SUMMARY
Protein kinase C (PKC) isozymes are the paradigmatic effectors of lipid signaling. PKCs translocate to cell membranes and are allosterically activated upon binding of the lipid diacylglycerol to their C1A and C1B domains. The crystal structure of full-length protein kinase C βII was determined at 4.0 Å, revealing the conformation of an unexpected intermediate in the activation pathway. Here, the kinase active site is accessible to substrate, yet the conformation of the active site corresponds to a low-activity state because the ATP-binding side-chain of Phe629 of the conserved NFD motif is displaced. The C1B domain clamps the NFD helix in a low activity conformation, which is reversed upon membrane binding. A low resolution solution structure of the closed conformation of PKCβII was derived from small angle x-ray scattering. Together, these results show how PKCβII is allosterically in two steps, with the second step defining a novel protein kinase regulatory mechanism.
Introduction
The formation of small molecule second messengers is one of the most important consequences of the activation of cell surface receptors. Many of the most prominent second messengers are lipids, and the paradigmatic lipid second messenger is sn-1,2 diacylglycerol (DAG). DAG activates a variety of cellular effectors, including kinases and GAPs and GEFs for small G-proteins (Yang and Kazanietz, 2003). By far the most widely distributed class of DAG effectors are the protein kinase C (PKC) isozymes (Rosse et al., 2010). Intensively studied for the past three decades, PKCs are the archetypal allosteric transducer of lipid second messenger signaling (Newton, 1995; Nishizuka, 1992). In keeping with their widespread tissue distribution, PKCs regulate a remarkable range of physiological pathways, including, but not limited to, T-cell recognition, cell polarity, cell migration, proliferation and differentiation, neuronal signaling, and metabolism (Rosse et al., 2010).
PKCs are serine/threonine kinases of the AGC family (Pearce et al., 2010). The AGC family includes protein kinases A, B, C, D and G and is characterized by a C-terminal extension of the kinase domain that contains one or two regulatory phosphorylation sites, important for kinase activity (Pearce et al., 2010). Like other kinases, PKCs require phosphorylation of a conserved Ser/Thr in the activation loop for activity (Pearce et al., 2010). PKCs are grouped into subclasses based on the domain composition of the regulatory portion and their respective co-factor requirements (Hurley and Grobler, 1997; Mellor and Parker, 1998; Newton, 1995). The conventional PKCs (α, βI, βII, γ) are regulated via two DAG-binding C1 domains (Hurley et al., 1997) and a Ca2+ and phospholipid-binding C2 domain (Nalefski and Falke, 1996) (Figure 1A). The novel, Ca2+-independent PKCs (δ, ε, η, θ) have an N-terminal C2 domain that does not bind Ca2+ or phospholipids, and two typical DAG-binding C1 domains. The Ca2+/DAG-independent atypical isoforms (ζ, λ/ι) have a single atypical C1 domain that does not bind DAG and lack a C2 domain. All PKCs have a pseudosubstrate region, in which the phosphorylatable Ser/Thr is replaced by an Ala, which maintains the inactive state in the absence of an activating signal (Newton, 1995; Orr and Newton, 1994). PKCs are primed for activation by the phosphorylation of three residues (Tsutakawa et al., 1995). First, the activation loop Ser/Thr is phosphorylated by PDK1 (Chou et al., 1998; Le Good et al., 1998), and then the C-terminal turn and hydrophobic motif are phosphorylated by mTORC2 (Facchinetti et al., 2008; Ikenoue et al., 2008). Primed PKCs are activated to phosphorylate their substrates when their regulatory domains engage the appropriate combination of signals. Signal engagement triggers the release of the pseudosubstrate sequence from the active site, allowing access to substrates. In the case of the conventional PKCs, these signals are DAG, Ca2+, and phospholipids (Hurley and Grobler, 1997; Mellor and Parker, 1998; Newton, 1995).
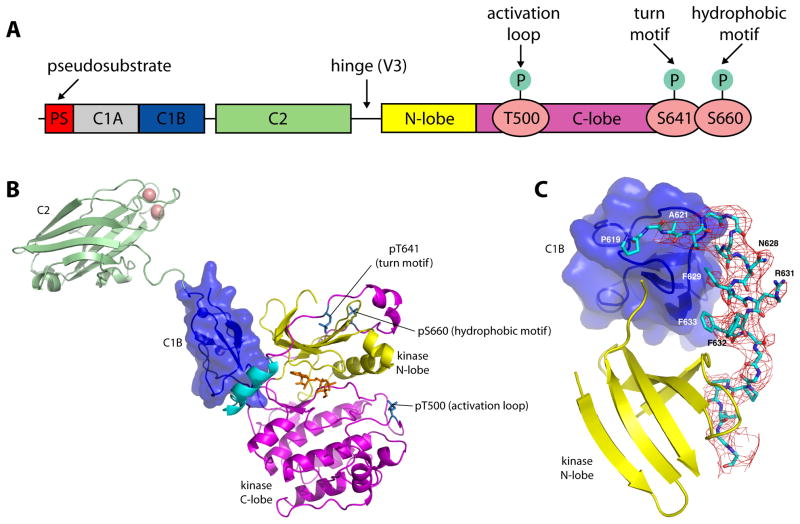
Figure 1. Structure of PKCβII.
(A) Schematic of the domain structure of PKCβII. (B) Structure of the ordered portion of full-length PKCβII, comprising the C1B (blue), C2 (green) and kinase domains. Magenta denotes the C-lobe, and yellow the N-lobe of the kinase domain. The NFD helix is colored cyan, calcium ions salmon, and AMPPNP orange. Phospho-Thr500, Thr641 and Ser660 are shown in a stick model. (C) Difference map (Fo−Fc) phased using a model subjected to a single pass of DEN-restrained refinement, prior to inclusion of residues 620-639. The map is contoured at 2σ.
The question of how signal engagement triggers kinase activation at the structural level has preoccupied many laboratories, and a large body of fragmentary structural information is available for the isolated domains of PKCs. Structures have been solved of the catalytic domains of PKCs βII, θ, and ι (Grodsky et al., 2006; Messerschmidt et al., 2005; Takimura et al., 2010; Xu et al., 2004), the C2 domains of PKCs α, βII, δ, η, and ε (Guerrero-Valero et al., 2009; Littler et al., 2006; Ochoa et al., 2001; Pappa et al., 1998; Sutton and Sprang, 1998; Verdaguer et al., 1999), and C1 domains from PKCα (Hommel et al., 1994), γ (Xu et al., 1997), and δ (Zhang et al., 1995) alone or bound to the DAG-mimetic phorbol ester. This information has been difficult to integrate into a high-resolution picture of the activation pathway because of various technical challenges in the crystallization of full-length PKCs. In addition to the usual challenges of crystallization of multi-domain proteins, suitable PKC samples for crystallization must be stoichiometrically phosphorylated at the activation loop, turn, and hydrophobic sites. Proteolysis in the highly labile V3 region connecting the regulatory and catalytic domains must be avoided. The Zn2+ ions required for the stability of the C1 domains must be retained. We optimized sample purification according to these criteria and were able to crystallize full-length rat PKCβII, an isozyme that has been the subject of especially intensive mechanistic analysis. Moreover, PKCβII is the target for the investigational diabetes drug ruboxistaurin (Das Evcimen and King, 2007).
