Abstract
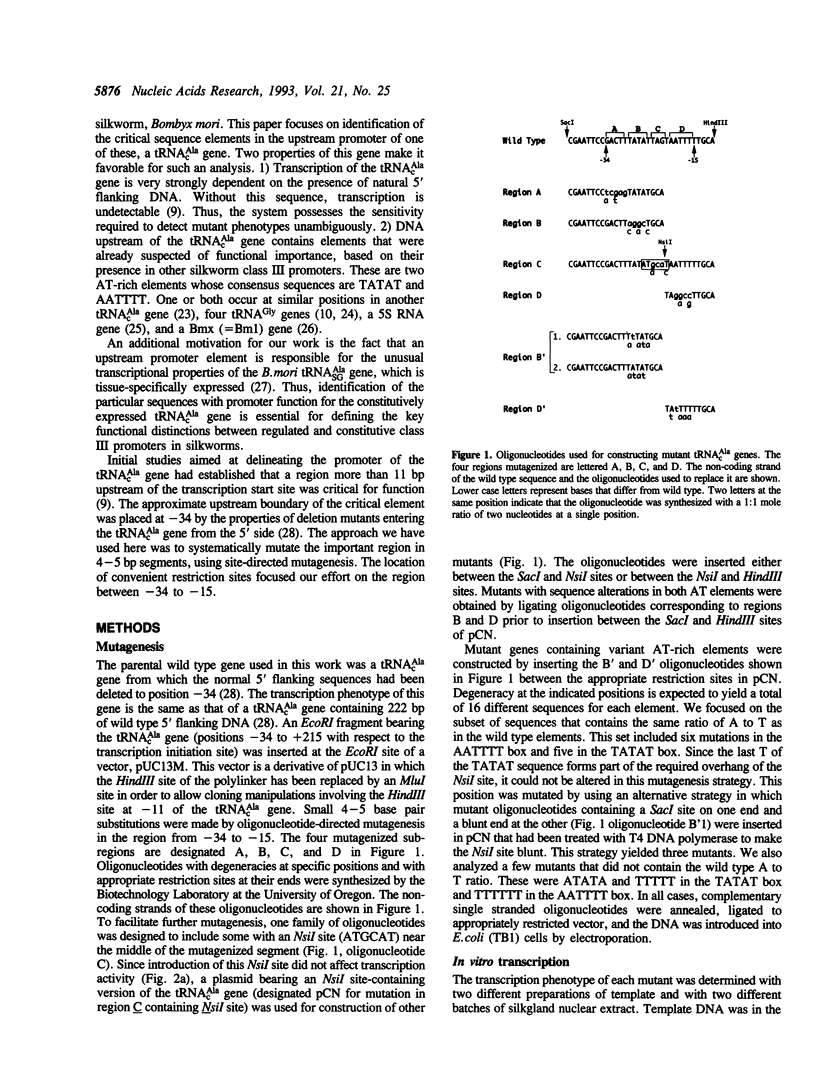
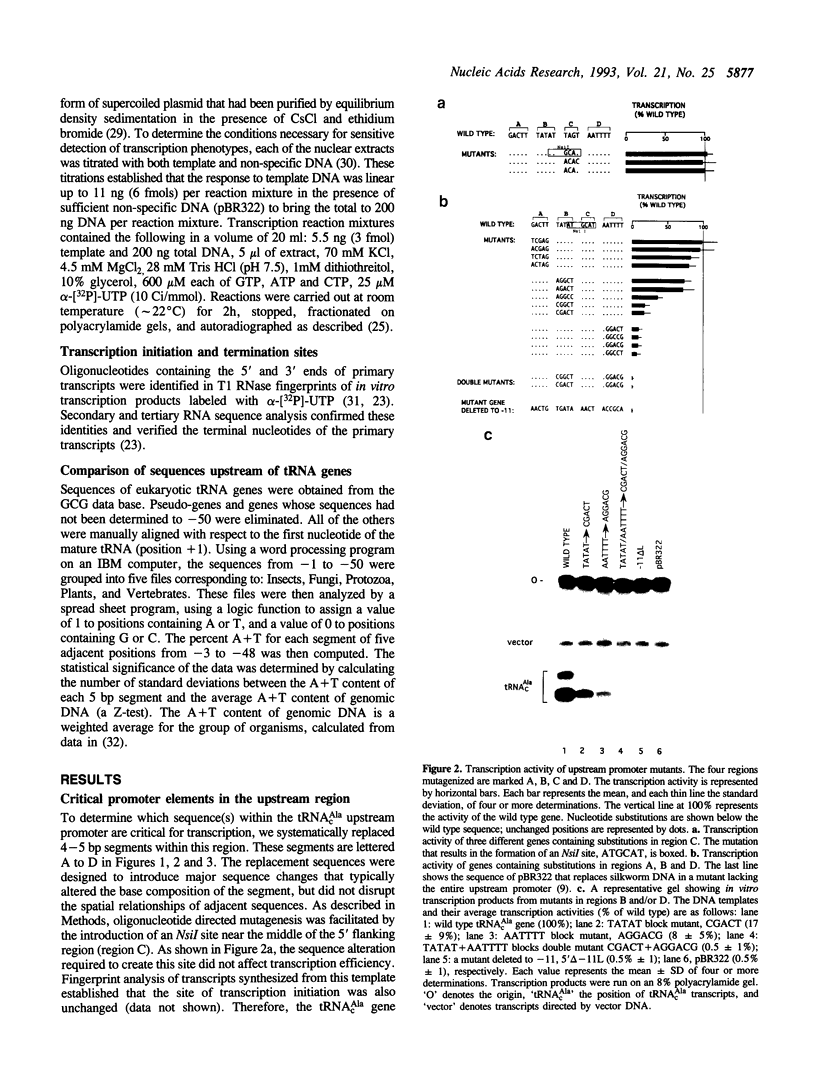
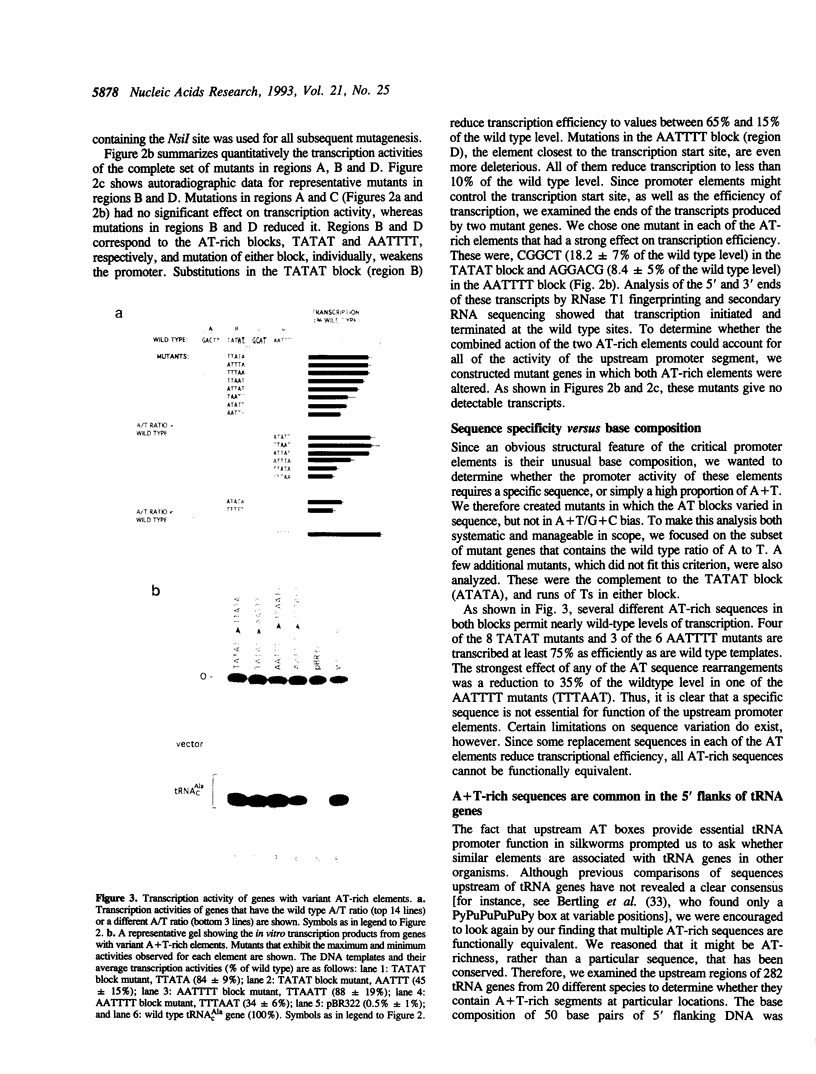
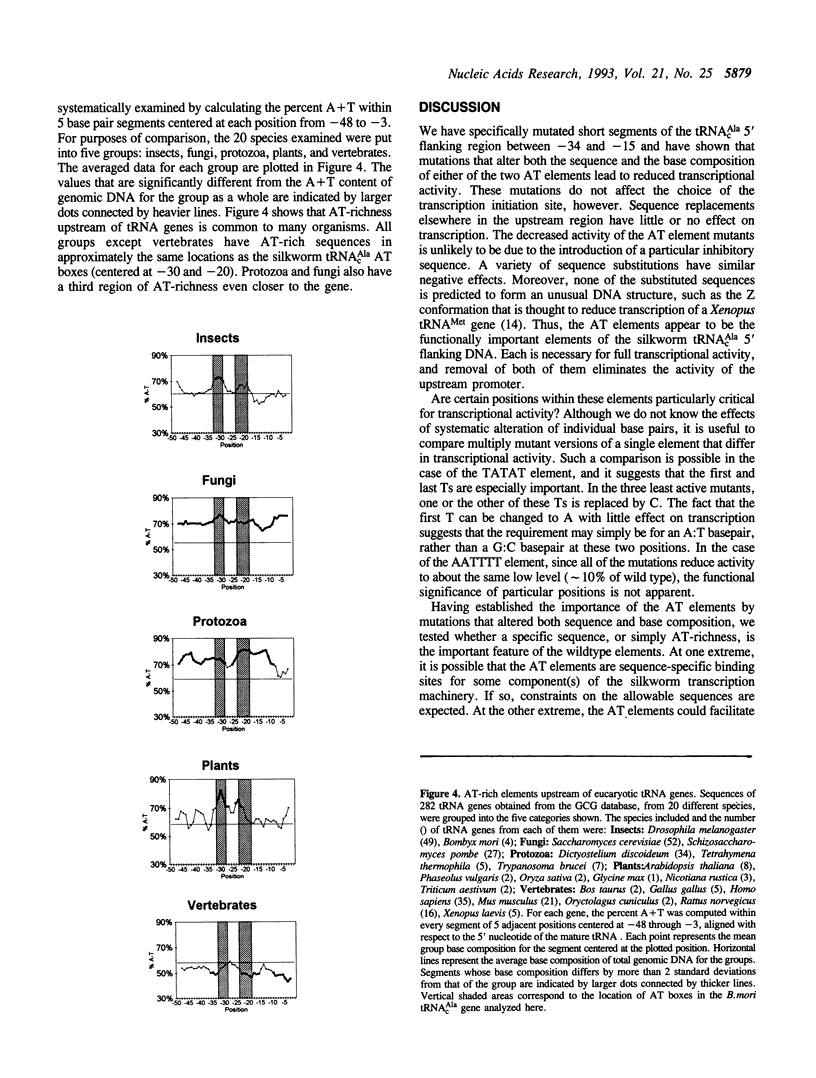
A region within 35 nucleotides upstream of the transcription initiation site of a variety of silkworm Class III templates is absolutely required for transcription in vitro. To determine whether the activity of this region can be attributed to a particular sequence element, we systematically replaced 4-5 bp segments of the region upstream of a silkworm tRNA(cAla) gene. We show that replacement of either of two AT-rich blocks markedly impairs promoter function, whereas replacement of other sequences has little or no effect. Additional mutants were constructed to test whether base composition or sequence is important for function of the AT blocks. We find that some sequences are more effective than others, but that various AT-rich sequences can direct transcription at a high level. Possible mechanisms by which such elements could act are discussed.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold G. J., Gross H. J. Unrelated leader sequences can efficiently promote human tRNA gene transcription. Gene. 1987;51(2-3):237–246. doi: 10.1016/0378-1119(87)90312-x. [DOI] [PubMed] [Google Scholar]
- Braun B. R., Kassavetis G. A., Geiduschek E. P. Bending of the Saccharomyces cerevisiae 5S rRNA gene in transcription factor complexes. J Biol Chem. 1992 Nov 5;267(31):22562–22569. [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992 May 15;69(4):685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Fournier A., Guérin M. A., Corlet J., Clarkson S. G. Structure and in vitro transcription of a glycine tRNA gene from Bombyx mori. EMBO J. 1984 Jul;3(7):1547–1552. doi: 10.1002/j.1460-2075.1984.tb02009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers D. L. Guanylyl cyclase receptors and their endocrine, paracrine, and autocrine ligands. Cell. 1992 Oct 2;71(1):1–4. doi: 10.1016/0092-8674(92)90258-e. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A. The TATA-binding protein participates in TFIIIB assembly on tRNA genes. Nucleic Acids Res. 1992 Dec 25;20(24):6451–6454. doi: 10.1093/nar/20.24.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D., Raymond G. J. Three regions of a yeast tRNALeu3 gene promote RNA polymerase III transcription. J Biol Chem. 1984 May 10;259(9):5990–5994. [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Brown D. D. Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell. 1978 Dec;15(4):1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. K., Horikoshi M., Roeder R. G. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991 Dec 20;67(6):1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- Lobo S. M., Tanaka M., Sullivan M. L., Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992 Dec 11;71(6):1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- Lofquist A., Sharp S. The 5'-flanking sequences of Drosophila melanogaster tRNA5Asn genes differentially arrest RNA polymerase III. J Biol Chem. 1986 Nov 5;261(31):14600–14606. [PubMed] [Google Scholar]
- Léveillard T., Kassavetis G. A., Geiduschek E. P. Saccharomyces cerevisiae transcription factors IIIB and IIIC bend the DNA of a tRNA(Gln) gene. J Biol Chem. 1991 Mar 15;266(8):5162–5168. [PubMed] [Google Scholar]
- Morry M. J., Harding J. D. Modulation of transcriptional activity and stable complex formation by 5'-flanking regions of mouse tRNAHis genes. Mol Cell Biol. 1986 Jan;6(1):105–115. doi: 10.1128/mcb.6.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D. G., Sprague K. U. In vitro transcription of a silkworm 5S RNA gene requires an upstream signal. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5519–5522. doi: 10.1073/pnas.81.17.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D. G., Sprague K. U. Silkworm 5S RNA and alanine tRNA genes share highly conserved 5' flanking and coding sequences. Mol Cell Biol. 1982 Dec;2(12):1524–1531. doi: 10.1128/mcb.2.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A. Bending and supercoiling of DNA at the attachment site of bacteriophage lambda. Trends Biochem Sci. 1990 Jun;15(6):222–227. doi: 10.1016/0968-0004(90)90034-9. [DOI] [PubMed] [Google Scholar]
