to be intrinsically unfolded with no defined Porod-Debye plateau (Figure 5A,B). Attaching this domain to a well-folded protein, maltose-binding protein (MBP) from E. coli, created a biphasic 80 kDa scattering particle for SAXS. Inspection of the Kratky and Porod plots reveal a particle with a defined Porod-Debye plateau and an associated V of 145,000 Å3, consistent with a 120 kDa protein using dprotein of 1.37 (Figure 5B). The attachment of the intrinsically unfolded domain to MBP had the effect of increasing the apparent macromolecular volume. Therefore, using a dprotein of 1.37 over-estimated the mass of the scattering particle, where the actual dprotein is 0.92 gm·cm−3
FIGURE 5.
SAXS with an exemplary intrinsically disordered domain Rad51 AP1. Data was collected for both rad51 AP1 (red) and a fusion construct with E. coli maltose binding protein (MBP) (black). A) Kratky plot demonstrating the parabolic shape for a partially folded particle (black) and hyperbolic shape for a full unfolded particle (red). B) Porod-Debye plot demonstrating a clear plateau for the partially folded MBP-Rad51 AP1 hybrid particle. In the absence of MBP, the fully unfolded domain is devoid of any discernible plateau. Data was kindly provided by Gareth Williams at the Lawrence Berkeley National Laboratory.
In addition, the determination of V for 12 previously reported proteins varying in size from 8 to 118 kDa with known flexible or 11 amino acid His-tagged N-terminal extensions (Table 1), determines a mean protein density that decreased by 23% from 1.37 to 1.09 gm·cm−3. In these protein samples, the presence of the Porod plateau and increased partial specific volume suggests the protein can be characterized by at least two scattering volumes in solution: 1) a well defined homogenous element composed of a folded domain, and 2) a diffuse scattering volume created by the flexible extension. This increase in the particle’s apparent macromolecular volume would grossly overestimate the particle’s true mass when assuming either the standard dprotein of 1.35 to 1.37 gm·cm−3 or when standardizing the I(0) measurements using a protein standard such as glucose isomerase38. Consequently, an overestimated mass would lead to erroneous hypotheses regarding the protein’s multimeric or thermodynamic state.
Table 1.
SAXS Derived Particle Volumes Using the Porod-Debye Law
| BID | V (Å3) |
mass (kDa) |
Rg (Å) |
dprotein (gm·cm−3) |
|
|---|---|---|---|---|---|
| glucose isomerase | n.d. | 210000 | 172.2 | 33.1 | 1.36 |
| PF0699 | 1KINHP | 50333 | 32.1 | 23 | 1.06 |
| NADH oxidase | 1NADHP | 52829 | 42.7 | 22.9 | 1.34 |
| Acyl-CoA synthetase | 1ACSHP | 129680 | 80.4 | 34.2 | 1.03 |
| Y/S phosphatase | 1TSPHP | 27990 | 19.7 | 16.9 | 1.17 |
| phosphoribosyltransferase | 1XGTHP | 77047 | 51.6 | 25.3 | 1.11 |
| PF1372 | 3HYPHP | 61972 | 39.5 | 23.4 | 1.06 |
| PF1291 | 2HYPOP | 174720 | 117.5 | 36.1 | 1.12 |
| hypothetical | 1HYP0P | 13945 | 8.34 | 19.5 | 0.99 |
| arsR transcriptional regulatory protein | 1ARSDP | 26217 | 18 | 20.2 | 1.14 |
| amidotransferase | 1AMIGP | 38165 | 22.8 | 19.9 | 0.99 |
| PF0863 | 1HYPHP | 73405 | 42.6 | 27.4 | 0.96 |
BID: Bioisis ID code
n.d. not deposited in Bioisis
DISTINGUISHING FLEXIBILITY AND DISCRETE CONFORMATIONAL CHANGE
Previous SAXS studies on the abscisic acid binding (ABA) protein PYR1 demonstrated conformational differences between the bound and apo forms of the protein in solution39. In the absence of ABA, PYR1 in solution is nearly identical to the X-ray crystal structure (PDB: 3K3K). Upon binding ABA, PYR1 experiences a decrease in Rg (23.71 to 22.72 Å) suggesting compaction of the protein (Figure 6A). Thus, it is expected that a protein undergoing a discrete conformational change must display a clearly defined Porod plateau for both states, whereas it is predicted that destabilization of the particle would subsequently invalidate the application of the Porod-Debye law resulting in a loss of the plateau. Examination of the Porod plots for PYR1 (Figure 6B) show distinct plateaus that correspond to a decreased volume (from 74,000 to 59,000 Å) in the presence of ABA. Calculations of dprotein reveal PYR1 goes from 0.99 to 1.18 gm·cm−3 consistent with compaction of the protein, where it is expected the extended loops surrounding the binding pocket collapse during the conformational rearrangement.