Crystals of PKCβII diffracted only to 4.0 Å, but by taking advantage of improved methodology for low resolution crystallographic refinement, the data proved adequate to map a previously unobserved conformation of a helix encompassing the conserved NFD motif of the AGC kinase family. Unexpectedly, lattice contacts between the C2 and catalytic domains led to the observation of what appeared to be a partially activated conformation. We were able to confirm the structural inference that the observed conformation is part of the physiological activation pathway by mutational analysis of PKCβII translocation. Thus, this structure provides a snapshot of an intermediate in the lipid activation pathway of PKCβII. The analysis identified an unexpected mechanism of allosteric regulation through plasticity of the NFD motif region. In order to fill out the structural picture of the PKCβII activation pathway, small angle x-ray scattering (SAXS) was used, in conjuction with constraints provided by the crystal structure, to determine a low-resolution structure of the closed, autoinhibited conformation. Together, these structural analyses allow us to map out a conformational activation pathway that is more complex than anticipated.
RESULTS
Structure of PKCβII
PKCβII samples were confirmed for integrity with respect to intact primary structure, stoichiometric Zn binding, phorbol ester-dependent membrane binding and kinase activity, phosphorylation at the three canonical sites on the catalytic domain, and monodispersity (Figure S1). Crystals of a maximum dimension of 20 μm were grown, and diffraction data were collected to 4.0 Å on the GM/CA-CAT dedicated microbeam (ID23-B) at the Advanced Photon Source (APS) and the structure was solved by molecular replacement using the coordinates of the PKCβII kinase domain (Grodsky et al., 2006) (PDB: 2I0E) and the PKCβII C2 domain (Sutton and Sprang, 1998) (PDB: 1A25). Electron density was visible for the kinase domain, the C2 domain and a single C1 domain (Figure 1B, C, Figure S2A). The identity and orientation of the C1 domain was confirmed using an anomalous difference Fourier synthesis to locate the two native Zn atoms (Figure S2B). Following refinement, density for the C1-C2 linker became evident. The short, direct connection between the C1 and C2 domains identified the single ordered C1 domain as the C1B domain and confirmed that these domains belonged to the same PKCβII molecule. The use of deformable elastic network (DEN) (Schroder et al., 2010) restraints for the C2 and catalytic domains, representing the majority of the scattering matter, led to electron density maps of a substantially higher quality than expected at 4.0 Å (Figure 1C).
The initial map calculated following DEN refinement with a model lacking residues 620–639 revealed a novel and well-ordered conformation for this entire region (Figure 1C). The sequence could be assigned to this region, in spite of the limited resolution. Clear side-chain density was visible for Phe629, Phe632, and Phe633, providing an unambiguous set of markers to assign the primary sequence to the model. Residues 624–634 are helical, and the register of these residues was therefore defined with high confidence. Because these residues include the conserved motif NFD (residues 628–630; see Figure 2E), we refer to it as the “NFD helix”. Side-chain features were less obvious for the sections 620–623 and 635–639. However, the main-chain density was nearly continuous and the start and end points of these sections were anchored either to landmarks in the catalytic domain, or to the assigned sequence in the new helix. Therefore these assignments also have a high confidence level. The C1B-C2 connector 151–158 has poorer density and its register is not assigned with high confidence. As described below, the configuration of the C1B-C2 linker in this structure is probably not physiologically relevant, so this limitation of the model does not affect its functional interpretation. The DEN refinement led to an excellent free R-factor of 0.25. Nevertheless, the pseudosubstrate and C1A domains and the V3 (C2-kinase) linker were completely absent from the electron density.
Figure 2. The kinase catalytic domain and the NFD helix.
(A) Comparison of the NFD helix in the isolated PKCβII catalytic domain structure (grey) and the present full-length PKCβII structure, where the NFD helix and adjacent regions are colored cyan. (B) Close-up of the NFD region. (C) Comparison of the NFD region in full length PKCβII (cyan) and the ATP-bound kinase domain of PKCι (orange) (Takimura et al., 2010). (D) Sequence alignment of the NFD region of mammalian PKCs, PKCs from C. elegans and C. albicans (yeast), compared to AGC kinase family members PKA and Akt. An invariant proline corresponding to PKCβII Pro619 is at the start of the AGC-specific tail. The inactive NFD helix of our structure is indicated on the alignment (cyan box corresponds to helix in C), as are the rearranged, active NFD motifs of PKCι (orange boxes correspond to helices in C–D) and Akt2 (orange boxes).
The kinase domain adopts the intermediate open conformation, as also observed in the bisindolymaleimide-bound isolated kinase domain structure (Figure 2A–B) (Grodsky et al., 2006), with minor exceptions noted below. The lattice is stabilized by contacts between the C2 domain of one PKC and two different instances of the catalytic domain (Figure S2A). The Ca2+-binding CBRs of the C2 span the two lobes of the first instance of the catalytic domain (Figure S2A, contact 1). The burial of the C2-bound Ca2+ ions in an interface with the catalytic domain would be inconsistent with the physiological function of these ions in bridging PKC to the membrane (Nalefski and Falke, 1996), therefore we dismissed this lattice contact as functionally irrelevant. The other major lattice contact involves non-conserved polar residues of both the C2 domain and catalytic domain (Figure S2A, contact 2). Because the C2 domain is a conserved and functionally important feature of conventional and novel PKCs, we anticipated that functionally relevant contacts should include at least some conserved residues. This second contact is formed by some of the least conserved regions on both the C2 and catalytic domains and therefore we judged its functional relevance to be implausible. Moreover, the conformation of the C2 domain in this structure is not judged to represent the closed, fully autoinhibited conformation of PKCβII. As described below, this conformation is not consistent with SAXS analysis of PKCβII in solution. Rather, it probably represents a snapshot of the behavior of the C2 domain in the activated state as trapped by lattice contacts. Strong difference density in a phosphate omit map is observed for the activation loop, confirming the phosphorylation of Thr500 (Figure S2C). Difference omit density is also observed for the C-terminal tail of the kinase, confirming the presence of the canonical turn and hydrophobic motif phospho-Thr641 and phospho-Ser660 (Figure S2D, E). The active site contains bound AMPPNP and Mg2+, however, the Mg2+ ions could not be positioned accurately into the model given the limitation of 4.0 Å resolution.
