Abstract
Transcription intermediary factor 1γ (TIF1γ) was suggested to play a role in erythropoiesis. However, how TIF1γ regulates the development of different blood cell lineages and whether TIF1γ is involved in human hematological malignancies remain to be determined. Here we have shown that TIF1γ was a tumor suppressor in mouse and human chronic myelomonocytic leukemia (CMML). Loss of Tif1g in mouse HSCs favored the expansion of the granulo-monocytic progenitor compartment. Furthermore, Tif1g deletion induced the age-dependent appearance of a cell-autonomous myeloproliferative disorder in mice that recapitulated essential characteristics of human CMML. TIF1γ was almost undetectable in leukemic cells of 35% of CMML patients. This downregulation was related to the hypermethylation of CpG sequences and specific histone modifications in the gene promoter. A demethylating agent restored the normal epigenetic status of the TIF1G promoter in human cells, which correlated with a reestablishment of TIF1γ expression. Together, these results demonstrate that TIF1G is an epigenetically regulated tumor suppressor gene in hematopoietic cells and suggest that changes in TIF1γ expression may be a biomarker of response to demethylating agents in CMML.
Introduction
TIF1γ (also known as tripartite motif protein TRIM33) is an ubiquitous nuclear protein that belongs to the transcriptional intermediary factor 1 family (1). Four TIF1 family members (α to δ) have been identified in mammals, and orthologs are present in organisms such as Drosophila (1–6). TIF1α (also known as TRIM24) interacts with nuclear receptors and modulates their transcriptional activity either positively or negatively in a ligand-dependent fashion (5, 7). In mice, TIF1α functions as a liver-specific tumor suppressor whose deletion reveals the deleterious effect of retinoic acid receptor α aberrant activation to liver homeostasis (8). TIF1β, a component of the histone deacetylase N-CoR1/HDAC3 complex (9), functions as a corepressor for the large family of Krüppel-associated box (KRAB) zinc finger transcription factors (3, 10) and is required for post-implantation embryogenesis and mesoderm induction (11). TIF1δ is involved in heterochromatin-mediated gene silencing (4). Human and mouse TIF1G is closely related to zebrafish moonshine (mon), a gene whose mutations disrupt embryonic and adult hematopoiesis with severe red blood cell aplasia (12). Targeted deletion of Tif1g is embryonic lethal in mice (13, 14). In zebrafish and human CD34+ cells, TIF1γ functionally links positive elongation factors such as p-TEFb and FACT to blood-specific transcription complexes (e.g., the SCL/TAL1 complex) to regulate elongation of genes by antagonizing RNA polymerase II (RNA Pol II) pausing (15). TIF1γ also affects the human hematopoietic progenitor cell response to the cytokines of the TGF-β superfamily through various mechanisms (14, 16–18).
To further explore the role of TIF1γ in hematopoiesis, we examined the effects of hematopoietic tissue–targeted deletion of Tif1g in mice. Tif1g deletion affects the transition from very primitive progenitors (i.e., LT-HSC population) to common myeloid progenitors and leads to a selective expansion of granulo-monocytic progenitors. This effect correlates with an inhibition of the hematopoietic progenitor cell response to TGF-β and provokes the age-dependent appearance of a cell-autonomous phenotype that recapitulates important features of human chronic myelomonocytic leukemia (CMML). Interestingly, a downregulation of TIF1G gene expression is observed in hematopoietic cells of approximately 35% of patients with CMML. While no inactivating mutations were identified, a low level of TIF1γ expression in CMML cells was related to the hypermethylation of the gene promoter, and the expression of TIF1γ was reestablished after treatment with the demethylating agent decitabine, suggesting that changes in TIF1γ expression may be a biomarker of CMML response to demethylating agents. Together, the results indicate that TIF1γ is a key regulator of HSC fate that behaves as a tumor suppressor gene.
Results
The Tif1g deletion affects hematopoietic progenitor populations in mice.
To obtain further insights into the contribution of TIF1γ to adult hematopoiesis, and in particular HSCs, we generated mice selectively deficient for Tif1γ by breeding floxed Tif1g mice (Tif1gfl/fl) (19) with cFES-Cre transgenic animals (ref. 20 and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45213DS1). In litters from crosses between Tif1gfl/fl mice and cFES-Cre transgenic animals, cFES-Cre;Tif1gfl/fl (Tif1gΔ/Δ) mice correspond to the hematopoietic tissue–restricted knockout mice, whereas Tif1g fl/fl mice represent controls (Supplemental Figure 1A). Although Tif1g-null mice invariably died perinatally (13, 14), Tif1gΔ/Δ mice reached adulthood and were fertile. As expected, the deletion of Tif1g floxed alleles was observed in hematopoietic organs and cells, including long-term HSCs (LT-HSCs) (Supplemental Figure 1, B and C, and ref. 20), and was associated with a low expression of Tif1g at both the RNA and protein levels in the hematopoietic organs (Supplemental Figure 1, D and E).
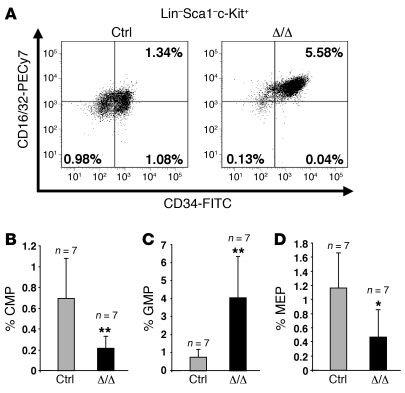
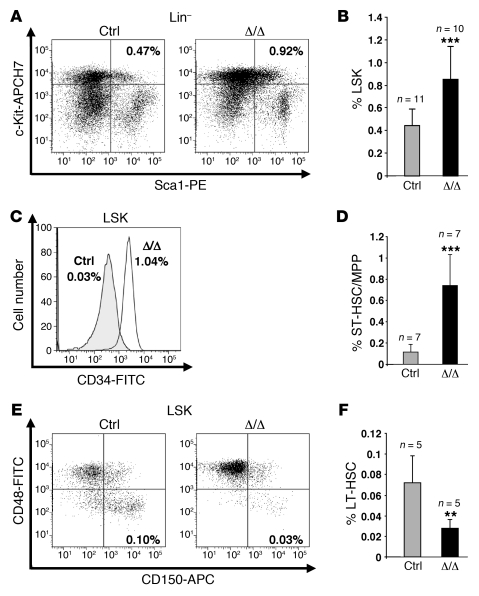
Mice younger than 6 months old did not display any macroscopic and blood peripheral abnormalities (data not shown). Nevertheless, the proportion of granulocyte/monocyte progenitors (GMPs; Lin–Sca-1–c-Kit+CD34+CD16/32+) was increased (~400%) at the expense of common myeloid progenitors (CMPs; Lin–Sca-1–c-Kit+CD34+CD16/32–; ~50%) and megakaryocyte-erythroid progenitors (MEPs; Lin–Sca-1–c-Kit+CD34–CD16/32–; ~60%) (Figure 1). A significant increase in the Lin–Sca-1+c-Kit+ (LSK) fraction was also observed (Figure 2, A and B), including an increase in the proportion of short-term HSCs/multipotent progenitors (ST-HSCs/MPPs; LSK CD34+) (Figure 2, C and D) and a decrease in the fraction of primitive LSK (long-term reconstituting HSCs [LT-HSCs]) identified on the “SLAM code” (signaling lymphocyte activation molecule: CD150+CD48–) (ref. 21 and Figure 2, E and F). These data demonstrate that TIF1γ is a key regulator of HSC fate.