FIGURE 6.
Metabolite induced structural changes monitored by SAXS. A) SAXS data for the 42 kDa abscisic acid binding domain PYR1 bound (black) and apo (red). B) Porod-Debye plot demonstrating discrete plateaus in bound and apo states suggesting each state can be characterized by discretely folded particles with sharp scattering contrasts. X-ray crystal structure (PDB: 3K3K) characterizes the apo state where it is expected the loop regions reorganize collapsing the structure in the bound state. C) SAXS data for the SAM-1 riboswitch bound (black) and apo (red). Crystal structure of the bound form (blue) changes in the apo state. D) Porod-Debye plot illustrating loss of the Porod plateau in the unbound state suggesting enhanced flexibility of the riboswitch. Transforming apo SAXS data by q3 I(q) vs. q3 (inset) verifies the intensity decay is no longer q−4 but q−3. Data adapted from Nishimura, N.; Hitomi, K.; Arvai, A. S.; Rambo, R. P.; Hitomi, C.; Cutler, S. R.; Schroeder, J. I.; Getzoff, E. D. Science 2009, 326, 1373–1379.
In contrast, SAXS studies on the S-adenosylmethionine (SAM)-1 riboswitch demonstrated conformational differences between the bound and unbound states due to an increased flexibility40. The SAM-1 riboswitch binds the small molecule SAM and solution SAXS studies demonstrated the bound form is nearly identical to the bound crystallographic structure (PDB: 2GIS). In the absence of the metabolite SAM, the riboswitch experiences an increase in Rg (22.4 to 24.1 Å) (Figure 6C) leading to the hypothesis that the riboswitch becomes less compact41. Is this necessarily due to an alternate conformational state or an increase in flexibility? Inspection of the Porod plot shows a plateau for the bound form that is subsequently destroyed in the absence of the metabolite (Figure 6D). The loss of the plateau implies the scattering contrast has become diffuse suggesting an increase in the flexibility of the particle. More specifically, the flexibility is localized as the maximum dimension of particle changes by only a small distance (from 76 to 81 Å) between the two states40 (Figure 6D).
Identifying flexibility by visual inspection of the Kratky plot may not always be effectively accomplished, especially with limited or weakly collected SAXS data as it may confound the baseline convergence necessary for assessing flexibility. Recent SAXS studies on the ATP dependent DNA repair complex, Mre11-Rad50, reveal distinct changes in the complex upon binding ATP42 (Figure 7A). Kratky analysis of the SAXS data suggested the bound and apo forms of Mre11-Rad50 are likely compact particles leading to the hypothesis that the particle is switching between two distinct conformational states, similar to PYR1. However, inspection of the Porod plot uncovers a different story. In the presence of ATP, the complex forms a distinct particle with a sharp scattering contrast, as evidenced by the Porod plateau (Figure 7B). The bound particle has a packing density of 1.04 gm·cm−3 that decreases significantly to 0.89 gm·cm−3 in the absence of ATP suggesting the particle becomes more flexible. In fact, inspection of the Porod-Debye region demonstrates a loss of the plateau supporting the assertion that Mre11-Rad50 is flexible in the absence of ATP.
