In nature, three dimensional (3D) chemical patterns are generated and sustained with precisely controlled spatial and temporal profiles, on a variety of length and time scales.[1,2] Several studies have outlined the need for the development of in vitro methodologies that replicate the 3D spatio-temporal chemical patterns associated with chemotaxis, cell signaling, angiogenesis, homeostasis and immune surveillance.[3–7] There are a number of in vitro microfluidic systems that have been developed to mimic in vivo chemical micro-environments such as the creation of interleukin-8 gradients to study neutrophil chemotaxis.[8] However, microfluidic systems are inherently planar (2D) and their overall size and dependency on external equipment to enable active flow restricts their applicability.[9–11] Hence, the development of passive systems that enable diffusion-based 3D chemical pattern formation is attractive since they can be readily utilized to generate and sustain patterns within cell culture, homogeneous gels and other stationary media. Existing microparticles and reservoirs[12] can be utilized to create chemical patterns in 3D environments, however, the pre-dominant spatial release profile is one that is spherically symmetric[13] (Figure 1a).
Figure 1. Schematic of the proposed methodology for generation of three dimensional chemical patterns.
a–b) Schematic diagrams of conceptual spherical or cylindrical containers with uniform wall porosity. c) Further control over the shape and duration of the chemical distribution is afforded by selective patterning of the pores on the surface of the container. d–f) Schematic diagram of our proposed polyhedral containers that can be designed in a variety of shapes and sizes that approximate, for example, spherical or cylindrical containers. g–i) Precisely patterned 2D panels that can self-fold via surface tension forces into the polyhedral containers; numerical simulations guide pore designs.
Here, we argue that 3D spatio-temporal patterns can be achieved when chemicals are allowed to diffuse out from precisely shaped and patterned hollow containers placed in stationary media. For example, one can vary the shape and symmetry of the overall container as well as the wall porosity pattern to generate a large number of symmetric and asymmetric chemical release profiles. Conceptually, in stationary media, chemical patterns can be generated from spherical or cylindrical geometries by engineering the release rate via control in the wall porosity characteristics (Figure 1a–c). However, it is challenging to fabricate spherical or cylindrical containers with precisely patterned sidewalls (such as that shown in Figure 1c). Alternatively, these geometries can be approximated by polyhedral containers with precisely patterned side-walls. Hollow polyhedral containers (Figure 1d–f) can be constructed using the self-folding[14–17] of patterned 2D panels (Figure 1g–i).
In order to generate a specific 3D chemical pattern we first design a polyhedron with approximately the same size and symmetry as the desired pattern. Numerical simulations modelling the diffusion of the specific chemical release from this polyhedron are carried out with different pore sizes and patterns to determine precise pore dimensions and placement on the side-walls of the polyhedron. The side-wall pore placement is mapped onto 2D panels (Figure 1g–i) that are interconnected with hinges and utilized to self-fold the polyhedron. Since the pores are patterned in 2D, it is possible to utilize extremely precise and well developed patterning methods such as photolithography, electron beam lithography and soft-lithography. Our patterning technique also allows pore patterns designed by numerical simulations to be directly transferred to the computer-aided design (CAD) software used to generate the lithography masks. The fabrication approach is highly parallel, precise, and affords considerable versatility in the shape, size, density and pattern of pores. Elsewhere, we have demonstrated that containers can be constructed with sizes ranging from 100 nanometers[18] to several millimeters, and a variety of material compositions[14,16] Hence, this versatility in fabrication coupled with our present approach could be used to generate chemical patterns on a range of size scales.
To enable diffusion-based chemical pattern release, containers can be easily loaded by immersing them in the desired chemical which diffuses into the container through the pores. The containers are also reusable (the containers can be washed out by immersion in solvents and then reused again by immersion in the desired chemical), mobile, mechanically robust, and can be manipulated with tweezers or pipettes without any breakage. We note that our surface-tension based self-folding method also utilizes liquefying hinges at the periphery of the 2D panels, which results in robust sealing of the edges and corners of the polyhedral containers.[14] In contrast to porous polymer particles[2,19–22] which soak up the chemical within a cross-linked matrix, our containers physically entrap the chemicals.[23] Hence, chemical encapsulation is less susceptible to chemical fouling and less dependent on the molecular properties of the chemical or details of the matrix synthesis process. As a result, the containers can be used to generate patterns with a wide range of chemicals.
