Abstract
Computer-based genome-wide screening of the DNA sequence of Escherichia coli strain K12 revealed tens of thousands of tandem simple sequence repeat (SSR) tracts, with motifs ranging from 1 to 6 nucleotides. SSRs were well distributed throughout the genome. Mononucleotide SSRs were over-represented in noncoding regions and under-represented in open reading frames (ORFs). Nucleotide composition of mono- and dinucleotide SSRs, both in ORFs and in noncoding regions, differed from that of the genomic region in which they occurred, with 93% of all mononucleotide SSRs proving to be of A or T. Computer-based analysis of the fine position of every SSR locus in the noncoding portion of the genome relative to downstream ORFs showed SSRs located in areas that could affect gene regulation. DNA sequences at 14 arbitrarily chosen SSR tracts were compared among E. coli strains. Polymorphisms of SSR copy number were observed at four of seven mononucleotide SSR tracts screened, with all polymorphisms occurring in noncoding regions. SSR polymorphism could prove important as a genome-wide source of variation, both for practical applications (including rapid detection, strain identification, and detection of loci affecting key phenotypes) and for evolutionary adaptation of microbes.[The sequence data described in this paper have been submitted to the GenBank data library under accession numbers AF209020–209030 and AF209508–209518.]
Escherichia coli is a species of Gram-negative bacterium composed of numerous strains and serotypes (Ochman and Selander 1984; Ahmed et al. 1987; Jay 1996). Although certain strains comprise an important element of the normal intestinal microflora (Johnson 1991; Hays 1992), other strains produce toxins and are pathogenic (Johnson 1991; Olsivik et al. 1992; Yu and Kaper 1992). In environmental monitoring studies, coliform bacteria provide a presumptive indictor of fecal contamination of surface waters or food (European Economic Community 1980; American Public Health Association et al. 1985; Hays 1992). Food-safety studies routinely include monitoring for contamination by pathogenic E. coli (Vanderzant and Splittstoesser 1992), particularly in meat processing (Padhye and Doyle 1992; Witham et al. 1996). E. coli is an important model organism for study of gene expression in prokaryotes (Niedhardt 1996). Rapid detection and characterization of E. coli strains poses important scientific and practical applications.
Simple sequence repeats (SSRs, or microsatellites) are a class of DNA sequences consisting of simple motifs of 1–6 nucleotides that are tandemly repeated from two or three up to a few dozen times at a locus (Vogt 1990). SSRs long have been known to be distributed throughout the genomes of eukaryotes and to be highly polymorphic (Tautz 1989; Weber 1990). There is accumulating evidence that SSRs serve a functional role, affecting gene expression, and that polymorphism of SSR tracts may be important in the evolution of gene regulation (Rosenberg et al. 1994; Kunzler et al. 1995; Kashi et al. 1997; King et al. 1997; Kashi and Soller 1998; Tonjum et al. 1998; Moxon and Wills 1999; van Belkum 1999). The sequencing of prokaryotic genomes allows screening of entire genomes for the existence of SSRs (Field and Wills 1996, 1998), revealing large numbers of SSR tracts not detected in earlier studies that focused on particular loci. Recent publication of the complete genome sequence for E. coli (Blattner et al. 1997) provides the basis for characterizing SSR tracts in this organism, both genome-wide and at particular loci.
In this study we screen the entire E. coli genome for the presence, locations, and composition of SSR tracts. We test our observations against the null hypotheses that SSRs are randomly distributed among coding and noncoding regions and that they collectively have the same composition as the genome. We show that SSRs are differentially distributed among coding and noncoding regions. We also show that SSRs are polymorphic among E. coli strains, providing potential marker loci for rapid detection and characterization. To our knowledge, this is a first analysis of the E. coli genome for such purposes and represents a general approach for analysis of other prokaryotic genomes.