FIGURE 7.
Detecting conformational flexibility. A) SAXS data for the Mre11-Rad50 complex in the presence (black) and absence (red) of ATP. SAXS scattering profiles transformed as a Kratky plot does not confidently demonstrate flexibility (inset). B) Porod-Debye plot illustrating changes in the Porod-Debye region. Loss of the plateau suggests Mre11-Rad50 complex becomes more flexible in the absence of ATP. Transforming apo SAXS data by q3 I(q) vs. q3 (inset) verifies the intensity decay is q−3. Data was adapted from Williams, G.J., Williams, R.S., Williams, J.S., Moncalian, G., Arvai, A., Limbo, O., Guenther, G., SilDas, S., Hammel, M., Russell, P., and Tainer, J. A., (2011) ABC ATPase signature helices in Rad50 link its nucleotide state to the Mre11 interface for DNA double-strand break repair, Nature SMB, accepted.
The Porod-Debye law predicts well defined behaviors of the scattering particle that can be used to understand the nature of the structural changes observed in SAS experiments. In the first example with PYR1, the observed changes in the SAXS data were due to PYR1 switching between two distinct conformational states, as evidenced by the two distinct Porod plateaus for both the bound and free SAXS datasets. In contrast, SAXS experiments with the SAM-1 riboswitch and Mre11-Rad50 complex suggested the particles are flexible in the absence of their respective small molecules. In the bound states, a clear Porod plateau becomes established with a concomitant decrease in both Rg and V. Unlike the Kratky plot which requires SAXS data collected over a large angular range (at least 0.01 < q < 0.3 Å−1), the Porod-Debye plot requires a limited set of data just outside the Guinier region for reliable and consistent interpretation.
PARTICLE DENSITY AS A COEFFICIENT FOR ESTIMATING SCATTERING MASS
Establishing the correct scattering mass of the particle has not been a strict prerequisite for structural modeling of SAXS data using current ab-initio or atomistic modeling methods. More importantly, under- or over-estimating the scattering mass will not necessarily generate worrisome values in the method’s scoring function. In the most rigorous experiments, SAXS and multi-angle light scattering experiments are performed in tandem in order to accurately assess the polydispersity and mass of the scattering particle41. Such detailed analysis is especially important for multimeric particles where an accurate determination of mass can help choose the correct symmetry of the particle or guide the composition of the starting atomistic model. Since V is a SAXS derived parameter, which reflects the particle’s macromolecular volume, a statistical survey of empirical SAXS-based protein densities can determine if a proposed particle mass is consistent with the observed SAXS data.