Experimentally, we lithographically patterned and self-assembled nickel (Ni) based containers. Subsequently, containers could be coated with gold (Au) to render them chemically and biologically inert[24,25] and to reduce the dimensions of the pores (by varying the thickness of the coating). Elsewhere, we have shown that these containers can also be fabricated with alternate materials such as biocompatible polymers.[16] At the present time, however, metallic containers offer the greatest precision in terms of pore definition and edge sealing while seamless edge sealing of polymeric containers is still challenging. Hence, we utilized metallic containers for this demonstration.
In order to quantify chemical gradient formation, we loaded the containers by immersing them in an aqueous fluorescein solution. Fluorescein, a fluorescent water-soluble chemical with low molecular weight, allowed us to visualize the pattern and also quantify its concentration using fluorescence microscopy. After carefully rinsing the containers to remove excess fluorescein from their exterior, the containers were positioned in a stationary gel and we observed that fluorescein was released from these containers. Comparisons of numerical simulations with observations for the case of planar (2D) chemical patterns (such as linear gradients) were in excellent agreement with experimental data (see Supplementary Note 1 and Figure S1 for complete details).
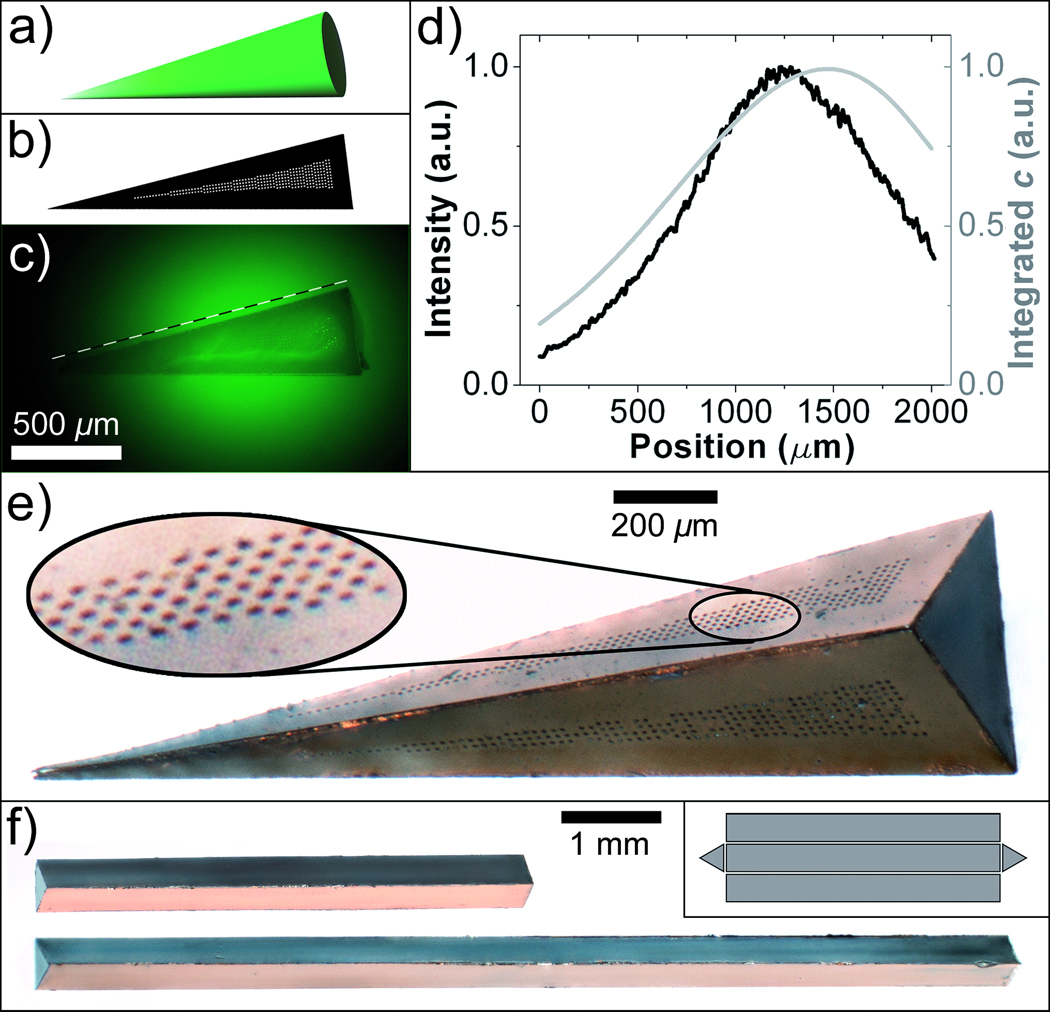
In order to generate 3D spatial patterns, we utilized two conceptually different strategies. In one strategy, the overall shape of the container was chosen to closely match the desired 3D spatial profile. For example, conical gradients were formed when chemicals were released from triangular pyramid shaped containers (Figure 2a–e), and experimentally realized gradients were in agreement with numerical simulations. We note that the container fabrication methodology can be utilized to assemble containers with alternate polyhedral shapes such as triangular prisms with different lengths (Figure 2f) to enable these gradients to be generated over small or large length scales.