The NFD helix
One of the unexpected features of this structure is the unusual positioning of the residues of the NFD motif (Figure 2), a conserved catalytic element of the AGC kinases. In the present structure, the NFD helix preceding residues in the C-terminal tail of PKCβII are in what we refer to as the “clamped” conformation (Figure 2A–B). The clamp keeps the NFD Phe629 out of the active site, but it is in intimate and extensive contact with both the C1B and catalytic domains. The Phe residue of this motif is normally part of the active site and binds the adenine of ATP, on the basis of the ATP-PKCι complex (Takimura et al., 2010), and it is not part of a helix. We refer to the active conformation of the NFD motif as the “in” conformation (Figure 2C). Grodsky et al., (2006) first reported that this sequence could form a helix, in the context of the isolated PKCβII catalytic domain. In the isolated catalytic domain structure (Grodsky et al., 2006), the Phe is remote from the active site and forms part of a novel helix. We will refer to this as the “out” conformation (Figure 2A). The out conformation has no known regulatory relevance, but does illustrate that this region has an intrinsic propensity to form a helix under appropriate conditions.
C1B contacts with the catalytic domain mimic membrane and phorbol ester
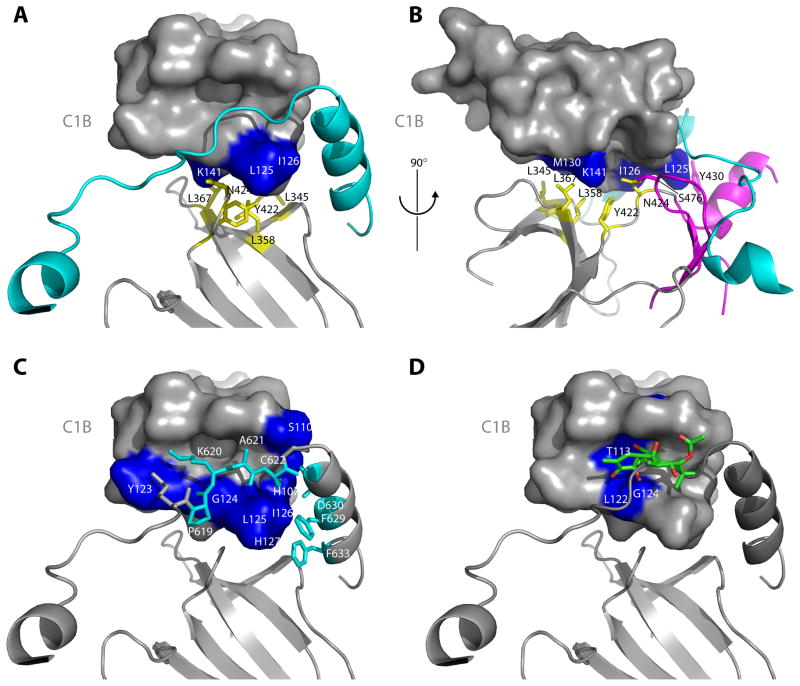
The C1B and catalytic domain have an extensive interface made up mainly of conserved hydrophobic residues. On the catalytic domain side, the contact involves a conserved and mostly non-polar region on the outer face of the N-lobe (Figure 3A–B). On the C1B side, residues from the conserved hydrophobic tip of the domain, and adjoining residues, make contacts with the catalytic domain and NFD helix. In particular, C1B membrane-penetrating side-chains of residues Leu125 and Ile126 make extensive contacts with Phe629 of the NFD adenine-binding motif. Finally, residues 619–622, immediately preceding the start of the NFD helix, fill the DAG binding site of the C1B domain (Figure 3C). The Pro619 carbonyl appears to accept a hydrogen bond from the main-chain amide of Gly124, replacing the 3-oxygen of phorbol ester as seen bound to the C1B domain of PKCδ (Zhang et al., 1995). The Ala621 main chain NH appears to donate a hydrogen bond to the main-chain carbonyl of Gly124, serving as a counterpart of the 4-hydroxyl of phorbol ester (Figure 3D). The C1B domain buries a total solvent accessible area of 1073 Å2 in all contacts with the rest of PKCβII.
Figure 3. The C1B clamp.
(A–B) The interface between the C1B domain and the N-lobe of the catalytic domain stabilizes the clamp by positioning the C1B appropriately with respect to the NFD helix. This interface is predominantly hydrophobic, with the N-lobe side chains of Leu345, Leu358, Leu367 and Y422 (yellow, stick model) packing against Leu125, Ile126 and K141 (blue, surface model) of the C1B domain. Y430 is contributed by the C-lobe of the kinase domain (magenta). (C) The C1B domain clamps the NFD helix and residues immediately N-terminal to the helix. Residues Pro619 – Cys622 (cyan, stick model) are in an extended conformation that makes intimate contacts with the phorbol ester binding cleft of the C1B domain. (D) Comparison of the C1B domain of PKCδ bound to PMA (Zhang et al., 1995) with that of PKCβII shows a steric clash between phorbol ester and residues 619–622 running through the cleft.
C1B clamps the novel NFD helix of the catalytic domain in a low-activity conformation
In the isolated PKCβII catalytic domain structure, residues 627–636 form the NFD helix, which is in the “out” conformation, and has not been visualized in the structures of other kinase catalytic domains. Residues 625–626 are disordered in the isolated kinase domain. In the present full-length PKCβII structure, the NFD helix comprises a slightly different set of residues, 624–634, and the helix shifts by up to 16 Å to occupy the clamped conformation (Figure 2A, B). In the closed, catalytically competent conformation of AGC protein kinases, the central Phe of the NFD motif directly contacts the adenine of ATP (Figure 2C–D). In the present structure, Phe629 is 12 Å away from the adenine moiety (Figure 2C). Phe629 corresponds to Phe327 in PKA (Figure 2D). This Phe is conserved throughout the AGC kinases. In PKA, Phe327 contacts the adenine of ATP. For PKA, mutation of Phe327 weakens the interaction with ATP and lowers catalytic efficiency 50-fold by increasing the Km for ATP (Yang et al., 2009). The only structure available of an isolated PKC catalytic domain bound to ATP is that of PKCι, where the equivalent residue, Phe543 (Figure 2C–E), directly contacts the adenine (Takimura et al., 2010). In the PKCι-ATP complex, the NFD helix is unwound in the middle, with the Phe and Asp of the NFD motif both in an extended conformation. The residues immediately preceding them, 539–542, are in a 310-helical conformation oriented at a roughly 90° angle to the novel helix in full length PKCβII (Figure 2D). Residues 534–542 are in a conformation that sterically overlaps with the PKCβII C1B conformation such that simultaneous occupancy of the fully active ATP-bound and C1B-engaged conformation is not possible. Downstream of the key ATP-binding Phe, one turn of the NFD helix persists in the ATP-bound conformation, but its path is different enough that the counterpart of the anchoring Phe633 in PKCβ a completely different set of contacts.