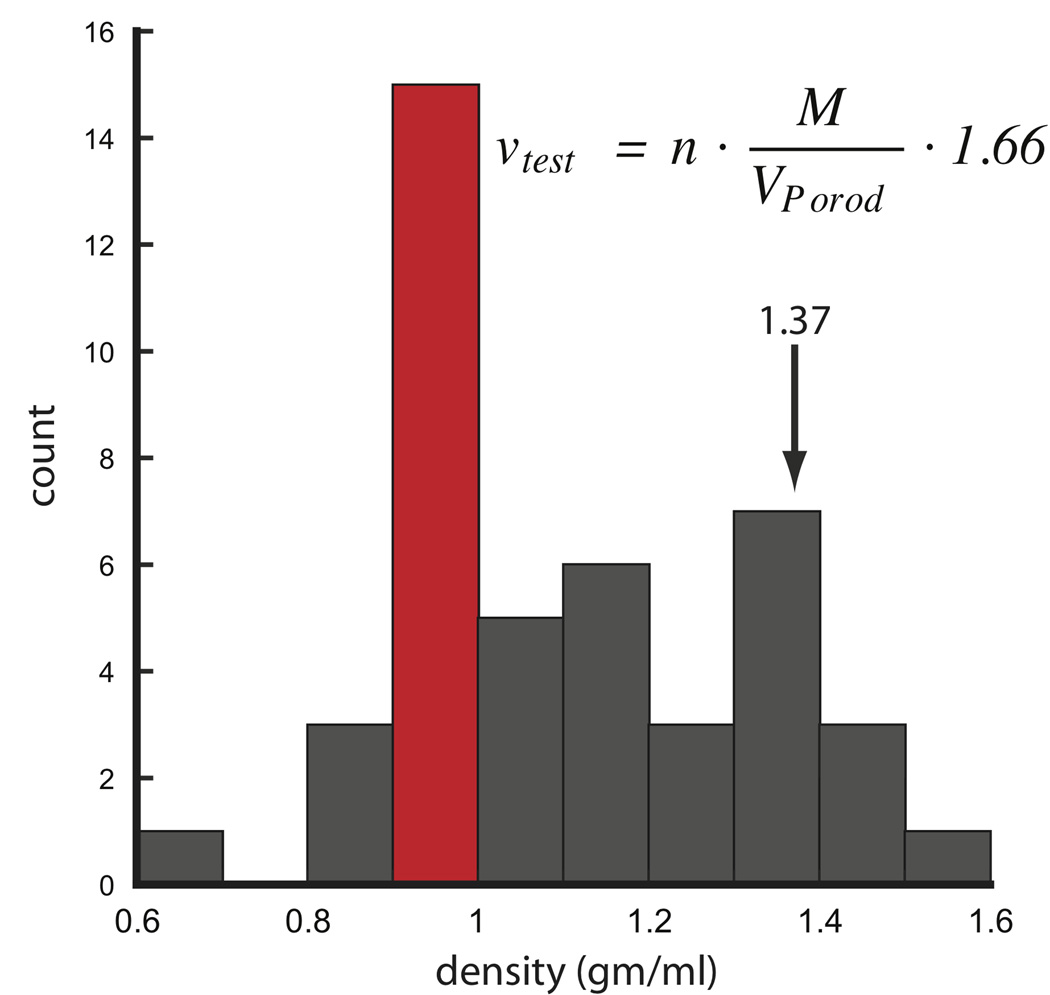
Using existing SAXS data from 31 different protein samples, a histogram of protein densities show the most commonly observed values are within 0.9 and 1.0 gm·cm−3, well below the canonical value of 1.37 (Figure 8). This is most likely due to the larger number of proteins with flexible or His-tagged extensions in our dataset. The next most likely observed density occurs within 1.3 and 1.4 gm·cm−3 encompassing the expected canonical value. With the exception that hollow-like structures may lower the packing densities below the empirically observed values, protein densities below 0.8 and above 1.5 are unlikely and may suggest under or over-modeling of the particle’s expected mass.
FIGURE 8.
Histogram analysis of protein densities calculated from SAXS data using 31 different proteins and conditions. For each protein, V was calculated using PRIMUS limiting the data to the Porod-Debye region. Distribution shows the most likely density is 0.9 to 1.0 gm·cm−3. The equation can be used to calculate a test density where n is the number of subunits in multimeric complexes. For hetero-complexes, n is 1. Units of M, VPorod and 1.66 are in Da, Å3 and Å3·gm·cm−3·Da−1, respectively. SAXS data was taken from http://www.bioisis.net.