Figure 2. Generation of 3D spatial patterns by varying container shape.
a) An idealized illustration of a conical gradient. b) Schematic of a single triangular panel with pores enclosed in a wedge-shaped area that was found in numerical simulations to generate a linear gradient along its length. c) Experimental realization of the gradient using a pyramid-shaped container with triangular faces patterned according to b). d) Plot of the experimentally measured chemical concentration (thick black line) compared with the numerical calculation (thin gray line) along the dashed line shown in (c). e) A magnified optical image of the container used to generate the conical gradient pattern. f) Triangular prism-shaped containers with two different lengths. The inset illustrates the 2D geometry of interconnected panels.
Our second strategy was to engineer the pattern of pores on one or many faces of the container. Here, we utilized the fact that the combination of 2D lithographic patterning and self-folding allowed the container faces to be precisely patterned in all three dimensions. Moreover, this patterning can be achieved in an asymmetric manner. We describe an example of chemical release in the shape of a helix, which is a representative 3D space curve. Since a helix can be thought of as a curve wrapped around a cylinder, we utilized a parallelepiped shaped container. In simulations, one pattern of slits that was found to release chemicals in a helix is shown in Figure 3a. A container with this slit arrangement was fabricated and assembled (Figure 3b). Fluorescein was loaded within this container and when placed in a gel, a helical pattern (Figure 3c) was observed whose spatial profile was in good agreement with the simulations (Figure 3d). Hence, this strategy directs local pattern formation while the previous strategy dictates the global boundaries of the pattern. Together, these two strategies enable the generation of 3D spatial patterns of virtually any shape consistent with the diffusion process in stationary media.
Figure 3. Generation of 3D spatial patterns by varying pore placement.
a) CAD layout showing the 2D panel geometry along with the patterned slits. b) Optical image of a container fabricated with the panel design shown in a). In the assembled container, the slits line up along a helical path. c) Experimentally realized and d) numerical simulation result of the helical spatial pattern of fluorescein.
Our demonstrations provide convincing proof-of-concept that a variety of 3D patterns can be generated and sustained by releasing chemicals from a container. We anticipate that these containers can be utilized for the in vitro generation of chemical patterns that are of importance in cell and tissue functions such as in microbial quorum sensing, organ and embryonic development, angiogenesis, and workings of the immune system. To demonstrate applicability of our concept to in vitro organization of living systems in specific 3D geometries, we present a demonstration of chemotactic organization of bacteria in a well-defined geometric space curve. Our hypothesis was that chemotactic cells could be guided by a chemoattractant pattern generated by the container (Figure 4a–b). Escherichia coli (E. coli) bacteria which are well characterized prokaryotes often used in chemotaxis studies, were chosen for the demonstration. We utilized the chemical helix-generating container (Figure 3b) that was loaded with L-serine, a known chemoattractant for E. coli.[26] The initial chemoattractant concentration inside the container, shape of the container and slit geometry determined the spatio-temporal characteristics (such as the time it would take E. coli to self-organize into a helix as well as cell density variations in the resulting bacterial distribution)[26] of the E. coli pattern. The container was placed in a chamber filled with motility medium and green fluorescent protein (GFP) expressing bacteria were seeded at a low density uniformly around the container. A series of time-lapse fluorescence images (Figure 4c1–c4, Supplementary Video S1) show transitions from a uniform bacterial distribution (Figure 4c1) into a helical bacterial distribution in the vicinity of the container (Figure 4c2–c4). This study demonstrates the applicability of these chemical pattern generating containers for in vitro cell studies.