Figure 1. The Tif1g deletion affects hematopoietic progenitor populations in mice younger than 6 months.
(A) Representative FACS staining profiles of the progenitor populations, including CMPs (lower right panel), MEPs (lower left panel), and GMPs (upper right panel), from the respective control (Ctrl) or Tif1gΔ/Δ (Δ/Δ) mice. Calculation of the numbers and percentages of each of the populations was based on the number of living cells. The number of CMPs and MEPs decreased, whereas the number of GMPs increased. Analysis of control and Tif1gΔ/Δ mice demonstrated a decrease in CMPs (B), an increase in GMPs (C), and a decrease in MEPs (D) in Tif1gΔ/Δ mice. *P < 0.05, **P < 0.01.
Figure 2. The Tif1g deletion affects HSCs in mice younger than 6 months.
(A and B) Analysis of LSK cells from representative control and healthy Tif1gΔ/Δ mice demonstrated an increase in the LSK population in Tif1gΔ/Δ mice. (C and D) Analysis of ST-HSCs/MPPs from representative control and Tif1gΔ/Δ mice demonstrated an increase in ST-HSCs/MPPs in Tif1gΔ/Δ mice. (E and F) Analysis of LT-HSCs from representative control and Tif1gΔ/Δ mice demonstrated a decrease in the LT-HSC population in Tif1gΔ/Δ mice. **P < 0.01, ***P < 0.001.
Tif1gΔ/Δ mice develop a CMML-like myeloproliferative disease with monocytic features.
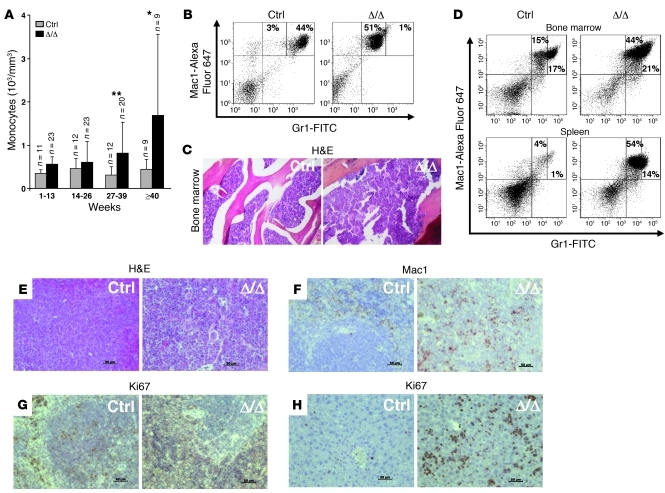
Mice older than 6 months developed a progressive hyperleukocytosis (data not shown). Although the number of MEPs was decreased, there were no obvious signs of anemia before or after the onset of the disease (Supplemental Figure 2A). The number of Ter119+CD71+ erythroid progenitors was slightly decreased in the bone marrow, while it is increased in the spleen (Supplemental Figure 2B). Morphologic analysis of peripheral blood identified Howell-Jolly bodies and stomatocytes in mice older than 6 months (Supplemental Figure 2C). The progressive hyperleukocytosis observed was due to the accumulation of monocytes (Figure 3A), which was confirmed by FACS analysis (Gr1loMac1+ cells) (Figure 3B). Bone marrow examination showed an accumulation of monocytes (Figure 3, C and D), which was also observed in the spleen and in the liver (Figure 3, D–H), leading to a severe hepatosplenomegaly (Supplemental Figure 3A). The splenic organization was destroyed (Figure 3E) by mature highly proliferative Mac1+ cells (Figure 3, F and G) that invaded the red pulp (Supplemental Figure 3B). Immature hematopoietic cells including erythroblast cells were observed in the spleen (not shown), and the liver was also infiltrated by highly proliferative cells (Figure 3H). Kinetics of hematopoietic recovery after sublethal irradiation of control or Tif1gΔ/Δ mice did not reveal another lineage specificity of Tif1g deletion, i.e., these mice recovered normally and accumulated monocytes in their blood and spleen when progressing in age (data not shown).
Figure 3. Tif1gΔ/Δ mice older than 6 months develop a CMML-like myeloproliferative disease with monocytic features.
(A) Peripheral monocyte counts in control and Tif1gΔ/Δ mice. The results are shown as mean ± SD (n = 9–23 mice). (B) FACS analysis of peripheral blood cells demonstrated an increase in the monocytic (Gr1loMac1+) population in Tif1gΔ/Δ mice. (C) Representative H&E-stained histopathologic sections of bone marrow demonstrated hypercellularity in Tif1gΔ/Δ mice. (D) FACS analyses of the cell populations from bone marrow or spleen demonstrated an increase of the monocytic (Gr1loMac1+) population in Tif1gΔ/Δ mice. (E) Representative H&E-stained histopathologic sections of spleen demonstrated that the splenic organization was destroyed in Tif1gΔ/Δ mice. (F) Immunohistochemical (IHC) staining for Mac1 in paraffin-embedded spleen sections indicated an expanded monocytic population. (G and H) IHC staining for Ki67 in paraffin-embedded spleen (G) or liver (H) revealed a highly proliferative population. Original magnification, ×76. *P < 0.05, **P < 0.01.
Tif1gΔ/Δ myeloproliferative disease is transplantable into secondary recipients.