Mutational analysis of the C1B clamp
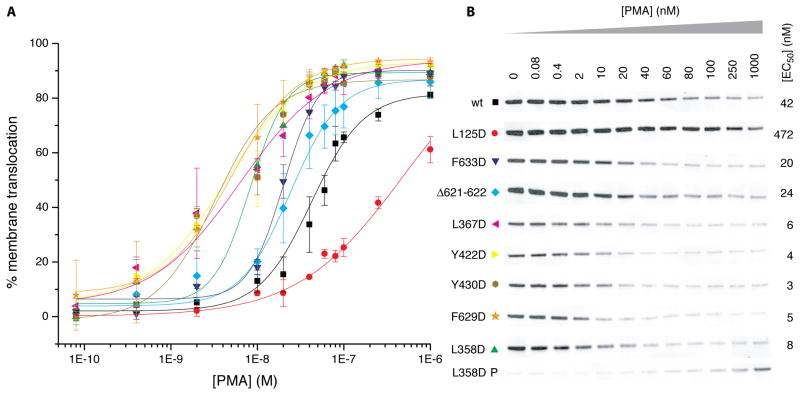
The physiological significance of the C1B clamp was investigated by analyzing the in vivo membrane translocation of wild type PKCβII and mutants designed to destabilize the interfaces between the kinase N-lobe, C1B domain, and AGC-specific C-terminal tail containing the NFD helix. HEK293 cells expressing PKCβII and PDK1 were treated with phorbol 12-myristate 13-acetate (PMA). PMA-dependent membrane translocation was quantitated by the decrease in the cytosolic pool of PKCβII (Figure 4A, B). Deletion of 2 residues in the AGC linker that passes through the cleft of the C1B domain (Δ621–622) results in a two-fold decrease in the concentration of PMA required to mediate half maximal translocation (EC50, Figure 4B). Mutation of residues in the hydrophobic interface between the kinase N-lobe and the C1B domain (L358, L367D, Y422D, Y430D) result in 5- to 14-fold decreases in the EC50 value. Within the NFD helix, F629D and F633D led to 2- to 5-fold decreases in EC50 (PMA). Mutation of Leu125 in the C1B domain, which is part of the C1B clamp interface, was the only mutation that increased EC50 (PMA). Indeed, the increase was dramatic, more than 10-fold. This is consistent with its primary role in membrane binding (Medkova and Cho, 1999; Xu et al., 1997). The destabilization of PKCβII by mutations that disrupt the C1B clamp confirms the physiological significance of this interface in maintaining a secondary mode of autoinhibition in addition to the known mechanism of pseudosubstrate engagement in the active site of the kinase domain.
Figure 4. Mutational analysis of the C1B clamp in PKCβII translocation.
(A) Plot of in vivo membrane translocation of PKCβII against [PMA]. Data points are the mean of two measurements, and errors bars indicate the standard deviation from the mean. (B) PKCβII translocation was quantitated as the reduction in the cytosolic pool of PKC following exposure to PMA. Shown are the Western blots for wild type PKCβII and the indicated mutants. The intensity of the normalized bands is plotted in A. EC50 values are reported for each mutant. Both soluble cytosolic (S) and membrane pellet (P) fractions are shown for L358D.
Solution Structure of PKCβII from Small Angle X-Ray Scattering
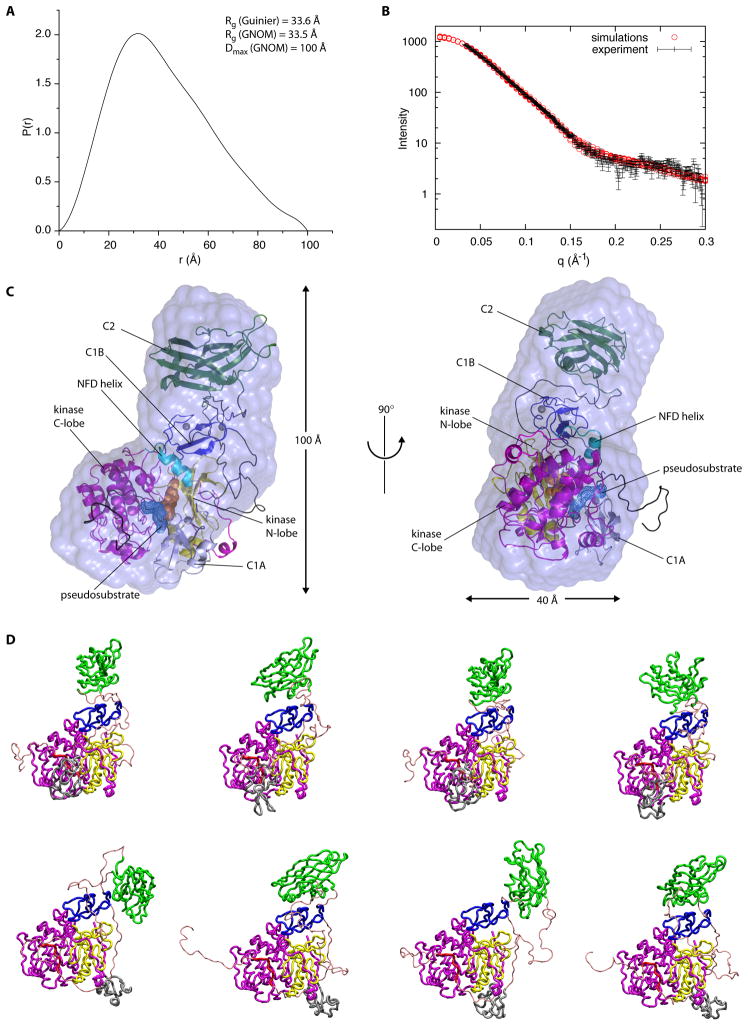
The solution structure of full-length PKCβII was analyzed using small angle x-ray scattering (SAXS; Figure 5). Ab initio determination of the molecular envelope converged on a radius of gyration (RG) of 33 Å and a maximum particle dimension of ~100 Å (Figure 5A). The envelope did not, on its own, contain sufficient detail to position the four structural domains (C1A, C1B, C2, and kinase). Therefore, a new computational procedure, ensemble refinement of SAXS (Rozycki et al., in press) was applied in order to determine the best fit of the structural domains and inter-domain linkers of PKCβII to the experimental SAXS curve, subject to the constraints imposed by the position of the C1B domain in the crystal structure. The pseudosubstrate was modeled in the conformation seen for the interaction between the catalytic and regulatory subunits of PKA (Kim et al., 2007; Kim et al., 2005). The position of the C1A was constrained by the shortness of the 8-residue pseudosubstrate-C1A and 15-residue C1A-C1B linkers. The C1B and kinase domains were held fixed in their crystallographic conformation. The C2 domain N-terminus was constrained by the 6-residue C1B-C2 connector. The C2 domain C-terminus was essentially unconstrained by the 46-residue V3 loop connecting it to the catalytic domain. The conformational space occupied by these linked domains was exhaustively sampled and the conformations that best fit the SAXS data were selected (Figure 5D). The conformations that provided acceptable fits to the SAXS I(q) curve (Figure 5B) also led to acceptable fits within the ab initio molecular envelope (Figure 5C). All had in common a conformation in which the C2 domain projected away from the rest of PKCβII. In each case, the C2 domain made contact with the C1B domain and/or associated linker regions, but not with the catalytic or C1A domains. More compact conformations of the C2 domain relative to the rest of PKCβII did not lead to acceptable fits, nor did attempts to fit a single compact conformation combined with an ensemble of highly open structures. In solution, the conformation of PKCβII is best described as a single closed, autoinhibited state in which the C2 domain projects away from, and has limited contact with, the rest of the structure.