Figure 4. Directing the chemotactic self-organization of E. coli in the shape of a helix.
a–b) A conceptual schematic of the desired chemotactic self-organization. At the start of the experiment, a) the chemoattractant is confined to the container and the E. coli cells (represented by green ellipsoids) are distributed uniformly throughout the medium. b) E. coli cells self-organize in a helical pattern based on the underlying chemical pattern once the chemo-attractant (yellow) is allowed to diffuse out of the container. c1–c4) Experimental realization of the concept. Time-lapse images of green fluorescent E. coli as they self-organized in a helical pattern around a container with slits as shown in Figure 3. The number indicated in the left corner of each figure is the time lapse after the container was placed into the E. coli containing medium. The underlying chemical pattern was not visualized in this experiment.
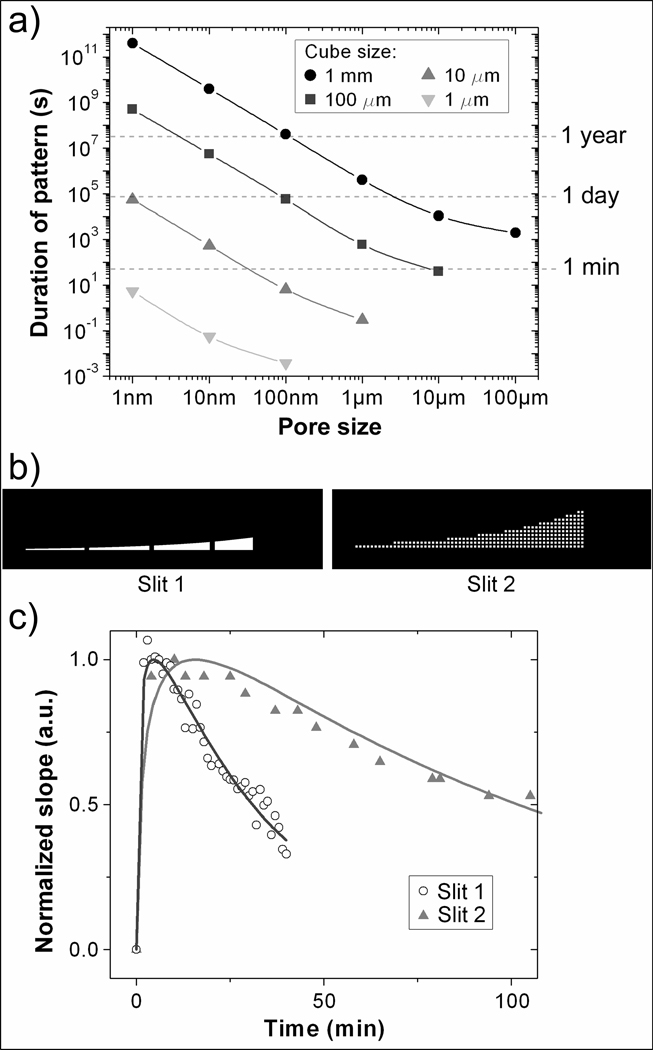
We note that other applications such as studies involving mammalian cells may necessitate the generation of 3D chemical patterns over longer time scales. The temporal stability of the chemical pattern depends on the duration of chemical release from the containers by diffusion. Analytical estimates (see Supplementary Note 2) show that the most straightforward way to alter the maximal duration of chemical release (τ) is by varying the ratio of the container size (d) to the total area of the pores (A): τ~(π/6)×(d3w)/(AD); here w is the container wall thickness and D is the diffusion coefficient whose magnitude depends on the specific chemical and medium. By increasing the container size d to 1 mm and decreasing the total pore area to A=6×104 nm2 the characteristic time scale τ of pattern formation in stationary media, for a molecule with a diffusion constant of D=4.2×10−10 m2/s (corresponding to that of fluorescein dye[27], assuming w=1 µm) is approximately 2×109 s, i.e. comparable to human lifetime. Alternate methods can be utilized to speed up loading of such containers (see Supplementary Note 3).