RESULTS
Genomic Content, Distribution, and Composition of SSRs
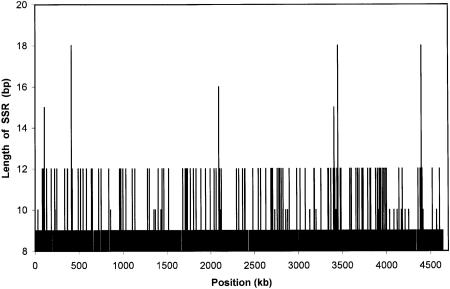
Although the existence and abundance of SSRs in eukaryotes are well documented, SSRs are not well studied in prokaryotes. Using computer software that we developed, we conducted a genome-wide scan of the DNA sequence of E. coli. A total of 235,495 SSR tracts were found (Table 1). These tracts were distributed rather evenly throughout the genome (Fig. 1). Total lengths of particular SSR tracts in E. coli were small (Table 1; Fig. 1). Those with mononucleotide repeats seldom exceeded 9 bp in length, and higher-order SSRs (i.e., those with di-, tri-, or tetranucleotide repeats) rarely exceeded 12 bp. SSR tracts of 6 or more bp in length comprised 2.4% of the E. coli genome (a total of 109 kb).
Table 1.
Numbers of Loci Exhibiting Simple Sequence Repeats of Given Structure in the Genome of E. coli Strain K12
| No. of repeats per locus | Motif length in nucleotides | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mono- | di- | tri- | tetra- | penta- | hexa- | ||||||
| 3 | 163,345 | (163,134) | 7,081 | (9,538) | 2,353* | (778) | 51 | (52) | — | 3 | (0) |
| 4 | 42,901* | (40,795) | 465 | (590) | 64* | (12) | 1 | (0) | — | — | |
| 5 | 13,837* | (10,262) | 28 | (39) | 2 | (0) | — | — | — | ||
| 6 | 4,123* | (2,570) | 1 | (2) | — | — | — | — | |||
| 7 | 1,000* | (625) | — | — | — | — | — | ||||
| 8 | 217* | (158) | — | — | — | — | — | ||||
| 9 | 22 | (39) | — | — | — | — | — | ||||
| 10 | 1 | (11) | — | — | — | — | — | ||||
Genome-wide frequencies of SSR arrays of given motif length and repeat number were compared to mean frequencies in computer-generated randomized genomes (shown in parentheses). (*) Significant deviations from normal distribution-based expectations at the 0.001 level are indicated by asterisks.
Figure 1.
Abundance, distribution, and lengths of SSR tracts in the E. coli genome, shown as overall length of an SSR tract at a given position in the genome.
Analysis of genome-wide frequencies of SSR arrays of given motif length and repeat number showed a significant (P < 0.001) excess of mono- and trinucleotide SSRs relative to expectations (Table 1). Expected frequencies of SSRs of given motif length and repeat number were determined by observing those in 10 computer-generated genomes constructed by random ordering of nucleotides according to their overall frequencies in the genome, with departures tested using parametric statistics. A few significant test results for tract lengths larger than 10 bp (data not shown) likely were attributable to the small numbers expected.
In eukaryotes, SSRs are most abundant in noncoding areas that have little or no effect on gene expression. To determine whether this also was the case for E. coli, its complete DNA sequence first was subjected to a computerized screening for the gross locations of SSRs relative to open reading frames (ORFs). The E. coli K-12 genome of 4.64 × 106 nucleotides includes 79.5% of the genome in ORFs and 20.5% of the genome in noncoding regions (Table 2). Mononucleotide SSRs 3 bp in length were distributed among coding and noncoding regions at very nearly the same proportions, 78.0% and 22.0%, respectively. However, as mononucleotide repeat number became higher, the tracts became more and more under-represented in ORFs. The regression of proportion of mononucleotide SSRs in noncoding regions on tract length was positive and significant (see Table 2). In contrast, the distribution of SSR tracts with higher-order motifs among ORFs and noncoding regions approximated the overall proportion of these regions in the genome.
Table 2.
Distribution of SSR Tracts Among Coding and Noncoding Regions for Escherichia coli
| Genome-wide | Coding | Noncoding | |||
|---|---|---|---|---|---|
| no. | no. | % | no. | % | |
| Mononucleotidesa (bp) | |||||
| 3 | 163,345 | 127,407 | 78.0 | 35,938 | 22.0 |
| 4 | 42,901 | 31,777 | 74.1 | 11,124 | 25.9 |
| 5 | 13,837 | 9,830 | 71.0 | 4,007 | 29.0 |
| 6 | 4,123 | 2,696 | 65.4 | 1,427 | 34.6 |
| 7 | 1,000 | 544 | 54.4 | 456 | 45.6 |
| 8 | 217 | 92 | 42.4 | 125 | 57.6 |
| 9 | 22 | 12 | 54.5 | 10 | 45.5 |
| 10 | 1 | 0 | 0.0 | 1 | 100.0 |
| Dinucleotides >6 | 7,575 | 5,779 | 76.3 | 1,796 | 23.7 |
| Trinucleotides >9 | 2,419 | 1,989 | 82.2 | 430 | 17.8 |
| Tetranucleotides | 52 | 41 | 78.9 | 11 | 21.1 |
| Genome partition | — | — | 79.5 | 20.5 | |
The regression of proportion of mononucleotides SSR tracts in noncoding region on tract length is y = 6.8 x − 1.77, where y is the predicted proportion in noncoding and x is tract length. The correlation coefficient was ρ = 0.96, with a = 6.8, which is significant with p < 0.005.