Figure 5. SAXS of PKCβII.
(A) Pair distribution function (P(r)) for autoinhibited PKCβII. The radius of gyration is estimated to be 33.6 Å and 33.5 Å by Guinier analysis and the P(r) function respectively. The maximum dimension of the particle is estimated to be 100 Å. (B) Fit of the simulated scattering curves (I(q)) to the observed scattering of autoinhibited PKCβII. Experimental I(q) data points represent the mean of ten consecutive measurements of the same sample, and the error bars represent the standard error of the mean. (C) Superimposition of the best fit solution to the SAXS curve with an ab initio molecular envelope generated by DAMMIF, in two views rotated by 90 degrees from one another. The assembly of PKCβII kinase and regulatory domains fits inside the envelope. (D) A sample of the top solutions to the simulation of full length, autoinhibited PKCβII indicating the uncertainty in the placement of the C1A domain (shown in blue).
DISCUSSION
The central finding in this study is that the NFD helix is a linchpin of the regulation of PKCβII. The structural observation that the C1B domain clamps the NFD helix in an inactive conformation originated with the unexpected observation of a partially open conformation for PKCβII. The relevance of this mechanism for PKCβII in vivo was corroborated by mutational analysis of contact residues on both the C1B domain and NFD helix. It was expected that destabilizing the C1B clamp would promote its disengagement from the NFD helix and the rest of the catalytic domain, lowering the energy barrier for translocation, and therefore lowering EC50 (PMA). Changing the register of the AGC linker in the cleft of the C1B domain, mutation of the hydrophobic side chains of Ph629 and Phe633 of the NFD helix, and mutations in the core of the N-lobe:C1B interface all serve to destabilize PKCβII to such an extent that the EC50 values 14-fold. These EC50 shifts provide strong corroboration for the structural mechanism of regulation by NFD helix clamping.
The phorbol ester and DAG binding site on the C1B domain is surrounded on three sides by an extremely hydrophobic rim formed by the exposed side chains of (PKCβII numbering) Pro112, Phe114, Tyr123, and Leu125. The counterparts of these residues in related PKCs have been shown to contact lipid tails by NMR in lipid micelles (Xu et al., 1997) and to penetrate the hydrocarbon core of membrane bilayers by surface pressure studies (Medkova and Cho, 1999). The burial of Tyr123 and Leu125 against the catalytic domain in the present partially open structure renders them unavailable for membrane penetration. This situation is similar to the burial of the membrane-binding surface of the C1 domain in the inactive conformation of β2-chimaerin (Canagarajah et al., 2004). The C1 domain of β2-chimaerin buries 1530 Å2 of solvent accessible surface area in contacts with the rest of the molecule, almost 50 % more than for the PKCβII C1B. The more extensive C1 domain burial in β2-chimaerin helps explain why its EC50 for PMA-induced translocation is so much higher than for PKCβII, 1.2 μM as compared to 42 nM under otherwise similar conditions. It also may explain in part why mutations that disrupt the β2-chimaerin C1 domain interface yield larger EC50 decreases of up to 80-fold, as compared to a maximum of ~ 14-fold for PKCβII. A full explanation of these differences will require the structure of the completely closed conformation of PKCβII and an assessment of the burial of the membrane-binding surface of the C1A domain in that conformation. It is notable that, in contrast to all other mutants tested, L125D shifts the PMA dose-response curve to the right. This mutant is expected to weaken membrane binding by the C1B domain at the same time as it destabilizes the clamp. The right-shift produced by L125D outweighs the potential left-shift caused by destabilization of the C1B clamp. The role of the membrane binding site residues in clamping the inactive state highlights that dual and opposing effects are to be expected upon mutating these residues. This duality probably underlies the sometimes contradictory conclusions of various studies employing mutations to dissect the relative roles of PKC C1A and C1B domains in membrane translocation (Bogi et al., 1999; Szallasi et al., 1996) and lipid activation (Medkova and Cho, 1999).
Upon activation, most of the NFD helix unwinds in order for Phe629 to insert itself into the active site. This highlights the NFD region as one of the most dynamic parts of the catalytic domain and one of the most important for regulation of PKCβII. The conservation of key residues involved in the clamp, such as Phe633, suggests that NFD helix regulation extends in evolution as far as the aPKCs and Akts. The aPKCs and Akts lack a C1B domain, and indeed the residues that insert into the phorbol ester binding site of the C1B are not as well conserved. It is unclear what other domain or partner might take the place of the C1B domain of the cPKCs and nPKCs in this context. Conservation of the clamp residues is weaker still in PKA, where the present conformation of the NFD helix is not observed, even in the inactive holoenzyme (Kim et al., 2005). Observation of the NFD helix has only been reported once before (Grodsky et al., 2006), at which time its significance was not clear. These results beg the question as to why NFD helix-mediated regulation has not been reported in other kinases, and whether it is unique to the PKCs and perhaps their closest relatives. The αC helix of the PKCs appears to be unusually rigid, having been observed in the catalytically active conformation in all PKC structures to date (Grodsky et al., 2006; Messerschmidt et al., 2005; Takimura et al., 2010; Xu et al., 2004). Perhaps NFD helix regulation of PKC evolved as a substitute for what appears to be an inoperative αC helix mechanism in this kinase subfamily. There have been three major structural mechanisms described for regulation of kinase activity: steric blockage of the active site; modulation of the activation loop; and positioning of the αC helix (Huse and Kuriyan, 2002). To this list, we propose adding a fourth mechanism for the PKCs and perhaps extending to the Akt/PKBs, regulation by clamping of the NFD helix.
Solution structural analysis by SAXS is consistent with the long-standing model for PKC activation, in which displacement of the pseudosubstrate sequence is coupled to lipid binding by the C1 domains and their displacement from contacts with the catalytic domain (Hurley and Grobler, 1997; Newton, 1995; Orr and Newton, 1994). In models for the activation of the Ca2+-dependent cPKCs α, βI, βII, and γ, the C2 domain rapidly engages with the membrane following Ca2+ binding to the C2 domain CBRs (Nalefski and Newton, 2001; Oancea and Meyer, 1998). C2 binding initiates two-dimensional diffusion of the cPKC on the membrane surface until the cPKC dissociates or encounters a DAG molecule. The exposure of the Ca2+-binding CBRs of the C2 domain in the SAXS structure of PKCβII is consistent with such a function for the C2 domain. The current structural results suggest that the next step in cPKC activation is DAG engagement with the C1A domain, which both stabilizes membrane residency and pulls the pseudosubstrate out of the active site (Figure 6). However, this is still insufficient for full activation, which requires yet another step. This last step is the binding of the C1B domain to the membrane, unclamping of the NFD helix, and engagement of Phe629 into the active site (Figure 6). Taken together, the crystallographic and SAXS analysis leads to a picture of PKC activation that is fully consistent with long-standing models for pseudosubstrate displacement (Newton, 1995; Orr and Newton, 1994), while at the same time revealing a novel and unexpected role for the NFD motif region as an additional mode of allosteric regulation.