Numerical simulations corroborate and refine these estimates and enable quantitative temporal data sets to be plotted (Figure 5a) for different combinations of container and pore size. The data set is not restricted to chemicals with diffusion coefficient similar to that of fluorescein. We note that since the diffusion coefficients of most chemicals scale with their molecular weight (Mw) as, D ∝ (Mw)−n, with n~0.5 for water,[28] release times will be longer for molecules larger than fluorescein dye and shorter for smaller molecules. The temporal release characteristics are also affected by the pore distribution (Supplementary Figure S2c–d).
Figure 5. Numerical and experimental results showing temporal control over the chemical patterns.
a) Calculated chemical release times for cubic containers with varying cube and pore size releasing a chemical with D=4.2×10−10 m2/s. The data points plotted correspond to the period at which at least 90% of the maximal concentration is maintained at a distance d away from the center of the container. b) Schematic of two panels with slits which have a similar geometric pattern but with different individual pore size and porosity. c) Experimental (dots) and simulation (solid lines) data showing that duration for a linear spatial gradient is extended by roughly a factor of three for containers with slit 2 as compared to slit 1.
Simulations also confirm that the temporal characteristics can be readily varied while preserving the same spatial release pattern. Experimentally, in 2D, pore size on the panels is limited by the lithographic patterning resolution. However, after self-assembly of metallic containers, pores can be uniformly reduced by Au electrodeposition (Supplementary Figure S3). The technique is not limited to isolated pores, and complicated pore patterns can also be uniformly reduced in size. For example, the slit used to generate linear gradients (Supplementary Figure 1a–b) was broken in CAD into individual similarly sized pores 12 µm in size (Figure 5b; see also Figure 2b and e) which were subsequently shrunk to about ~2 µm by plating gold on the exterior of the assembled container; consequently the porosity was also reduced. Such a “pixilation with subsequent gold electroplating” process preserved the linear shape of the generated chemical pattern while increasing the time over which the gradient persists roughly by a factor of three (Figure 5c). Our experimental demonstration confirms that by utilizing smaller sized pores within the same pattern on the container, one can retain the spatial characteristics of the chemical profile while increasing the time over which the pattern persists.
In summary, we have described an in vitro methodology that provides unprecedented precision and versatility in generation of 3D chemical patterns in stationary media for a variety of applications in chemistry[23], microbiology, cell signalling and tissue development.
In biological applications, patterns can function as chemical scaffolds to study important cellular functions in vitro. The 3D chemical release can also be utilized with single or multiple containers to engineer spatio-temporal characteristics of chemical reactions and hence realize 3D reaction-diffusion based pattern formation. Our containers have relatively simple release kinetics typical of membrane controlled reservoir systems. In our present demonstration, the containers are composed of nickel, a ferromagnetic metal and coated with gold, an inert material. Due to the inertness of gold, the containers can be used to encapsulate a range of chemicals. Additionally, we have previously demonstrated that the containers are non-toxic to mammalian cells[29] and hence appropriate for in vitro applications. The ferromagnetic composition enables the containers to be remotely steered using magnetic fields and heated to alter chemical gradients[30,31] and thus provides a route for the generation of dynamical chemical patterns, if needed. Here, we have demonstrated the generation of one space curve, however we anticipate that chemicals could also be released in a variety of space curves by variation in geometric design parameters.