Detecting under- and over-estimation of mass is illustrated with homomeric multi-subunit proteins GI, PYR1, WRN and catalase (Table 2). dprotein calculations with a varying number of subunits, n, will show a set of values that fall within an acceptable range, indicating the likely number of subunits constituting the particle (Table 2). Consider GI, calculation of dprotein using a 1 and 2 subunits result in dprotein values that are unrealistically low suggesting the particle mass is under-estimated. Likewise, for 5 and 6 subunits, dprotein is exceedingly high leaving the possible number of subunits at 3 or 4. Therefore, a combined ab-initio modeling with DAMMIN/F43 using P1 symmetry and GASBOR44 with both P3 and P4 symmetries will show one model, P3, underestimating the particle volume leaving P4 as the correctly assigned symmetry. In addition, SAXS data for 1RECBP (Table 2) was previously modeled as a monomer using a previously determined X-ray crystal structure24. A poor model-data agreement and further protein density calculations from the SAXS data strongly suggest the protein exists as a dimer or a mixed monomer-dimer in solution where density calculations with either the monomeric and trimeric forms of the protein produced unrealistic values. Thus, a direct check against the frequency of empirically determined values can help prevent under- or over-modeling of the SAXS data similar to the application of the Matthews coefficient in X-ray crystallographic structure determination of biological macromolecules45.
Table 2.
Calculated Protein Densities Using Empirically Derived Volumes
| n | GI | PYR1 | WRN | CAT | 1RECBP |
|---|---|---|---|---|---|
| 1 | 0.34 | 0.49 | 0.45 | 0.32 | 0.62 |
| 2 | 0.68 | 0.99 | 0.9 | 0.63 | 1.24 |
| 3 | 1.02 | 1.48 | 1.34 | 0.95 | 1.85 |
| 4 | 1.36 | 1.98 | 1.79 | 1.27 | 2.47 |
| 5 | 1.7 | 2.47 | 2.24 | 1.58 | 3.09 |
| 6 | 2.04 | 2.97 | 2.69 | 1.90 | 3.71 |
n: number of subunits
GI: glucose isomerase (43 kDa)
PYR1: abscisic acid binding protein (21 kDa)
WRN: Werner’s exonuclease domain (47 kDa)
CAT: human catalase (57 kDa)
1RECBP: recombinase (27 kDa)
DISCUSSION
Together with NMR, X-ray crystallography and electron microscopy (EM), SAXS is becoming a powerful complementary biophysical technique because it deals robustly with flexible particles in the solution state10,44,46. A limitation to the value of SAS characterizations has been objective tools for the validation of flexibility. The Porod-Debye law is an under-appreciated but powerful tool for detecting flexibility in SAS experiments of biopolymers. We show that a useful application of this law requires a subset of SAS data for reliable analysis that is more robust and conclusive than the traditional Kratky analysis.
The Porod-Debye region fundamentally describes the nature of the scattering intensity decay. The location of this region or plateau will vary depending on the size of the particle. In general for folded to partially folded particles, the Porod-Debye region will occur after the q corresponding to the first major peak identified in a Kratky plot. For small particles (< 10 KDa), the plateau is expected to occur at moderately higher q (>0.2 Å−1) necessitating the collection of inherently weak SAXS data at higher q values (qmax ≈ 0.4 Å−1). Therefore, analysis must be made on data collected at sufficiently high concentrations with an acceptable signal-to-noise ratio. For larger particles (>10 KDa), the plateau is expected to occur within smaller q values compatible with most synchrotron beamline configurations.
Particularly for comparative SAS experiments, application of the law can distinguish between discrete conformational changes and localized flexibility allowing for a more insightful understanding of the biological particle. Furthermore, the ability to calculate the particle density using the Porod-Debye criteria allows for a direct quality assurance check during the modeling phase of the SAXS experiment. The practical implementation of the Porod-Debye law in SAS experiments of biopolymers is a tool for assessing flexibility and for validation of SAXS models.