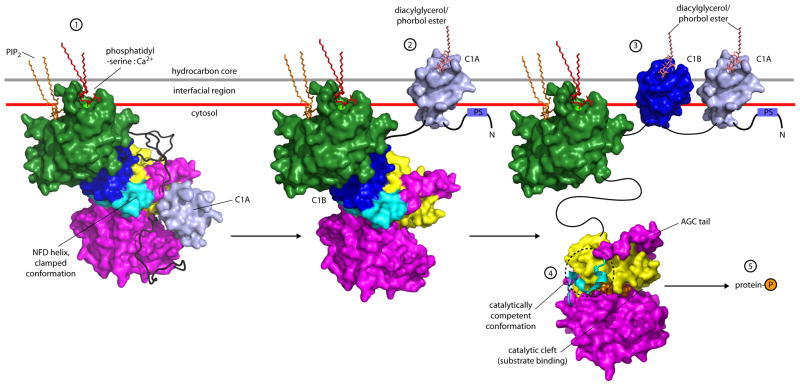
Figure 6. Model for multistep activation of PKCβII.
PKCβII translocates to the membrane upon Ca2+ release in the cell, where the calcium binding regions of its C2 domain mediate bridging to the anionic phospholipid phosphatidylserine, with an adjoining site on the C2 domain binding phosphatidylinositol (4,5)-bisphosphate (1). Subsequent binding of DAG to the C1A domain results in disengagement of the C1A domain (2), which in turn forces the removal of the pseudosubstrate (PS) from the catalytic cleft. Binding of a second molecule of DAG by the C1B domain results in unclamping of the kinase:C1B assembly (3) and rearrangement of the NFD helix into the catalytically competent state (4), triggering the phosphorylation of cellular targets (5) and subsequent downstream signaling. The C2 domain was docked to the membrane in the geometry described by (Landgraf et al., 2008), and the C1 domains as modeled by (Zhang et al., 1995).
EXPERIMENTAL PROCEDURES
Protein expression and purification
Homo sapiens PDK-1 and Rattus norvegicus PKCβII containing a TEV-cleavable N-terminal glutathione S-transferase (GST) tag were cloned into the P10 and PH cassettes respectively of pFastBac Dual (Invitrogen). Wild-type and a mutant PKCβII in which three surface-exposed Cys had been mutated to Ser (C70S/C217S/C622S, the “SEC” mutant) were subcloned. Recombinant bacmid was generated by transforming DH10 MultiBac cells (Berger et al., 2004), and the Sf9 cells were transfected with the bacmid to generate baculovirus. Hi5 cells were infected with the baculovirus and grown in Express Five (Invitrogen) medium supplemented with 40 μM ZnCl2 for 72 h at 28°C. For purification, cells were lysed in 50 mM Tris, pH 7.4, 300 mM NaCl, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1 mM TCEP, 2 mM benzamidine, 2 μg/ml leupeptin, 0.5 mM sodium orthovanadate, 0.5% (w/v) CHAPS, and 1:100 protease inhibitor cocktail (Sigma P8849). All steps were performed at 4°C. The lysate was spun at 43,152 × g, 45 min to pellet insoluble material. The supernatant was incubated with glutathione sepharose (GS-4B; GE Healthcare), 2 h, then washed with 100 ml lysis buffer, 1 h, followed by 100 ml of TEV cleavage buffer (50 mM Tris, pH 7.4, 50 mM NaF, 5 mM sodium pyrosphophate, 10 mM β-glycerophosphate, 1 mM TCEP, 0.5 mM sodium orthovanadate, 0.25 % (w/v) CHAPS. GST- PKCβII was cleaved on-resin with TEV protease overnight. The supernatant from the TEV cleavage reaction was applied to a Q-sepharose anion exchange column (GE Healthcare) and PKCβII eluted with a linear NaCl gradient in 50 mM Tris, pH 8.0, 1 mM TCEP. Fractions containing PKCβII were pooled, concentrated, and loaded onto a Superdex 200 (10/300 GL; GE Healthcare) gel filtration column equilibrated in 20 mM Tris, pH 8.0, 100 mM NaCl, 2 mM MgCl2, 1 mM TCEP. PKCβII eluted as a single peak with a retention time consistent with a globular, monomeric protein of 78 kDa.
Crystallization and structure determination
Crystals of wild-type and SEC mutant PKCβII grew over two days from fresh protein preparations. SDS-PAGE analysis of the drops from which crystals were harvested showed no indication of PKCβII degradation (Figure 1A). PKCβII was combined with 1 mM AMPPNP (Sigma) and concentrated to 2 mg/ml. Both wild-type and SEC proteins crystallized, however the best diffractors were obtained for the SEC mutant. Hexagonal crystals of the SEC mutant were grown in 1–4 % (w/v) PEG 8000, 100 mM Tris, pH 8.5 either attached to the plate or to a skin of denatured protein on the surface of the drop. Multiple crystals were mounted in a loop, cryoprotected in 20% ethylene glycol and drop frozen in liquid nitrogen. Data were collected on the microfocus beamline GM/CA-CAT ID23-B at the Advanced Photon Source (APS). Crystals in the loop were detected and centered in the beam by using the RASTER software to scan the loop for diffraction. A complete data set from a single crystal with unit cell dimensions a = b =114.27 Å, c = 170.84 Å was collected to 4.0 Å resolution in spacegroup P3221. The structure of PKCβII was solved by molecular replacement using the program Phaser (McCoy et al., 2007). The catalytic domain of PKCβII (PDB ID: 2I0E) and the C2 domain of PKCβII (PDB ID: 1A25) were used as search models. The C1B domain was located in the map using the interatomic distance between the zinc ions to help position it, and this was followed by rigid body refinement. No density was observed for the N-terminal pseudosubstrate segment and C1A domain (residues 1–100) and the V3 linker (residues 293–338). Refinement was carried out using the deformable elastic network (DEN) model as implemented in CNS 1.3 (Schroder et al., 2010) using the catalytic and C2 domain as separate restraint groups. The DEN parameters γ and κ were set at 0.5 and 0.1, respectively. Residues 353 and 619–639 of the catalytic domain were omitted from the restraint group. Density syntheses following DEN refinement were instrumental in determining the position of the novel helix. The refined structure has no residues in disallowed regions of the Ramachandran plot and only ten residues in generously allowed regions (Laskowski et al., 1993). The MolProbity (Davis et al., 2007) all atom clash score was 41.6, placing the structure in the 64th percentile (with 100th best) of structures refined at 3.0 Å or lower resolution. The MolProbity protein geometry score was 3.5, placing the structure in the 70th percentile of structures at 3.25–4.25 Å resolution. Clashes and stereochemical outliers were more common in the C1B domain, the C1B-C2 linker, and the NFD helix and adjacent residues, which were not constrained by the DEN model.