Experimental Section
Numerical simulations
Numerical simulations were carried out using COMSOL Multiphysics (COMSOL, Inc.). Solutions of the time-dependent diffusion equation were sought in the geometry corresponding to the container with a pattern on it. The container was surrounded by a stationary medium with a volume ~ 100 times larger than that of the container. We assumed that the chemical concentration at the outer boundaries of the medium was maintained at zero at all times. The accuracy of the model was independently verified by numerical calculations of the diffusion coefficient of fluorescein from the experimental data shown in Figure5c. This value was found to be 4.2×10−10 m2/s and is in excellent agreement with the published data.[27]
Container fabrication
The patterns and slit shapes obtained in our numerical simulations were exported to AutoCAD (Autodesk, Inc.) and printed on masks at 40,000 dpi. The masks were used for fabrication of 2D panels and solder hinges following the procedures published previously.[14,23] The panels with hinges were then released from the wafers and heated above melting point of solder at which point they spontaneously folded into 3D containers. The self-folding process was driven by a minimization of surface energy of the molten hinges and is described in detail elsewhere.[14] The hinges were designed to completely seal the edges and the corners of the containers. Gold (Au) was coated on the inside and outside of the cubes by electrodeposition after assembly. The final size of the pores was determined by the duration and current density of electrodeposition.[25]
Chemical loading
All containers were loaded with the appropriate chemical by soaking them in aqueous solutions overnight. We utilized solutions of ~ 10−4 M, 7mM and 1 mM of fluorescein, uranine (sodium salt of fluorescein, which has a much higher solubility in water) and L-serine (in chemotaxis buffer) respectively.
Imaging and diffusion studies
Imaging of diffusion from fluorescein-filled containers was achieved by submerging the containers into either a 1 mm tall polydimethylsiloxane (PDMS) chamber or glass capillaries approximately 1 mm to 5 mm in diameter, that were filled with either with 1% or 3% (w/v) agarose gel (to suppress convection and undesired flows). Previous studies have shown that the fluorescent intensity from fluorescein dyes depends linearly on its concentration.[11,26,32] Linearity of the camera was verified before conducting any experiments.
Bacterial culturing and bacterial 3D pattern formation via chemotaxis
Escherichia coli strain RP437 transformed with plasmid pTrc-GFP (a kind gift of M. DeLisa, Cornell University), were grown following the protocol published previously.[11,26] Cultures grown overnight were diluted 1:17 into fresh media, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM (to induce the expression of fluorescent proteins) and bacteria were grown in the shaker bath for 3 more hours. Prior to experiments, cells were centrifuged for 2 min at 1500 g, the supernatant was removed and the dense cell pellet resuspended in 10 ml of chemotaxis buffer (10 mM phosphate buffer–0.1 mM EDTA–1 µM methionine–10 mM lactic acid [pH 7.3]) to the final optical density OD600~0.05.
To enable chemotactic organization, bacteria were injected in a rectangular PDMS chamber with dimensions 1 mm (height) by 7 mm (width) by 7 mm (depth). A helical pattern generating container measuring 2 mm × 500 µm × 500 µm (shown in Figure 3) was placed at the center of the chamber.
Supplementary Material
Footnotes
This work was supported by the NIH Director's New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD004346-01. Information about the NIH Roadmap can be found at http://nihroadmap.nih.gov. We thank Prof. Peter Devreotes, Elizabeth Cha, Kai-Wen Pai, Christina Randall, Rohan Fernandes and Svetlana Ratner for valuable suggestions and discussions, Prof. Mingming Wu and Kasyap Vasudevan for RP437 strain of E. coli.
Supporting information for this article is available on the WWW under http://www.angewandte.org.