Mice transplanted with Tif1gΔ/Δ bone marrow cells from 4-month-old mice (phenotypically normal) survived lethal irradiation, indicating the reconstitution of hematopoiesis. Two months after transplantation, Tif1g-deleted cells were detected by quantitative PCR (Q-PCR) in the peripheral blood of the transplanted mice (Supplemental Figure 4A), this deletion being correlated to the decreased expression of Tif1g mRNA (data not shown). These observations suggested that the deletion occurred in HSCs. Two months after transplantation, the mice developed the same myeloproliferative disease (i.e., monocytosis, hepatosplenomegaly) (data not shown), supporting the notion that the phenotypic effects of the Tif1g deletion were cell autonomous. We observed an increased number of Gr1loMac1+ cells in the bone marrow and in the spleen (Supplemental Figure 4C) and an augmentation of the LSK fraction in Tif1gΔ/Δ mice compared with control littermates (Supplemental Figure 4D). The abnormal distribution and frequency of progenitors were also reproducible (Supplemental Figure 4E). A second transplantation into lethally irradiated recipients reproduced the same disorder after 2 months (data not shown). We performed reciprocal transplant experiments in which wild-type donor cells were transplanted into either control or Tif1gΔ/Δ mutant recipients. We did not observe any alteration in the distribution of LSK (Supplemental Figure 4F) and progenitors (CMPs, GMPs, MEPs) (Supplemental Figure 4G) in the Tif1gΔ/Δ mutant recipients. These observations indicate that the disease generated by the deletion of Tif1g is initiated from the HSC compartment and is cell autonomous.
The hematopoietic cell response to TGF-β is inhibited in Tif1gΔ/Δ mice.
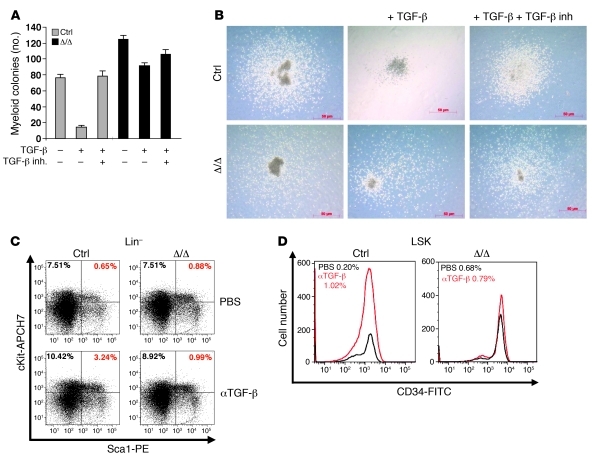
TIF1γ was identified as part of the TGF-β signaling pathway, including its E3 ubiquitin ligase activity on Smad4 (16–18, 22). Tif1g-deficient LSK CD34+ cells (ST-HSCs/MPPs) were cultured in the presence or absence of TGF-β and TGF-β inhibitor (Figure 4, A and B). Stimulation with TGF-β dramatically (~80%) reduced the formation of myeloid cells from control LSK CD34+ cells as compared with untreated cultures, whereas it only slightly decreased (~25%) myelopoiesis of Tif1g-deficient cells (Figure 4, A and B). We also treated i.p. 6-month-old Tif1gΔ/Δ or control mice with either PBS or 15 mg/kg of a neutralizing pan–TGF-β monoclonal antibody that blocks all 3 TGF-β mammalian isoforms (TGF-β1, -β2, and -β3) (23, 24). Treatment was administered once a week for 5 weeks, when the mice were sacrificed and evaluated for hematopoietic progenitor populations. In control mice, treatment with 2G7 induced an increase in the number of LSK (Figure 4C) and ST-HSCs/MPPs (Figure 4D), whereas the antibody did not affect the distribution of these cells in Tif1gΔ/Δ mice. We conclude that Tif1gΔ/Δ HSCs escape to the effect of TGF-β1–β3 cytokines.
Figure 4. Alteration of the TGF-β pathway in aging Tif1gΔ/Δ mice.
(A) Defective TGF-β signaling pathway responsiveness in Tif1gΔ/Δ mice. Sorted ST-HSCs/MPPs were plated on methylcellulose medium in triplicate, with or without TGF-β and TGF-β inhibitor (inh.). The number of myeloid colonies was determined at day 8. The results are shown as the mean ± SD of triplicates. (B) Representative images (original magnification, ×138) of the resulting myeloid colonies from sorted ST-HSCs/MPPs, untreated or treated with TGF-β or with TGF-β plus TGF-β inhibitor. (C and D) TGF-β–neutralizing antibody (αTGF-β) does not affect hematopoietic progenitor cell distribution in Tif1gΔ/Δ mice. (C) Analysis of LSK cells from treated or untreated representative control and Tif1gΔ/Δ mice demonstrated an increase in the LSK population in control mice treated with the antibody, which was not observed in Tif1gΔ/Δ mice. (D) Analysis of ST-HSCs/ MPPs from treated or untreated representative control and Tif1gΔ/Δ mice demonstrated an increase in MPPs in control mice treated with the antibody, which was not observed in Tif1gΔ/Δ mice.
The Tif1g deletion affects Csf1r expression in mice.
We next tested whether Tif1γ could regulate expression of Csf1r (M-CSFR), Csf3r (G-CSFR), and Csf2ra (GM-CSFR), 3 genes known to participate in monocyte or granulocyte differentiation. Unlike Csf3r and Csf2ra, Csf1r expression was significantly decreased by the lack of Tif1γ (Supplemental Figure 5, A and B), which could account for the altered production of peritoneal macrophages observed in Tif1gΔ/Δ mice (Supplemental Figure 5C). These Tif1gΔ/Δ macrophages did not adhere to the plastic and were morphologically abnormal in vitro (Supplemental Figure 5D). The plasma levels of M-CSF (CSF-1) and G-CSF remained unchanged in Tif1gΔ/Δ compared with control mice (Supplemental Figure 5E).
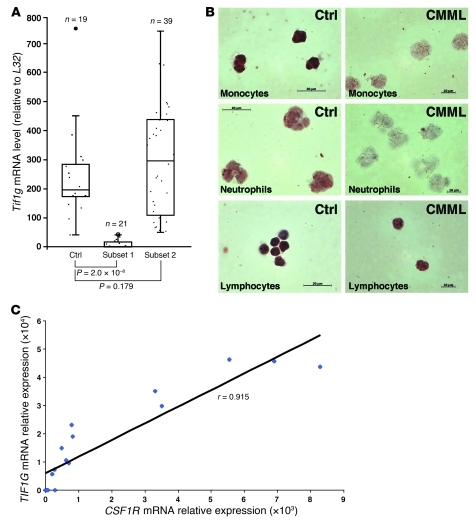
TIF1G is downregulated in a subset of chronic myelomonocytic leukemia patients.