Many biological systems have evolved around exploiting changes in flexibility. In DNA repair, enhanced flexibility of the DNA is being used as a test for DNA damage. For some DNA repair enzymes, which exhibit open to closed conformational changes upon damaged DNA binding, DNA damage recognition is coupled to specific deformation of the DNA that promotes the damage excision pathway47–49. Conformational flexibility can regulate the XPB helix recognition of DNA damage versus DNA transcription sites12. Further conformational changes of repair complexes utilize delocalized flexibility to capture open assemblies to load onto DNA or to remodel stalled RNA polymerases to expose damaged DNA for repair50,51. In other cases, DNA repair proteins use structured surfaces to sculpt damaged DNA to create structural intersections of different DNA repair pathways52. SAXS has been particularly efficient at detecting and defining the types of flexibility exhibited by the DNA repair proteins53. This knowledge has enhanced our mechanistic insights and expands the experimental framework for understanding these systems in vivo.
Knowledge of localized flexibility can be used practically to design anti-peptide antibodies that will bind intact protein targets54–56. Similarly, the ability to experimentally define the flexibility of linked functional domains has many applications in biology and medicine, as seen for the creation of linked enzymes with long serum half-lives in vivo. For the proper functioning of biological pathways, unstructured biopolymers can undergo disorder to order transitions that can create large, yet dynamic interfaces. Identifying localized flexibility can provide the basis for inhibitor specificity by designing compounds that can further open the flexible regions of a binding pocket57,58. Thus, biopolymer flexibility ranges across the spectrum from unstructured proteins to localized flexibility, which creates challenges for most biophysical methods, including crystallography and electron microscopy. As molecular modeling seeks to provide increasingly accurate models of protein or RNA domains at the atomic level, the concept of the structural model must include in solution information regarding conformational states and flexibility59.
Looking forward, validation of SAS characterizations using parameters, such as the one presented here, add to our supramolecular understanding that links proteins or RNA to assemblies and pathways to biological outcomes. We can expect synergistic integration of SAXS structural results in solution with biochemical, genetic, and bioinformatics approaches to effectively create a structural interpretation of how shape and conformation join macromolecules in biological networks.
Materials and Methods
Except for glucose isomerase, all SAXS data used in this study was taken from BioIsis.net. The SAXS data in BioIsis.net defines q as the momentum transfer function: (4π sinθ)/λ with λ and θ describing the wavelength (Å) and angle of the scattered radiation. For glucose isomerase, sample was prepared by dissolving 33 uL of protein crystals (Hampton Research, Aliso Viejo, CA) in 1400 uL of buffer A containing 40 mM HEPES (pH 8.0), 1% glycerol, 6 mM MgCl2, 50 mM KCl, and 50 mM NaCl. Sample was incubated at 37 °C on a nutator for 30 minutes and concentrated to 180 uL before injecting on a KW-803 size exclusion column (Shodex, San Diego, CA) equilibrated in buffer A. Sample purification was monitored inline with an 18-angle light scattering instrument (Wyatt Technology, Santa Barbara, CA) where detector 12 was replaced with a quasi-elastic light scattering detector. The light scattering was connected in tandem to the Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA). Peak fraction at 2.3 mg/mL was taken for direct SAXS analysis (Beamline 12.3.1, Advanced Light Source, Berkeley, CA).
The determination of the Porod-Debye region and Porod volume was performed in several steps using the software package PRIMUS35. For the determination of the Porod volume, it is essential that the initial analysis starts with the accurate determination of the Guinier region since the determination of the particle volume relies on I(0). Furthermore, SAXS profiles should be truncated of erroneous data from both the beginning of the lowest q region and highest q region (q·Rg < 1.3 ) such that visual inspection of the residuals during Guinier analysis produce an unbiased distribution. Any “smiling” or “frowning” of the residuals is indicative of an improper fit. Following the definition of the Guinier region, higher q data is added back incrementally and visualized in the Porod plot (q4 I(q) vs. q) of PRIMUS until an asymptotic region is found. Verification of the asymptotic region can be achieved by plotting the selected data in SASPLOT35. An asymptotic plateau will be observed for folded particles in a q4 I(q) vs. q4 plot and for completely flexible particles in a q2 I(q) vs. q2 plot. In addition, for partially folded particles, an asymptote may be visible in a q3 I(q) vs. q3 plot. The maximum q value that maintains the asymptotic region in the Porod plot defines the upper limit for which the determination of the particle volume should be made in PRIMUS. For further information regarding the determination of the particle’s volume from SAXS data we refer to Glatter, O., and Kratky, O.26, Konarev, P.V., et al35, and Feigin L.A. et al60.