- Nietfeld W., Digweed M., Mentzel H., Meyerhof W., Köster M., Knöchel W., Erdmann V. A., Pieler T. Oocyte and somatic 5S ribosomal RNA and 5S RNA encoding genes in Xenopus tropicalis. Nucleic Acids Res. 1988 Sep 26;16(18):8803–8815. doi: 10.1093/nar/16.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oei S. L., Pieler T. A transcription stimulatory factor binds to the upstream region of Xenopus 5 S RNA and tRNA genes. J Biol Chem. 1990 May 5;265(13):7485–7491. [PubMed] [Google Scholar]
- Okamoto H., Ishikawa E., Suzuki Y. Structural analysis of sericin genes. Homologies with fibroin gene in the 5' flanking nucleotide sequences. J Biol Chem. 1982 Dec 25;257(24):15192–15199. [PubMed] [Google Scholar]
- Platt T., Yanofsky C. An intercistronic region and ribosome-binding site in bacterial messenger RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2399–2403. doi: 10.1073/pnas.72.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney R. J., Harding J. D. Transcriptional activity and factor binding are stimulated by separate and distinct sequences in the 5' flanking region of a mouse tRNAAsp gene. Nucleic Acids Res. 1988 Mar 25;16(6):2509–2521. doi: 10.1093/nar/16.6.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjadi F. G., Spiegelman G. B. Modulation of a Drosophila melanogaster tRNA gene transcription in vitro by a sequence TNNCT in its 5' flank. Gene. 1987;60(1):13–19. doi: 10.1016/0378-1119(87)90208-3. [DOI] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Burke D. J., Cooley L., Söll D. The extent of a eukaryotic tRNA gene. 5'- and 3'-flanking sequence dependence for transcription and stable complex formation. J Biol Chem. 1984 Feb 10;259(3):1461–1467. [PubMed] [Google Scholar]