In vivo translocation assay
Wild-type and mutant PKCβII proteins were analyzed for their ability to translocate to membranes in a phorbol ester dependent manner in vivo. HEK293 GnTI cells were co-transfected with mammalian expression vectors containing PDK-1 and PKCβII and grown at 37°C for 48 h. Cells were stimulated with 80 pM – 1 μM phorbol 12-myristate 13-acetate (PMA) (Sigma) for 15 min at 37°C, harvested and snap frozen on dry ice. Cells were resuspended in 500 μl PKC lysis buffer, sonicated on low power on ice, 15 sec, 1 sec pulses followed by 1 sec off. The lysate was spun at 900 × g, 5 min, 4°C to discard the organelles and enrich plasma membrane. The supernatant was then transferred to a fresh tube and spun at 100,000 × g, 30 min, 4°C. The supernatant (cytosolic fraction) was transferred to a fresh tube on ice. 10 μl of a 3:1 mixture of cytosolic or membrane fraction with 4× LDS gel loading buffer (Invitrogen) containing 50 mM β-mercaptoethanol was loaded on a 4–12 % polyacrylamide gel (Invitrogen). The gel was blotted onto PVDF and PKCβII detected using an anti- PKCβII primary antibody (BD Biosciences BD610128) and Alexa Fluor 488 conjugated goat anti-rabbit IgG secondary antibody (Invitrogen). Cytosolic PKCβII fractions were quantitated using a Typhoon scanner at 519 nm.
Small angle X-ray scattering (SAXS)
Wild-type PKCβII samples were prepared with and without the non-hydrolyzable nucleotide analog AMPPNP. Data were collected on 25 μM samples in 20 mM Tris, pH 8.0, 100 mM NaCl, 2 mM MgCl2, 1 mM TCEP ± 1 mM AMPPNP at SSRL beamline BL4-2. Data reduction and analysis was performed using the beamline software SAStool. The program AutoGNOM of the ATSAS suite (Petoukhov et al., 2007) was used to generate P(r) curves and to determine Dmax and Rg from the scattering intensity curve (I(q) vs. q) in an automatic, unbiased manner, although rounds of manual fitting in GNOM (Svergun, 1992) were used to verify these values. Ab initio molecular envelopes were computed by the programs DAMMIF (Franke and Svergun, 2009) and GASBOR (Svergun et al., 2001). Multiple iterations of DAMMIF and GASBOR were averaged using DAMAVER (Volkov and Svergun, 2003), the core residues fixed and the model subjected to a further cycle of DAMMIN (Svergun, 1999) refinement.
SAXS structural refinement
Monte Carlo simulations of PKCβII were performed using an energy function and simulation model developed to study multiprotein complexes (Kim and Hummer, 2008; Kim et al., 2008; Ren et al., 2009; Rozycki et al., in press). In this model, folded protein domains are represented as rigid bodies. Here, the catalytic domain together with the bound C1B domain and the pseudosubstrate peptide modeled on the basis of PKA (pdb entries 3FHI and 2QCS) (Kim et al., 2007; Kim et al., 2005) is treated as one rigid body. C1A and C2 domains are modeled as two separate rigid bodies. The interactions between the domains are treated at the residue level with amino-acid-dependent pair potentials and Debye-Hückel-type electrostatic interactions. Flexible linker peptides connecting the rigid domains are represented as amino acid beads on an excluded-volume random-coil polymer. We adapted the CRYSOL algorithm (Svergun et al., 1995) to calculate small angle X-ray scattering (SAXS) intensity I(q) for all simulated structures. CRYSOL default parameters were used in the calculation. To estimate the uncertainty of computed intensity I(q), we varied the hydration layer electron density between 0.015 e/Å3 and 0.03 e/Å3 (Yang et al., 2009). To gain insights into the protein relevant configurations, we selected several structures whose SAXS profiles I(q) best fit the experimental data (Figure 5B–D). We used chi-square as a measure of discrepancy between the computed SAXS curves and experimental data. The error term that enters the χ2 formula contains the statistical error of experiment and the uncertainty of computed intensity I(q). Only structures with χ2 < 1.5 were considered.
Supplementary Material
Table 1.
Crystallographic Data Collection and Refinement Statistics
| Data collection | |
| space group | P3221 |
| unit cell (a, b, c in Å) | 114.27, 114.27, 170.84 |
| resolution range (Å) | 57.1 – 4.0 |
| observations | 46215 |
| unique reflections | 10117 |
| completeness (%)a | 95.0 (88.0) |
| I/σI | 3.5 (1.8) |
| Rsym (%) | 19.2 (39.3) |
| Structure refinement | |
| resolution range | 57.1 – 4.0 Å |
| reflections used | 10670 |
| R-factor, Rfreeb (%) | 19.4 (25.0) |
| model residues | 101–292, 339–669 |
| rms deviations | |
| bond lengths (Å) | 0.012 |
| bond angles (deg) | 0.878 |
Values in parentheses refer to the highest resolution shell.
Rfree = free R-factor based on random 5% of all data.