The particle volume (Å3) from homogeneous SAXS sample of known mass can be
used to calculate the particle’s corresponding density (in gm·cm−3) using: where Na is Avogadro’s number at 6.022 × 1023 molecules per mole. The ratios on the
right reduce to a convenient representation: All graphs were prepared with Kaleidagraph (Synergy Software, Reading, PA).
Acknowledgements
The SIBYLS SAXS beamline (BL12.3.1) at the Advanced Light Source is supported by United States Department of Energy program Integrated Diffraction Analysis Technologies DE-AC02-05CH11231. SAXS research on microbial systems and pathways in the Tainer laboratory is supported by supported by the ENIGMA Program of the Department of Energy, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory. SAXS efforts on eukaryotic DNA repair complexes is supported by the National Institutes of Health (NIH) Structural Cell Biology of DNA Repair Machines P01 grant CA92584. We thank SIBYLS staff members G. Williams, G. Hura and M. Hammel for discussions.
REFERENCES
- 1.Rambo RP, Tainer JA. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 3.Dunker AK, Silman I, Uversky VN, Sussman JL. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Proc Natl Acad Sci U S A. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Receveur V, Czjzek M, Schulein M, Panine P, Henrissat B. J Biol Chem. 2002;277:40887–40892. doi: 10.1074/jbc.M205404200. [DOI] [PubMed] [Google Scholar]
- 6.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pretto DI, Tsutakawa S, Brosey CA, Castillo A, Chagot ME, Smith JA, Tainer JA, Chazin WJ. Biochemistry. 49:2880–2889. doi: 10.1021/bi9019934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Biochemistry. 49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. J Biol Chem. 2009;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagata A, Tainer JA. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, Volkmann N, Tsuruta H, Blundell TL, Venkitaraman AR, Tainer JA. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 15.Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, 2nd, Jenney FE, Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, Trauger SA, Kalisiak E, Apon JV, Siuzdak G, Yannone SM, Tainer JA, Adams MW. Nature. 2010;466:779–782. doi: 10.1038/nature09265. [DOI] [PubMed] [Google Scholar]
- 16.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, Chance MR. J Mol Biol. 2004;343:1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 17.DiDonato M, Craig L, Huff ME, Thayer MM, Cardoso RM, Kassmann CJ, Lo TP, Bruns CK, Powers ET, Kelly JW, Getzoff ED, Tainer JA. J Mol Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 18.Shin DS, Didonato M, Barondeau DP, Hura GL, Hitomi C, Berglund JA, Getzoff ED, Cary SC, Tainer JA. J Mol Biol. 2009;385:1534–1555. doi: 10.1016/j.jmb.2008.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karunatilaka KS, Solem A, Pyle AM, Rueda D. Nature. 467:935–939. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. Nat Struct Mol Biol. 2009;16:509–516. doi: 10.1038/nsmb.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selmer M, Al-Karadaghi S, Hirokawa G, Kaji A, Liljas A. Science. 1999;286:2349–2352. doi: 10.1126/science.286.5448.2349. [DOI] [PubMed] [Google Scholar]
- 22.Putnam CD, Tainer JA. DNA Repair (Amst) 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Petoukhov MV, Svergun DI. Curr Opin Struct Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putnam CD, Hammel M, Hura GL, Tainer JA. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 26.Glatter O, Kratky O. Small angle x-ray scattering. London; New York: Academic Press; 1982. [Google Scholar]
- 27.Guinier A. Ann Phys. 1939;12:76. [Google Scholar]