- Schultz M. C., Reeder R. H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992 May 15;69(4):697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Starr D. B., Hawley D. K. TFIID binds in the minor groove of the TATA box. Cell. 1991 Dec 20;67(6):1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- Taneja R., Gopalkrishnan R., Gopinathan K. P. Regulation of glycine tRNA gene expression in the posterior silk glands of the silkworm Bombyx mori. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1070–1074. doi: 10.1073/pnas.89.3.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Jackson S. P., Rigby P. W. A role for the TATA-box-binding protein component of the transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1949–1953. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. W., Appelt K., Wilson K. S., Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5(4):281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- Wiley S. R., Kraus R. J., Mertz J. E. Functional binding of the "TATA" box binding component of transcription factor TFIID to the -30 region of TATA-less promoters. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5814–5818. doi: 10.1073/pnas.89.13.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. T., Condliffe D. P., Sprague K. U. Transcriptional properties of BmX, a moderately repetitive silkworm gene that is an RNA polymerase III template. Mol Cell Biol. 1988 Feb;8(2):624–631. doi: 10.1128/mcb.8.2.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. T., Larson D., Young L. S., Sprague K. U. A large region controls tRNA gene transcription. J Mol Biol. 1985 May 25;183(2):153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]
- Wobbe C. R., Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990 Aug;10(8):3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- Young L. S., Rivier D. H., Sprague K. U. Sequences far downstream from the classical tRNA promoter elements bind RNA polymerase III transcription factors. Mol Cell Biol. 1991 Mar;11(3):1382–1392. doi: 10.1128/mcb.11.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S., Takahashi N., Sprague K. U. Upstream sequences confer distinctive transcriptional properties on genes encoding silkgland-specific tRNAAla. Proc Natl Acad Sci U S A. 1986 Jan;83(2):374–378. doi: 10.1073/pnas.83.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]