References
- 1.Burdick JA, Vunjak-Novakovic G. Tissue Eng. Part A. 2009;15:205. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mapili G, Lu Y, Chen SC, Roy KJ. J. Biomed. Mater. Res. B. 2005;75B:414. doi: 10.1002/jbm.b.30325. [DOI] [PubMed] [Google Scholar]
- 3.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. J. Cell Biol. 1997;137:231. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. J. Cell Biol. 2003;160:267. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat. Methods. 2007;4:855. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 7.Nelson CM, VanDuijn MM, Inman JL, Fletcher DA, Bissell MJ. Science. 2006;314:298. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon NL, Baskaran H, Dertinger SKW, Whitesides GM, Van de Water L, Toner M. Nat. Biotechnol. 2002;20:826. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 9.Abhyankar VV, Toepke MW, Cortesio CL, Lokuta MA, Huttenlocher A, Beebe DJ. Lab Chip. 2008;8:1507. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosadegh B, Huang C, Park JW, Shin HS, Chung BG, Hwang SK, Lee KH, Kim HJ, Brody J, Jeon NL. Langmuir. 2007;23:10910. doi: 10.1021/la7026835. [DOI] [PubMed] [Google Scholar]
- 11.Kalinin Y, Neumann S, Sourjik V, Wu M. J. Bacteriol. 2010;192:1796. doi: 10.1128/JB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freiberg S, Zhu XX. Int. J. Pharm. 2004;282:1. doi: 10.1016/j.ijpharm.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhao XJ, Jain S, Larman HB, Gonzalez S, Irvine DJ. Biomaterials. 2005;26:5048. doi: 10.1016/j.biomaterials.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Leong TG, Lester PA, Koh TL, Call EK, Gracias DH. Langmuir. 2007;23:8747. doi: 10.1021/la700913m. [DOI] [PubMed] [Google Scholar]
- 15.Cho JH, Azam A, Gracias DH. Langmuir. 2010;26:16534. doi: 10.1021/la1013889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azam A, Laflin KE, Jamal M, Fernandes R, Gracias DH. Biomed. Microdevices. 2010 doi: 10.1007/s10544-010-9470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filipiak DJ, Azam A, Leong TG, Gracias DH. J. Micromech. Microeng. 2009;19:075012. doi: 10.1088/0960-1317/19/7/075012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho JH, Gracias DH. Nano Lett. 2009;9:4049. doi: 10.1021/nl9022176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YY, Chung TS, Bai XL, Chan WK. Chem. Eng. Sci. 2000;55:2223. [Google Scholar]
- 20.Batycky RP, Hanes J, Langer R, Edwards DA. J. Pharm. Sci. 1997;86:1464. doi: 10.1021/js9604117. [DOI] [PubMed] [Google Scholar]
- 21.Rosoff WJ, McAllister R, Esrick MA, Goodhill GJ, Urbach JS. Biotechnol. Bioeng. 2005;91:754. doi: 10.1002/bit.20564. [DOI] [PubMed] [Google Scholar]
- 22.Semino CE, Merok JR, Crane GG, Panagiotakos G, Zhang SG. Differentiation. 2003;71:262. doi: 10.1046/j.1432-0436.2003.7104503.x. [DOI] [PubMed] [Google Scholar]
- 23.Leong T, Gu ZY, Koh T, Gracias DH. J. Am. Chem. Soc. 2006;128:11336. doi: 10.1021/ja063100z. [DOI] [PubMed] [Google Scholar]
- 24.Randall CL, Gillespie A, Singh S, Leong TG, Gracias DH. Anal. Bioanal. Chem. 2009;393:1217. doi: 10.1007/s00216-008-2538-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randall CL, Leong TG, Bassik N, Gracias DH. Adv. Drug Delivery Rev. 2007;59:1547. doi: 10.1016/j.addr.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Kalinin YV, Jiang LL, Tu YH, Wu MM. Biophys. J. 2009;96:2439. doi: 10.1016/j.bpj.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Culbertson CT, Jacobson SC, Ramsey JM. Talanta. 2002;56:365. doi: 10.1016/s0039-9140(01)00602-6. [DOI] [PubMed] [Google Scholar]
- 28.Baker RW. Controlled Release of Biologically Active Agents. John Wiley and Sons; 1987. [Google Scholar]
- 29.Randall CL, Kalinin YV, Jamal M, Manohar T, Gracias DH. Lab Chip. 2011;11:127. doi: 10.1039/c0lc00368a. [DOI] [PubMed] [Google Scholar]
- 30.Gimi B, Artemov D, Leong T, Gracias DH, Bhujwalla ZM. Magn. Reson. Med. 2007;58:1283. doi: 10.1002/mrm.21431. [DOI] [PubMed] [Google Scholar]
- 31.Ye HK, Randall CL, Leong TG, Slanac DA, Call EK, Gracias DH. Angew. Chem. Int. Ed. 2007;46:4991. doi: 10.1002/anie.200604414. [DOI] [PubMed] [Google Scholar]
- 32.Walker DA. J. Phys. E Sci. Instrum. 1987;20:217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.