CMML is a heterogeneous disease with regard to molecular abnormalities. Our mouse data prompted us to investigate the expression of TIF1γ in peripheral blood monocytes sorted from a cohort of 60 patients with CMML (Supplemental Table 1). The transcript was almost undetectable in 21 of the 60 patients (35%) (Figure 5A). Immunocytochemical analysis identified a decrease in the expression of the protein in monocytes and neutrophils of these patients (Figure 5B). The decreased expression in Csf1r expression identified in Tif1gΔ/Δ mouse cells (Supplemental Figure 5) was confirmed in CMML cells, in which it was strongly correlated with a low level of TIF1G (r = 0.915) (Figure 5C).
Figure 5. TIF1γ is downregulated in 35% of CMML patients.
(A) RQ-PCR analysis of TIF1γ expression in the purified monocytes (CD14+ cells) of healthy donors (Ctrl) or CMML patients (subsets 1 and 2). (B) Immunocytochemistry staining for TIF1γ from the monocytes, neutrophils and lymphocytes (×340) of a representative control or CMML patient. (C) Correlation of TIF1G and CSF1R expression in monocytes of 17 individuals with CMML. The scatter plot shows TIF1G and CSF1R mRNA expression. Expression values are expressed in arbitrary units. A linear regression curve and the correlation coefficient are indicated. *P < 0.05.
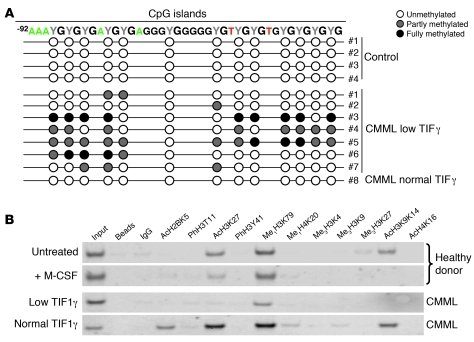
TIF1G is an epigenetically regulated tumor suppressor gene in chronic myelomonocytic leukemia.
Sequencing of all the TIF1G exons in the monocytes of a cohort of 66 CMML patients (Supplemental Table 2) with various TIF1γ mRNA and protein levels failed to identify any mutation in the coding sequence (data not shown). We analyzed by direct sequencing the bisulfite-modified TIF1G promoter sequence in CMML and health donor monocytes. The TIF1G promoter of CMML patients expressing low levels of TIF1γ was methylated, while none of the DNA from normal monocytes nor a CMML monocyte sample with normal TIF1G mRNA level showed any detectable methylation (Figure 6A and Supplemental Figure 6). Thus, low expression of TIF1γ in CMML cells correlates with TIF1G promoter hypermethylation. Histone modifications, which are highly related to the epigenetic status of DNA, may be correlated with gene expression levels (25). The decrease in TIF1G gene expression observed in normal human monocytes upon M-CSF exposure for 1 day was correlated with a specific pattern of histone modification (acetylations and methylations) on the TIF1G gene promoter (Figure 6B). A similar pattern that suggests the inactive form of the promoter was observed in CMML cells harboring very low levels of TIF1γ, whereas those with normal TIF1γ expression levels displayed a pattern similar to the active form of the promoter observed in untreated normal human monocytes (Figure 6B).
Figure 6. TIF1G is an epigenetically regulated tumor suppressor gene in CMML.
(A) Sequencing of the bisulfite modified TIF1G promoter sequence from normal monocytes (Control) or CMML monocytes. (B) ChIP analysis of the status of histone modifications on the TIF1G promoter. Chromatin was immunoprecipitated using specific antibodies directed against histone modifications. Ac, acetylated; Me1, methylated; Me3, tri-methylated.
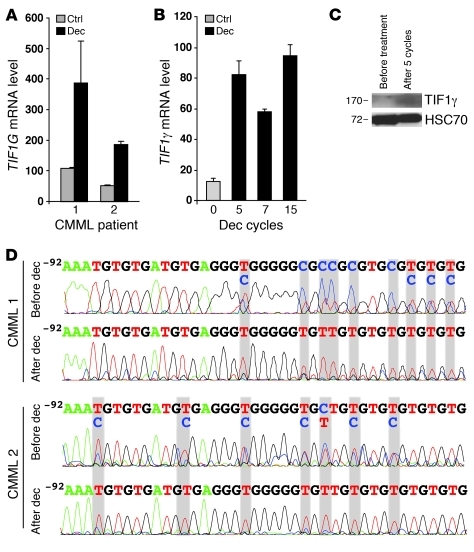
A 3-day exposure of the peripheral blood monocytes from CMML patients to decitabine, a demethylating agent that has demonstrated some efficacy in the treatment of CMML (26), restored TIF1G expression in the cells in which its levels were initially downregulated (Figure 7A). TIF1γ expression was also reestablished in the peripheral blood monocytes of a CMML patient who responded to decitabine in vivo (Figure 7, B and C). Cytosine methylation in the TIF1G promoter sequence identified in the monocytes of these patients before decitabine treatment (Figure 7D) was no longer detected after 7 cycles of decitabine (Figure 7D), which correlated with Tif1g mRNA reexpression (Figure 7B).
Figure 7. TIF1G is sensitive to decitabine in CMML.
(A) TIF1G expression (RQ-PCR) from the monocytes (CD14+) of two CMML patients (1 and 2) after 3 days of culture in the absence or presence of decitabine (dec). Data are the mean ± SD of the values from experiments performed in triplicate. (B) TIF1G expression (RQ-PCR) from the monocytes of one CMML patient before and during treatment with decitabine. Data are the mean ± SD of the values from experiments performed in triplicate. (C) Immunoblotting analysis for TIF1γ in human monocytes from the same patient after 5 cycles of decitabine. Equivalent loading of the lanes was controlled by anti-HSC70 antibody staining. (D) Sequencing of the bisulfite-modified TIF1G promoter sequence from the monocytes of two patients before and after 7 cycles of decitabine (CMML 1 is the same patient as in B).
