Abstract
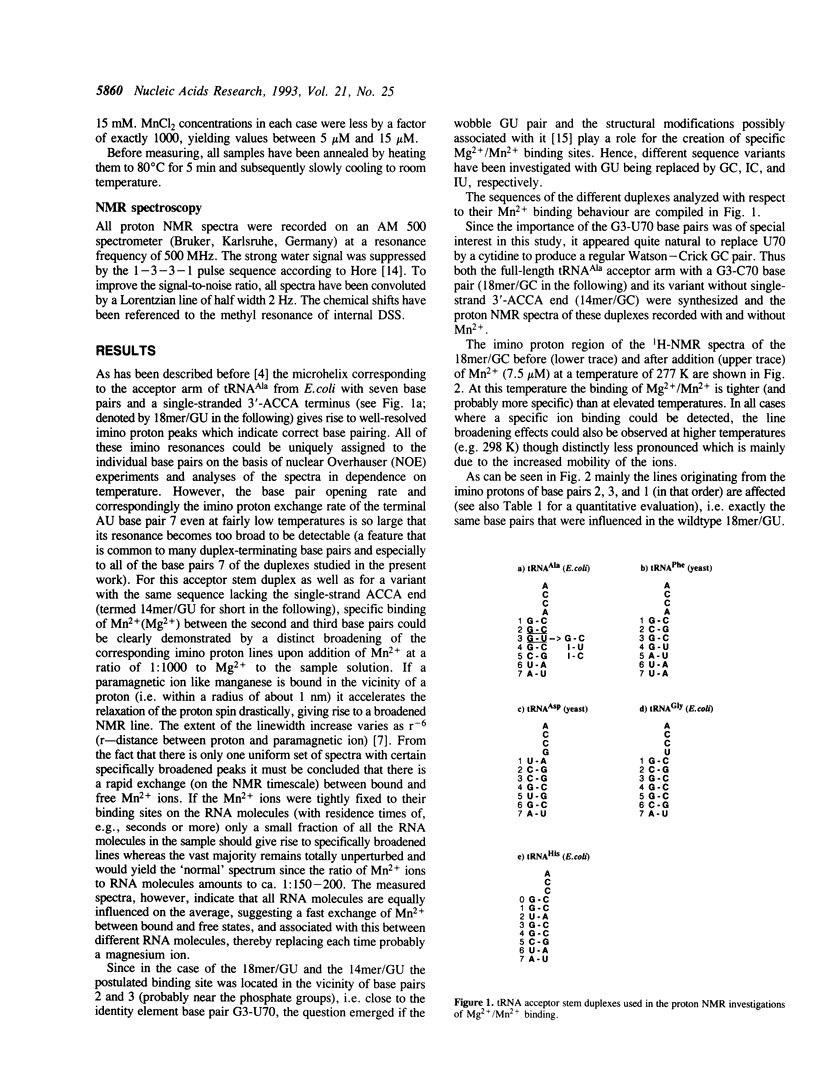
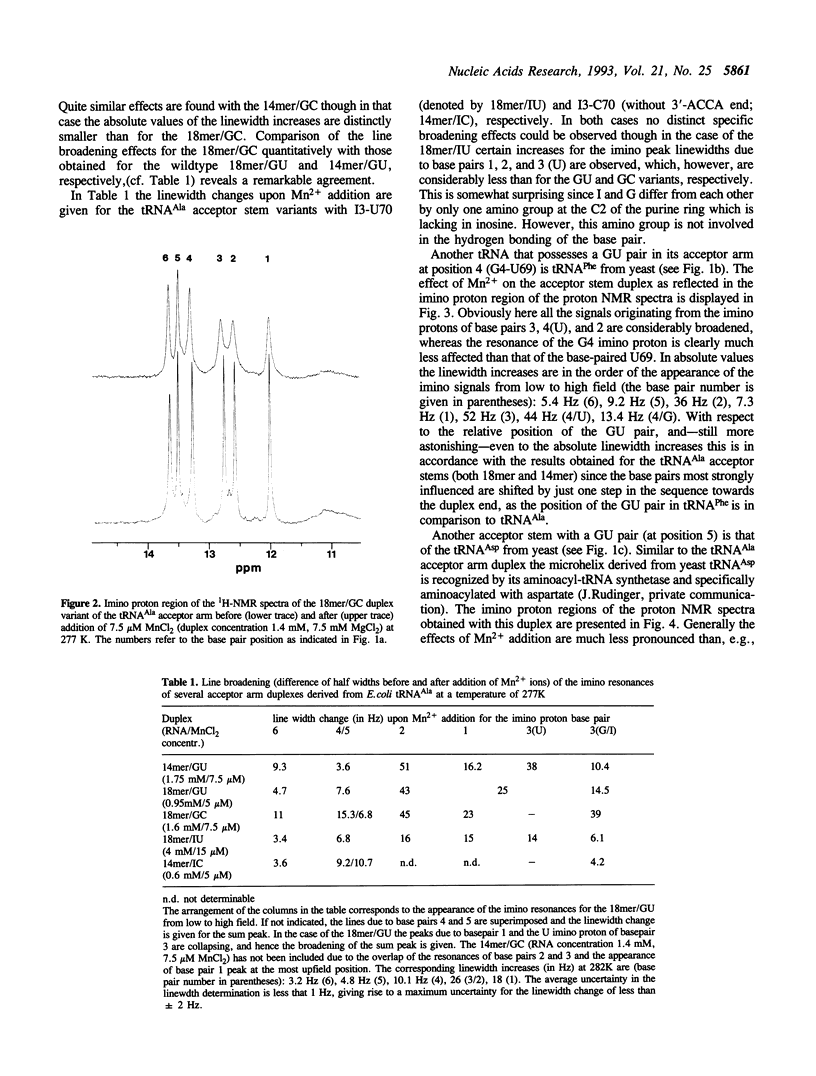
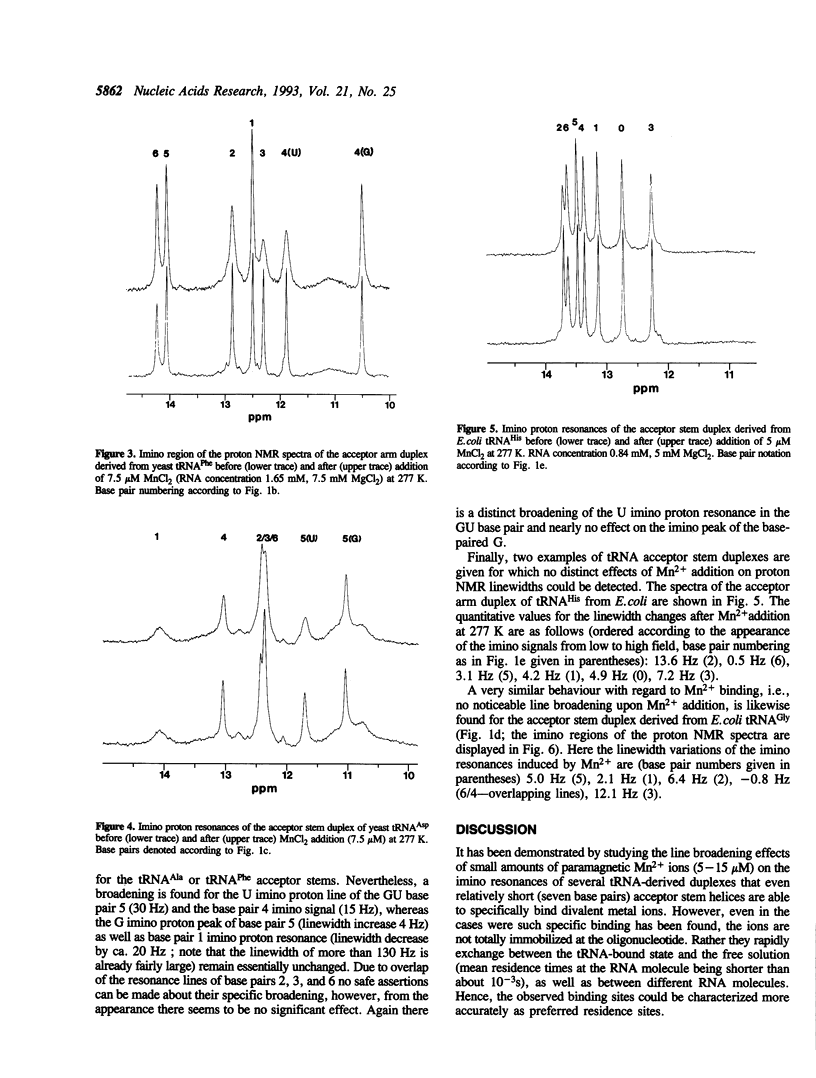
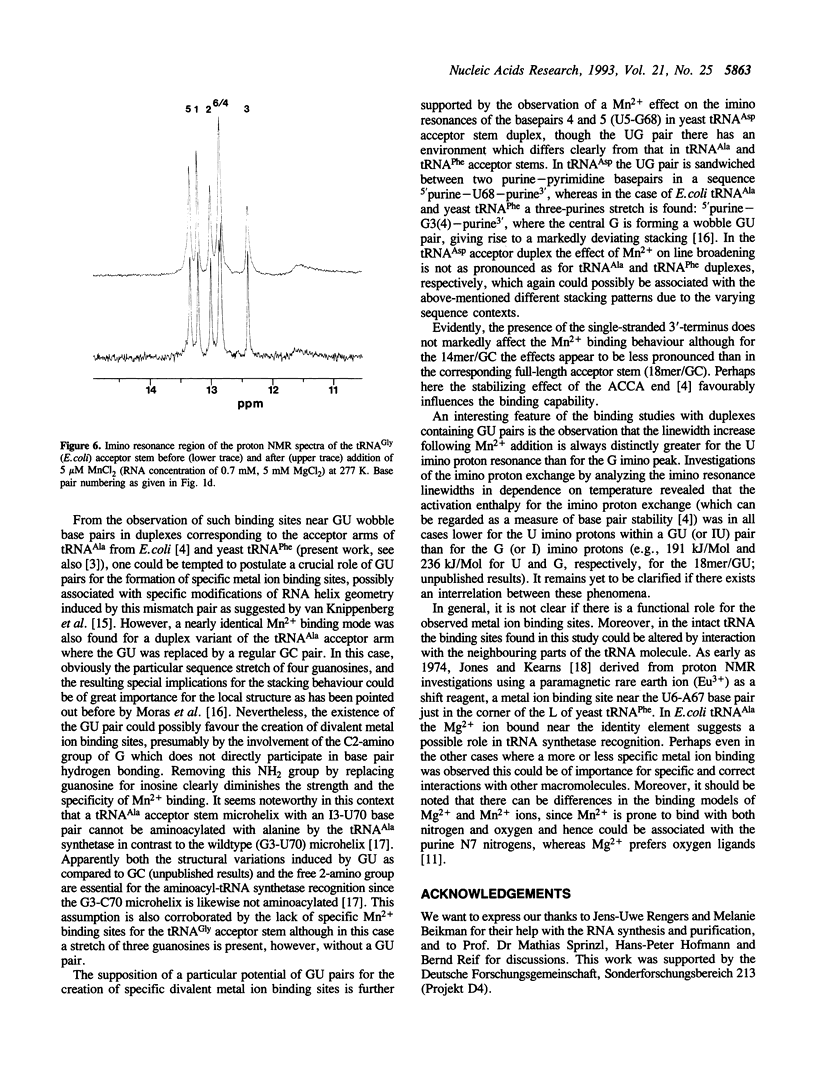
Several RNA duplexes corresponding to the acceptor arms of different tRNAs have been analyzed with respect to their divalent metal ion binding capability by means of proton NMR spectroscopy using paramagnetic Mn2+ ions as probes. In particular, the role of GU wobble base pairs has been analyzed with reference to their potential for creating metal ion binding sites. It is shown that both the structural modifications induced by GU pairs in the A-RNA geometry and the sequence context seem to affect the metal ion binding capabilities.