Acknowledgments
We thank J. Chang for technical assistance, J. Lloyd and D. E. Anderson for assistance with mass spectrometry, J. F. Mushinski and A. Toker for DNAs, E. Boura for assistance with SAXS data collection, H. Tsuruta and T. Weiss for user support at BL4-2, SSRL, M. Becker for user support at GM/CA-CAT, APS, H. Mischak and M. Pearson for early efforts, A. Newton for discussions, and A. Brünger for a prerelease version of CNS 1.3. GM/CA-CAT has been funded in whole or in part with Federal funds from the NCI (Y1-CO-1020) and the NIGMS (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. SAXS data were collected at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program. T. A. L. was supported in part by an EMBO long term fellowship. B. R. was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme. This research was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health to J. H.H. and G. H. Crystallographic coordinates have been deposited in the Protein Data Bank with accession code 3PFQ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- Bogi K, Lorenzo PS, Acs P, Szallasi Z, Wagner GS, Blumberg PM. Comparison of the roles of the C1a and C1b domains of protein kinase C alpha in ligand induced translocation in NIH 3T3 cells. FEBS Lett. 1999;456:27–30. doi: 10.1016/s0014-5793(99)00927-8. [DOI] [PubMed] [Google Scholar]
- Canagarajah B, Collucio Leskow F, Ho YSJ, Mischak H, Saidi L, Kazanietz MG, Hurley JH. Structural Mechanism for Lipid Activation of the Rac-Specific GAP, beta2-Chimaerin. Cell. 2004;119:407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Chou MM, Hou WM, Johnson J, Graham LK, Lee MH, Chen CS, Newton AC, Schaffhausen BS, Toker A. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr Biol. 1998;8:1069–1077. doi: 10.1016/s0960-9822(98)70444-0. [DOI] [PubMed] [Google Scholar]
- Das Evcimen N, King GL. The role of protein kinase C activation and the vascular complications of diabetes. Pharmacol Res. 2007;55:498–510. doi: 10.1016/j.phrs.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti V, Ouyang WM, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao YX, Miao RQ, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodsky N, Li Y, Bouzida D, Love R, Jensen J, Nodes B, Nonomiya J, Grant S. Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry. 2006;45:13970–13981. doi: 10.1021/bi061128h. [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S. Structural and mechanistic insights into the association of PKC alpha-C2 domain to PtdIns(4,5)P-2. Proc Natl Acad Sci U S A. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommel U, Zurini M, Luyten M. Solution Structure of a Cysteine-Rich Domain of Rat Protein- Kinase-C. Nat Struct Biol. 1994;1:383–387. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Grobler JA. Protein kinase C and phospholipase C: Bilayer interactions and regulation. Curr Opin Struct Biol. 1997;7:557–565. doi: 10.1016/s0959-440x(97)80122-4. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RI alpha) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- Kim YC, Hummer G. Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J Mol Biol. 2008;375:1416–1433. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Tang C, Clore GM, Hummer G. Replica exchange simulations of transient encounter complexes in protein-protein association. Proc Natl Acad Sci U S A. 2008;105:12855–12860. doi: 10.1073/pnas.0802460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf KE, Malmberg NJ, Falke JJ. Effect of PIP2 binding on the membrane docking geometry of PKC alpha C2 domain: An EPR site-directed spin-labeling and relaxation study. Biochemistry. 2008;47:8301–8316. doi: 10.1021/bi800711t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK - A PROGRAM TO CHECK THE STEREOCHEMICAL QUALITY OF PROTEIN STRUCTURES. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- Littler DR, Walker JR, She YM, Finerty PJ, Newman EM, Dhe-Paganon S. Structure of human protein kinase C eta (PKC eta) C2 domain and identification of phosphorylation sites. Biochem Biophys Res Commun. 2006;349:1182–1189. doi: 10.1016/j.bbrc.2006.08.160. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medkova M, Cho WH. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J Biol Chem. 1999;274:19852–19861. doi: 10.1074/jbc.274.28.19852. [DOI] [PubMed] [Google Scholar]
- Mellor H, Parker PJ. The extended protein kinase C superfamily. Biochem J. 1998;332:281–292. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt A, Macieira S, Velarde M, Badeker M, Benda C, Jestel A, Brandstetter H, Neuefeind T, Blaesse M. Crystal structure of the catalytic domain of human atypical protein kinase C-iota reveals interaction mode of phosphorylation site in turn motif. J Mol Biol. 2005;352:918–931. doi: 10.1016/j.jmb.2005.07.060. [DOI] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Newton AC. Membrane binding kinetics of protein kinase C beta II mediated by the C2 domain. Biochemistry. 2001;40:13216–13229. doi: 10.1021/bi010761u. [DOI] [PubMed] [Google Scholar]
- Newton AC. Protein-Kinase-C - Structure, Function, and Regulation. J Biol Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Intracellular Signaling by Hydrolysis of Phospholipids and Activation of Protein-Kinase-C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- Ochoa WF, Garcia-Garcia J, Corbalan-Garcia IFS, Verdaguer N, Gomez-Fernandez JC. Structure of the C2 domain from novel protein kinase C epsilon. A membrane binding model for Ca (2+)-independent C2 domains. J Mol Biol. 2001;311:837–849. doi: 10.1006/jmbi.2001.4910. [DOI] [PubMed] [Google Scholar]
- Orr JW, Newton AC. Intrapeptide Regulation of Protein-Kinase-C. J Biol Chem. 1994;269:8383–8387. [PubMed] [Google Scholar]
- Pappa H, Murray-Rust J, Dekker LV, Parker PJ, McDonald NQ. Crystal structure of the C2 domain from protein kinase C-delta. Structure. 1998;6:885–894. doi: 10.1016/s0969-2126(98)00090-2. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nature Reviews Molecular Cell Biology. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr. 2007;40:S223–S228. [Google Scholar]
- Ren X, Kloer DP, Kim YC, Ghirlando R, Saidi LF, Hummer G, Hurley JH. Hybrid structural model of the complete human ESCRT-0 complex. Structure. 2009;17:406–416. doi: 10.1016/j.str.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJM, Boeckeler K, Parker PJ. PKC and the control of localized signal dynamics. Nature Reviews Molecular Cell Biology. 2010;11:103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- Rozycki B, Kim YC, Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. doi: 10.1016/j.str.2010.10.006. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Sprang SR. Structure of the protein kinase C beta phospholipid-binding C2 domain complexed with Ca2+ Structure. 1998;6:1395–1405. doi: 10.1016/s0969-2126(98)00139-7. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szallasi Z, Bogi K, Gohari S, Biro T, Acs P, Blumberg PM. Non-equivalent roles for the first and second zinc fingers of protein kinase C delta - Effect of their mutation on phorbol ester-induced translocation in NIH 3T3 cells. J Biol Chem. 1996;271:18299–18301. doi: 10.1074/jbc.271.31.18299. [DOI] [PubMed] [Google Scholar]
- Takimura T, Kamata K, Fukasawa K, Ohsawa H, Komatani H, Yoshizumi T, Takahashi I, Kotani H, Iwasawa Y. Structures of the PKC-i kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533–551 in the C-terminal tail and their roles in ATP binding. Acta Crystallographica Section D-Biological Crystallography. 2010;66:577–583. doi: 10.1107/S0907444910005639. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Medzihradszky KF, Flint AJ, Burlingame AL, Koshland DE. Determination of in vivo phosphorylation sites in protein kinase C. J Biol Chem. 1995;270:26807–26812. doi: 10.1074/jbc.270.45.26807. [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca2+ bridges the C2 membrane-binding domain of protein kinase C alpha directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RX, Pawelczyk T, Xia TH, Brown SC. NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- Xu ZB, Chaudhary D, Olland S, Wolfrom S, Czerwinski R, Malakian K, Lin L, Stahl ML, McCarthy DJ, Benander C, et al. Catalytic domain crystal structure of protein kinase C-theta (PKC theta) J Biol Chem. 2004;279:50401–50409. doi: 10.1074/jbc.M409216200. [DOI] [PubMed] [Google Scholar]
- Yang CF, Kazanietz MG. Divergence and complexities in DAG signaling: looking beyond PKC. Trends Pharmacol Sci. 2003;24:602–608. doi: 10.1016/j.tips.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Yang J, Kennedy EJ, Wu J, Deal MS, Pennypacker J, Ghosh G, Taylor SS. Contribution of Non-catalytic Core Residues to Activity and Regulation in Protein Kinase A. J Biol Chem. 2009;284:6241–6248. doi: 10.1074/jbc.M805862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GG, Kazanietz MG, Blumberg PM, Hurley JH. Crystal-Structure of the Cys2 Activator-Binding Domain of Protein-Kinase C-Delta in Complex with Phorbol Ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.