Discussion
We demonstrate that hematopoietic tissue–targeted deletion of TIF1G gene, which encodes an E3 ubiquitin ligase that is also a transcription and elongation coregulator, deregulates the HSC compartment and leads in aging mice to a myeloproliferative disease with myelodysplastic features. Its epigenetically regulated downregulation in leukemic cells of a subset of patients with CMML also suggests a tumor suppressor function in human hematopoiesis.
cFES-Cre–mediated recombination of the floxed phosphatidylinositol glycan class A (Piga) allele induced the deletion of Piga in adult HSCs and all blood cell lineages (20), which prompted us to investigate the role of TIF1γ in adult hematopoiesis by crossing cFES-Cre with Tif1gfl/fl mice (19). Mutations in the zebrafish mon (tif1g) gene cause a disruption in both primitive embryonic and definitive adult hematopoiesis, resulting in a severe loss of erythroid cells (12). In zebrafish and human stem/progenitor CD34+ cells, TIF1γ functionally links positive elongation factors to blood-specific transcription complexes to regulate the erythroid commitment (15). Here, we show that targeted inactivation of Tif1g in stem cells does not affect the number of peripheral blood erythrocytes. The discrepancy with previous models may be related to compensatory mechanisms operating in the mouse model, i.e., the decrease in bone marrow erythroblast is compensated by splenic erythropoiesis, by a function of TIF1γ in zebrafish embryonic primitive erythropoiesis, or by the requirement of TIF1γ for in vitro erythroid commitment of human CD34+ cells while being dispensable for mammalian adult erythropoiesis in vivo. Based on the expression of c-myb, mon mutants were suggested also to have normal myeloid development (12). We show here that inactivation of Tif1g in mouse stem cells expanded the LSK population, in which LT-HSC number was decreased while ST-HSC/MPP number was dramatically increased. The Tif1γ defect also promotes the expansion of the GMP compartment at the expense of the CMPs and the MEPs. At an older age (>6 months), all Tif1g-deficient mice exhibit an accumulation of monocytes in peripheral blood, bone marrow, liver, and spleen, suggesting that aging favors the dysregulation of myelopoiesis. This phenotype recapitulates the human CMML that is observed in patients older than 50 (mean age, 70 years old) and associates proliferative and dysplastic features (27). Transplantation experiments demonstrate that hematopoietic cells are capable of propagating the Tif1g-deficient CMML-like phenotype in recipient mice, indicating a cell-autonomous process. In addition, similarly to mice in which Junb has been deleted (28), and in contrast to those deficient in Rarg (27) and Rb (29, 30), Tif1gΔ/Δ mice receiving control bone marrow transplants did not develop the phenotype.
TIF1γ was identified as a modulator of the TGF-β signaling pathway (22). Cytokines of the TGF-β family bind membrane heterodimeric serine/threonine kinases that in turn phosphorylate intracellular mediators known as Smads (31). Phosphorylated Smad2 and Smad3 accumulate into the nucleus and bind Smad4 to regulate target gene transcription. In hematopoietic, mesenchymal, and epithelial cells exposed to TGF-β, TIF1γ could selectively bind phosphorylated Smad2/3 in competition with Smad4 (17). TIF1γ could also antagonize Smad4 through its ubiquitin ligase properties (16). Here, we show that decreased expression of TIF1γ impairs the HSC response to TGF-β (32–34). Recent results indicate that the gene deletion also affects transcriptional elongation of blood genes such as SCL/TAL1 by antagonizing Pol II pausing (15). Hence, TIF1γ may act on cell fate and lineage commitment through transcription regulation, elongation, and protein degradation. These effects may account for the decrease in Csf1r gene expression observed in Lin– and total bone marrow cells of Tif1gΔ/Δ mice, which correlates with altered monocyte differentiation into macrophages.
TIF1G gene expression is almost undetectable in sorted leukemic cells of approximately 35% of patients with CMML, which, as in the mouse model, correlates with decreased expression of the CSF1R gene. In this disease, the most frequently identified somatic mutations involve TET2 (35), RUNX1 (36, 37), and ASXL1 (38); RAS and CBL mutations may be less frequent and mutually exclusive (36, 39); while mutations in several other genes encoding epigenetic regulators or signaling molecules (UTX, EZH2, IDH1, IDH2) were identified with a low frequency (40–47). It remains difficult to distinguish in these somatic events those that drive the disease pathogenesis from those acquired as a consequence of clonal evolution. The copy-neutral uniparental disomy that is frequently observed in CMML cells can affect the 1p chromosomal region containing the TIF1G gene (48), but we failed to detect any mutation in the coding sequence of the gene that could account for its decreased expression in patient cells.
TIF1G downregulation correlates with the hypermethylation of CpG islands and a specific pattern of histone modifications on the gene promoter in CMML samples (26, 49). The link between mutations in epigenetic genes such as TET2, ASXL1, EZH2, UTX, IDH1, and IDH2 and the downregulation of TIF1G has now to be explored (50–53). TIF1γ downregulation could also cooperate with mutated signaling molecules such as K-Ras, as recently demonstrated in a mouse model of pancreatic cancer (54). Epigenetic drugs including DNA methyltransferase inhibitors, histone deacetylase inhibitors, and their combination are currently tested as therapeutic approaches in CMML (55). Interestingly, the expression of TIF1γ was restored in monocytes of patients who responded to the demethylating agent decitabine, which could indicate either a demethylating effect of the treatment on the gene promoter or the selection of cells in which the gene was not repressed. Whatever the explanation, changes in TIF1γ expression may be a biomarker of response to demethylating agents in CMML (56).
Together, our results suggest that TIF1G is an epigenetically regulated tumor suppressor gene in hematopoietic cells. The decreased expression of this gene favors the proliferation of MPPs and leads to the age-dependent expansion of the monocyte population. Changes in TIF1G expression may be tested as a biomarker of response to demethylating agents and other chromatin structure modifiers such as HDAC inhibitors (57) that are currently being developed for the treatment of this disease.
Methods
Mice.