- 28.Mylonas E, Svergun DI. Journal of Applied Crystallography. 2007;40:s245–s249. [Google Scholar]
- 29.Fischer H, de Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. Journal of Applied Crystallography. 2009;43:101–109. [Google Scholar]
- 30.Koch MH, Vachette P, Svergun DI. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 31.Durand D, Vives C, Cannella D, Perez J, Pebay-Peyroula E, Vachette P, Fieschi F. J Struct Biol. 169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Porod G. Kolloid Zeitschrift. 1951;124:31. [Google Scholar]
- 33.Debye P, Anderson JHR, Brumberger H. Journal of Applied Physics. 1957;28:679–683. [Google Scholar]
- 34.Ruland W. Journal of Applied Crystallography. 1971;4:70–73. [Google Scholar]
- 35.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 36.Quillin ML, Wingfield PT, Matthews BW. Proc Natl Acad Sci U S A. 2006;103:19749–19753. doi: 10.1073/pnas.0609442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Squire PG, Himmel ME. Arch Biochem Biophys. 1979;196:165–177. doi: 10.1016/0003-9861(79)90563-0. [DOI] [PubMed] [Google Scholar]
- 38.Kozak M. Journal of Applied Crystallography. 2005;38:4. doi: 10.1107/S002188980502649X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambo RP, Tainer JA. RNA. 2010;16:638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai A, Limbo O, Guenther G, SilDas S, Hammel M, Russel P, Tainer J. A. Nat Struct Biol. 2011 doi: 10.1038/nsmb.2038. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franke D, Svergun D. Journal of Applied Crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun DI, Petoukhov MV, Koch MH. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews BW. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 46.Mareuil F, Sizun C, Perez J, Schoenauer M, Lallemand JY, Bontems F. Eur Biophys J. 2007;37:95–104. doi: 10.1007/s00249-007-0170-2. [DOI] [PubMed] [Google Scholar]
- 47.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Proc Natl Acad Sci U S A. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huffman JL, Sundheim O, Tainer JA. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Tsutakawa SE, Classen S, Chapados BR, Arvai A, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. Cell. 2011 doi: 10.1016/j.cell.2011.03.004. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Mol Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 51.Tainer JA, McCammon JA, Ivanov I. J Am Chem Soc. 132:7372–7378. doi: 10.1021/ja100365x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Curr Opin Struct Biol. 2010;20:283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tainer JA, Getzoff ED, Alexander H, Houghten RA, Olson AJ, Lerner RA, Hendrickson WA. Nature. 1984;312:127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- 55.Fieser TM, Tainer JA, Geysen HM, Houghten RA, Lerner RA. Proc Natl Acad Sci U S A. 1987;84:8568–8572. doi: 10.1073/pnas.84.23.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallewell RA, Laria I, Tabrizi A, Carlin G, Getzoff ED, Tainer JA, Cousens LS, Mullenbach GT. J Biol Chem. 1989;264:5260–5268. [PubMed] [Google Scholar]
- 57.Schnecke V, Swanson CA, Getzoff ED, Tainer JA, Kuhn LA. Proteins. 1998;33:74–87. [PubMed] [Google Scholar]
- 58.Garcin ED, Arvai AS, Rosenfeld RJ, Kroeger MD, Crane BR, Andersson G, Andrews G, Hamley PJ, Mallinder PR, Nicholls DJ, St-Gallay SA, Tinker AC, Gensmantel NP, Mete A, Cheshire DR, Connolly S, Stuehr DJ, Aberg A, Wallace AV, Tainer JA, Getzoff ED. Nat Chem Biol. 2008;4:700–707. doi: 10.1038/nchembio.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das R, Baker D. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 60.Svergun DI, Fe*igin LA, Taylor GW. Structure analysis by small-angle x-ray and neutron scattering. New York: Plenum Press; 1987. [Google Scholar]