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chao Y. Y., Kearns D. R. Manganese(II) as a paramagnetic probe of the tertiary structure of transfer RNA. Biochim Biophys Acta. 1977 Jul 5;477(1):20–27. doi: 10.1016/0005-2787(77)90157-5. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Hurd R. E., Azhderian E., Reid B. R. Paramagnetic ion effects on the nuclear magnetic resonance spectrum of transfer ribonucleic acid: assignment of the 15--48 tertiary resonance. Biochemistry. 1979 Sep 4;18(18):4012–4017. doi: 10.1021/bi00585a026. [DOI] [PubMed] [Google Scholar]
- Jones C. R., Kearns D. R. Investigation of the structure of yeast tRNAphe by nuclear magnetic resonance: paramagnetic rare earth ion probes of structure. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4237–4240. doi: 10.1073/pnas.71.10.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer S., Hofmann H. P., Ott G., Sprinzl M. The 3'-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6199–6202. doi: 10.1073/pnas.90.13.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991 Aug 16;253(5021):784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- Pyle A. M. Ribozymes: a distinct class of metalloenzymes. Science. 1993 Aug 6;261(5122):709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Redfield A. G. Transfer RNA in solution: selected topics. Annu Rev Biophys Bioeng. 1980;9:181–221. doi: 10.1146/annurev.bb.09.060180.001145. [DOI] [PubMed] [Google Scholar]
- van Knippenberg P. H., Formenoy L. J., Heus H. A. Is there a special function for U.G basepairs in ribosomal RNA? Biochim Biophys Acta. 1990 Aug 27;1050(1-3):14–17. doi: 10.1016/0167-4781(90)90134-n. [DOI] [PubMed] [Google Scholar]