Mice were housed in a temperature-controlled environment under a 12-hour light/12-hour dark cycle with free access to water and a standard rodent chow diet; all mice used were maintained under specific pathogen–free conditions according to animal study protocols reviewed and approved by the Animal Experiment Ethics Committee of the University of Burgundy. cFES-Cre mice were provided by P.P. Pandolfi (Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA). For genotyping, genomic DNA was prepared from blood, bone marrow, or tail biopsies using the Gentra Puregene Kit (QIAGEN). Floxed (531 bp), deleted (360 bp), and wild-type (498 bp) Tif1g alleles were identified by PCR with primers AFI 124 (5′-AGCTCTGGAACGTCGGTC-3′), AFI 125 (5′-GGTAAGTCAGCAAGAGCTCA-3′), and AFM 257 (5′-GGTAGTACTTGTATGGAGGT-3′). For genomic Q-PCR, a specific sequence located between exons 3 and 4 was amplified on a 7500 FAST thermocycler (Applied Biosystems) using the SYBR Green detection protocol as outlined by the manufacturer (Applied Biosystems). Mouse specific forward and reverse primers were: Moz, 5′-GAGGACCCAGGTTCAATTCTTAG-3′ and 5′-GAAGAAGCTGTCAGATCCTCTAGAACT-3′, used as a standardizing control, and Tif1g, 5′-GTGCCGCAGTGCCTATTTG-3′ and 5′-AATGCAGAGAGAGCCAGTTCCT-3′.
Bone marrow transplantation.
To generate bone marrow–reconstituted mice, we injected a total of 2 × 106 bone marrow cells from control or Tif1gΔ/Δ donor mice into the retro-orbital sinus of lethally irradiated recipient (11 Gy) BALB/c mice. For the second transplantation, the same protocol was followed. For reciprocal transplantations, we injected a total of 2 × 106 BM cells from control donor mice into the retro-orbital sinus of Tif1gΔ/Δ or control lethally irradiated recipient (16 Gy) C57BL/6 mice.
Blood cell analysis.
Adult control and Tif1gΔ/Δ mice were anesthetized with 1%–2% isoflurane. Blood was collected from the conjunctival vein in the eye using an EDTA-coated tube. Complete blood counts were performed using an automated hematology analyzer (MS Laboratories). Collected blood was also used to prepare blood smears, which were stained with May-Grünwald-Giemsa (MGG) stain.
Cell preparation.
Following sacrifice, mice were examined for the presence of abnormalities, and organs were collected for further cell and histopathology analyses. For the bone marrow cell preparations, femurs and tibias were removed aseptically and cells were flushed. For splenocytes, spleens were dilacerated, red blood cells were lysed, and cells were washed in 1× PBS (Lonza).
Responsiveness to TGF-β.
For myeloid progenitor assays studying the responsiveness to TGF-β ligand, we purified Lin– cells from bone marrow cells using the Lineage Cell Depletion magnetic isolation kit and AutoMACS separator according to the manufacturer’s instructions (Miltenyi Biotec). ST-HSCs/MPPs (LSK CD34+) were sorted from these cells. Then, 5 × 102 ST-HSCs/MPPs were plated in 1.1 ml serum-free methylcellulose (M3134; StemCell Technologies) supplemented with 15% FBS (Lonza), 1× BIT (StemCell Technologies), 100 μM MTG (Sigma-Aldrich), 2 mM glutamine (Invitrogen), 50 ng/ml recombinant murine SCF (rmSCF) (R&D Systems), 10 ng/ml rmIL-3 (R&D Systems), 10 ng/ml rmIL-6 (R&D Systems), and in the presence or absence of 10 ng/ml recombinant human TGF-β1 (R&D Systems) and/or 10 μM TGF-β inhibitor (SB431542; Sigma-Aldrich). Cells were plated in 35-mm culture dishes and incubated in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Myeloid colonies were scored on day 8 by light microscopy.
To block TGF-β in vivo, mice were treated with 2G7 neutralizing pan–TGF-β IgG2 (R&D Systems) delivered i.p. at a dose of 15 mg/kg once a week during 5 weeks. Mice were then sacrificed.
Histopathology and immunohistochemistry analyses.
Sectioned femoral bone marrow, spleens, and livers were fixed at least 72 hours in 10% neutral buffered formalin, dehydrated in alcohol, cleared in xylene, and infiltrated with paraffin on an automated processor (Leica TP1020). Tissue sections (4 μm thick) were placed on charged slides, deparaffinized in xylene, cleared through graded alcohol solutions, rehydrated in water and stained with H&E. Immunohistochemistry was performed on paraffin-embedded tissue sections using primary antibodies directed against Mac1 (BD Biosciences — Pharmingen) or Ki67 (Abcam).
Flow cytometry and cell sorting analysis.
FACS analysis was performed on blood cells, bone marrow cells, or splenocytes from mice. Briefly, single-cell suspension was prepared and stained with primary and secondary antibodies if necessary. The cells were washed twice in 1× PBS and resuspended in 1× PBS/30% FBS. Lin– cells were identified by absence of signal after staining with a biotinylated antibody cocktail containing B220, CD3, Ter119, Mac1, and Gr1 (BD Biosciences — Pharmingen) and development by streptavidin–Alexa Fluor 405 (Invitrogen). Antibodies Gr1-FITC, Mac1–Alexa Fluor 647, Ter119–Alexa Fluor 405, CD71-PE, Sca1-PE, c-Kit–APCH7, CD150-APC, CD48-FITC, CD34-FITC, and CD16/32-PECya7 were added and incubated on ice. Cell sorting analysis was performed on Lin– cells purified from bone marrow cells using the Lineage Cell Depletion magnetic isolation kit and AutoMACS separator according to the manufacturer’s instructions (Miltenyi Biotec). Flow cytometry was performed on LSRII (BD) and cell sorting on FACSAria (BD) using DIVA software (BD), and automatic compensations were applied. The results were analyzed using FlowJo software (Tree Star Inc.). For HSC and hematopoietic progenitor cell analysis and sorting, color compensation samples were produced by singly staining bone marrow cells or Lin– cells with one antibody of each fluorochrome or with a combination of cells stained by the Fluorescence Minus One (FMO) method, which was performed by sequentially adding fluorescence-labeled antibodies to the staining cocktail.
Real-time Q-PCR in mice.
Total RNA was isolated from bone marrow cells and splenocytes using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was obtained from 150 ng total RNA using M-MLV Reverse Transcriptase (Promega). Real-time Q-PCR (RQ-PCR) was performed in triplicate using TaqMan probes (Applied Biosystems) and analyzed in an Applied Biosystems 7500 Real-Time PCR System. The Tif1g TaqMan assay was Mm01308706_m1. Values for each PCR were normalized to Hprt levels (Mm03024075_m1). For the study of Csf1r, Csf3r, and Csf2ra expression, real-time PCR was performed in a 7500 FAST thermocycler (Applied Biosystems) using the SYBR Green detection protocol as outlined by the manufacturer. Mouse specific forward and reverse primers were: Csf1r, 5′-CATGGCCTTCCTTGCTTCTAAA-3′ and 5′-CAGCACGTTTCGAGCTGCTA-3′; Csf3r, 5′-GCGCCGACTGTCAGTACCA-3′ and 5′-GGAGCAGTTGTTCTGCCTCTTC-3′; and Csf2ra, 5′-ACGTGGCGCGATGCAT-3′ and 5′-TCACGACCAAGTAGGCCTCACT-3′. Hprt was used as internal control.
CMML sample collection and analysis.
Blood samples from patients with CMML were collected, and informed consent was obtained. CMML tissue studies were reviewed and approved by the institutional review board of Hospital Cochin (Paris, France). Peripheral blood mononucleated cells were isolated by Ficoll Hypaque (Eurobio), and monocytes were sorted using the CD14+ magnetic isolation kit and AutoMACS separator according to the manufacturer’s instructions (Miltenyi Biotec). Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was reverse transcribed by M-MLV reverse transcriptase with random hexamer primers (Promega). Real-time PCR was performed with AmpliTaq Gold polymerase in a 7500 FAST thermocycler (Applied Biosystems) using the SYBR Green detection protocol as outlined by the manufacturer. Briefly, 15 ng of total complementary DNA, 50 nM of each primer, and 1× SYBR Green mix were used in a total volume of 20 μl. Human specific forward and reverse primers were L32, 5′-TGTCCTGAATGTGGTCACCTGA-3′ and 5′-CTGCAGTCTCCTTGCACACCT-3′ used as a standardizing control, TIF1G, 5′-AGCAACGGCGACATCCA-3′ and 5′-TGCATTCTTGGCGGCATA-3′, and CSF1R, 5′-GCCCCCCATCACCTCACT-3′ and 5′-GTGTTTTGGAAGGTAGCGTTGTT-3′. L32 was used as internal control.
Treatment of CMML with decitabine.
Adults with a diagnosis of CMML were enrolled in “A Phase II Study of Decitabine in Patients with Chronic Myelomonocytic Leukemia” (GFM-DEC-LMMC-2007-02), and informed consent was obtained. CMML tissue studies were reviewed and approved by the institutional review board of Hospital Cochin. Monocytes were isolated, as described in CMML sample collection and analysis, after decitabine treatment and subjected to RQ-PCR analysis. One cycle of decitabine corresponds to 20 mg/m2/d i.v. for 5 days every 22 days. For in vitro culture, monocytes isolated from CMML patients were cultured in RPMI 1640 Glutamax medium (BioWhittaker) supplemented with 10% fetal calf serum (BioWhittaker), penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml) (BioWhittaker) at 37°C in a 5% CO2 humidified atmosphere. Cells were plated at 0.5 × 106 cells/ml and incubated for 3 days with M-CSF (100 ng/ml) in the presence or absence of decitabine (3 μM).
Bisulfite DNA treatment and sequencing.
Genomic DNA was isolated from monocytes of CMML patients before and after 7 cycles of decitabine using QIAGEN’s standard procedures. Two hundred nanograms of total genomic DNA was modified by bisulfite treatment according to the manufacturer’s instructions (MethylDetector, Active Motif). Converted TIF1G promoter was identified by PCR with converted primers forward (5′-GGCTTTAAAAAAAAAATCTCCCTT-3′) and reverse (5′-CCACCATATTTTCCTCTTTAAACCCG-3′), and direct sequencing reaction was performed using standard conditions according to the manufacturer’s instructions (Applied Biosystems).
ChIP procedure.
One million cells were fixed with 1% formaldehyde to crosslink DNA with proteins, then lysed and sonicated. The ChIP procedure was carried out according to the manufacturer’s instructions with modifications (Upstate Biotechnology) and as previously described (58). After pre-clearing with salmon sperm DNA/protein A/G agarose beads, the samples underwent immunoprecipitation with 3 μg of antibodies specific for acetylated H2B K5, phosphorylated H3 T11, acetylated H3 K27, phosphorylated H3 Y41, methylated H3 K79, methylated H4 K20 (Santa Cruz Biotechnology Inc.), tri-methylated H3 K4, tri-methylated H3 K9, tri-methylated H3 K27, acetylated H3 K9 K14, acetylated H4 K16 (Millipore), or rabbit IgG (Santa Cruz Biotechnology Inc.) at 4°C overnight. Beads were washed, protein/DNA complexes eluted, then cross-links reversed by heating at 65°C overnight. After RNA and protein digestions, DNA was purified on a spin column (NucleoSpin Extract II, Macherey-Nagel). Input corresponds to total sonicated DNA. DNA was amplified by PCR. PCR amplification was performed using Taq polymerase (Promega) and ChIP primers. The ChIP primers used to amplify regions of the promoter locus of TIF1G were 5′-CCCTCAGGCCCTTTGCA-3′ and 5′-GAAGGAACCTGTCTTGGAGAAGAG-3′ (Invitrogen).
Statistics.
Student 2-tailed t test and ANOVA were used for statistical analysis. Differences were considered significant when P was less than 0.05.
Supplementary Material
Acknowledgments
We gratefully acknowledge the clinicians (Groupe Francophone des Myélodysplasies) who provided human CMML samples. We thank O. Kosmider, A. Chlemaire, A. Bouchot, M. Ciudad, C. Racoeur, J. Guy, L. Desoche, and A. Aznague for technical assistance, and V. Saint-Giorgio and N. Hoffmann (Oncodesign) for their assistance in the mouse studies. We also thank W. Vainchenker and F. Louache for technical advice and helpful discussions, as well as P.-H. Roméo and S. Kusy for helpful discussions. This work was supported by the Conseil Régional de Bourgogne (FABER to L. Delva and N. Droin), the Ligue Contre le Cancer (CCIR-GE to L. Delva), the Institut National du Cancer (INCa; to E. Solary), the Agence nationale de la recherche (ANR; to E. Solary and L. Delva), the Association pour la Recherche sur le Cancer (ARC to L. Delva, Equipment of Gustave Roussy Institute), the national Programmes hospitaliers de recherche clinique (PHRC) (MAD06), the Ligue Nationale Contre le Cancer (to E. Solary), the Fondation de France (Comité Leucémie) (to N. Droin), and the Association Laurette Fugain (to N. Droin). R. Aucagne was supported by fellowships from the Ligue Contre le Cancer (Saône-et-Loire committee), the ARC, and the Société Française d’Hématologie (SFH); J. Paggetti by fellowships from the Ministère de l’Enseignement Supérieur et de la Recherche of France (MESR) and the ARC; B. Lagrange by fellowships from the MESR and the ARC; and A. Largeot by fellowships from Inserm and the Région Bourgogne.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(6):2361–2370. doi:10.1172/JCI45213.
Jérôme Paggetti’s present address is: CRP-Santé Laboratoire d’Hémato-Cancérologie Expérimentale, Luxembourg.
Kai-Ping Yan’s present address is: Gene Expression and Regulation, The Wistar Institute, Philadelphia, Pennsylvania, USA.
References
- 1.Venturini L, et al. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18(5):1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 2.Beckstead R, et al. Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit betaFTZ-F1-dependent transcription. Mol Cell. 2001;7(4):753–765. doi: 10.1016/S1097-2765(01)00220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman JR, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 4.Khetchoumian K, et al. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J Biol Chem. 2004;279(46):48329–48341. doi: 10.1074/jbc.M404779200. [DOI] [PubMed] [Google Scholar]
- 5.Le Douarin B, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14(9):2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24(24):4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong S, et al. A RA-dependent, tumour-growth suppressive transcription complex is the target of the PML-RARalpha and T18 oncoproteins. Nat Genet. 1999;23(3):287–295. doi: 10.1038/15463. [DOI] [PubMed] [Google Scholar]
- 8.Khetchoumian K, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39(12):1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- 9.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275(51):40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 10.Abrink M, et al. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc Natl Acad Sci U S A. 2001;98(4):1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127(13):2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- 12.Ransom DG, et al. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2004;2(8):E237. doi: 10.1371/journal.pbio.0020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Kaartinen V. Generation of mice with a conditional allele for Trim33. Genesis. 2008;46(6):329–333. doi: 10.1002/dvg.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morsut L, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137(15):2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 15.Bai X, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142(1):133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupont S, et al. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121(1):87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 17.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125(5):929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 18.Dupont S, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 19.Doisne JM, et al. iNKT cell development is orchestrated by different branches of TGF-beta signaling. J Exp Med. 2009;206(6):1365–1378. doi: 10.1084/jem.20090127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller P, et al. FES-Cre targets phosphatidylinositol glycan class A (PIGA) inactivation to hematopoietic stem cells in the bone marrow. J Exp Med. 2001;194(5):581–589. doi: 10.1084/jem.194.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8(12):970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 23.Arteaga CL, Hurd SD, Winnier AR, Johnson MD, Fendly BM, Forbes JT. Anti-transforming growth factor (TGF)-beta antibodies inhibit breast cancer cell tumorigenicity and increase mouse spleen natural killer cell activity. Implications for a possible role of tumor cell/host TGF-beta interactions in human breast cancer progression. J Clin Invest. 1993;92(6):2569–2576. doi: 10.1172/JCI116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas S, et al. Inhibition of TGF-beta with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. J Clin Invest. 2007;117(5):1305–1313. doi: 10.1172/JCI30740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107(7):2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantarjian H, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 27.Emanuel PD. Juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia. Leukemia. 2008;22(7):1335–1342. doi: 10.1038/leu.2008.162. [DOI] [PubMed] [Google Scholar]
- 28.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119(3):431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Walkley CR, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129(6):1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129(6):1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 32.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6(3):265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langer JC, Henckaerts E, Orenstein J, Snoeck HW. Quantitative trait analysis reveals transforming growth factor-beta2 as a positive regulator of early hematopoietic progenitor and stem cell function. . J Exp Med. 2004;199(1):5–14. doi: 10.1084/jem.20030980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capron C, et al. A major role of TGF-beta1 in the homing capacities of murine hematopoietic stem cell/progenitors. Blood. 2010;116(8):1244–1253. doi: 10.1182/blood-2009-05-221093. [DOI] [PubMed] [Google Scholar]
- 35.Kosmider O, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94(12):1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gelsi-Boyer V, et al. Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1 genes. BMC Cancer. 2008;8:299. doi: 10.1186/1471-2407-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo MC, et al. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23(8):1426–1431. doi: 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- 38.Gelsi-Boyer V, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 39.Tyner JW, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009;113(8):1749–1755. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77(2):307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 41.Steensma DP, et al. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106(4):1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jelinek J, et al. JAK2 mutation 1849G>T is rare in acute leukemias but can be found in CMML, Philadelphia chromosome-negative CML, and megakaryocytic leukemia. Blood. 2005;106(10):3370–3373. doi: 10.1182/blood-2005-05-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee BH, et al. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell. 2007;12(4):367–380. doi: 10.1016/j.ccr.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jankowska A, et al. Mutational Spectrum In Chronic Myelomonocytic Leukemia Includes Genes Associated with Epigenetic Regulation Such as UTX and EZH2.ASH Annual Meeting 2010 . December 6, 2010. Orlando, Florida, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szpurka H, et al. UTX Mutations and Epigenetic Changes In MDS/MPN and Related Myeloid Malignancies.ASH Annual Meeting 2010 . December 5, 2010. Orlando, Florida, USA. [Google Scholar]
- 46. Grossmann V, et al. Analyses of 81 chronic myelomonocytic leukemia (CMML) for EZH2, TET2, ASXL1, CBL, KRAS, NRAS, RUNX1, IDH1, IDH2, and NPM1 revealed mutations in 86.4% of all patients with TET2 and EZH2 being of high prognostic relevance.ASH Annual Meeting 2010 . December 6, 2010. Orlando, Florida, USA. [Google Scholar]
- 47.Kosmider O, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24(5):1094–1096. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- 48.Dunbar AJ, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68(24):10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenaux P, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AE, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116(19):3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 53.Kohlmann A, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28(24):3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 54.Vincent DF, et al. Inactivation of TIF1gamma cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5(7):e1000575. doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Candelaria M, et al. Hydralazine and magnesium valproate as epigenetic treatment for myelodysplastic syndrome. Preliminary results of a phase-II trial [published online ahead of print October 5, 2010]. Ann Hematol. doi: 10.1007/s00277-010-1090-2. doi:10.1007/s00277-010-1090-2. [DOI] [PubMed] [Google Scholar]
- 56.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24(1):88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 57.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 58.Paggetti J, et al. Crosstalk between leukemia-associated proteins MOZ and MLL regulates HOX gene expression in human cord blood CD34+ cells. Oncogene. 2010;29(36):5019–5031. doi: 10.1038/onc.2010.254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.