The nucleotide composition of the E. coli genome and its SSR tracts, including breakdowns for coding and noncoding regions, is presented in Table 3. The composition of mono- and dinucleotide SSRs differed from that of the genomic regions in which they occurred. The composition of mononucleotide SSRs exhibited a strong over-representation of A and T, 93% overall (Table 3b). Of the six possible dinucleotide motifs, the 49.1% frequency of CG/GC in ORFs clearly exceeded the 17.3% expected. In noncoding regions, AT/TA was over-represented relative to expectation (24.4% vs. 17.9%), as was CG/GC (23.1% vs. 15.4%). The frequencies of SSRs with particular motifs of 3 or 4 bp did not represent all possible combinations equally. Most notably, of 52 tetranucleotide SSRs, TGGC occurred 12 times and its complement, GCCA, 9 times in coding sequences. The finding that the E. coli genome is rich in TGGC has been attributed to the activity of VSP (very short patch) repair that corrects T:G mismatches to C:G (Bhagwat and McClelland 1992; Gutierrez et al. 1994). The occurrence of three repeats of the tetranucleotide TGGC has been identified as a mutation hot spot in the promoter of the lacI gene (Sedgwick et al. 1986; Murata-Kamiya et al. 1997).
Table 3.
Nucleotide Composition of the Genome, and of SSR Tracts, Distinguishing Among Coding and Noncoding Regions, for E. coli
| Nucleotide | Genome-wide | Coding | Noncoding | |||
|---|---|---|---|---|---|---|
| no. | % | no. | % | no. | % | |
| A. Nucleotide composition of genome | ||||||
| A | 1141 × 103 | 24.6 | 881 × 103 | 24.0 | 260 × 103 | 26.9 |
| C | 1178 × 103 | 25.4 | 955 × 103 | 26.0 | 224 × 103 | 23.1 |
| G | 1176 × 103 | 25.4 | 951 × 103 | 26.0 | 225 × 103 | 23.1 |
| T | 1139 × 103 | 24.6 | 879 × 103 | 24.0 | 261 × 103 | 26.9 |
| Total | 4636 × 103 | 100.0 | 3666 × 103 | 100.0 | 970 × 103 | 100.0 |
| B. Numbers of SSR tracts of given composition | ||||||
| Mononucleotide SSRs ≥6 bp | ||||||
| A | 2473 | 46.2 | 1521 | 45.5 | 952 | 47.3 |
| C | 191 | 3.6 | 125 | 3.7 | 66 | 3.3 |
| G | 186 | 3.5 | 111 | 3.3 | 75 | 3.7 |
| T | 2508 | 46.8 | 1587 | 47.5 | 921 | 45.7 |
| Total | 5358 | 100.0 | 3344 | 100.0 | 2014 | 100.0 |
| Dinucleotide SSRs ≥6 bp | ||||||
| AC/CA | 776 | 10.2 | 565 | 9.7 | 211 | 12.3 |
| AG/GA | 928 | 12.3 | 696 | 11.9 | 232 | 13.5 |
| AT/TA | 911 | 12.0 | 492 | 8.4 | 419 | 24.3 |
| CG/GC | 3274 | 43.2 | 2876 | 49.1 | 398 | 23.1 |
| CT/TC | 939 | 12.4 | 682 | 11.7 | 257 | 14.9 |
| GT/TG | 747 | 9.9 | 542 | 9.3 | 205 | 11.9 |
| Total | 7575 | 100.0 | 5853 | 100.0 | 1722 | 100.0 |
Fine Positions of SSRs Relative to ORFs
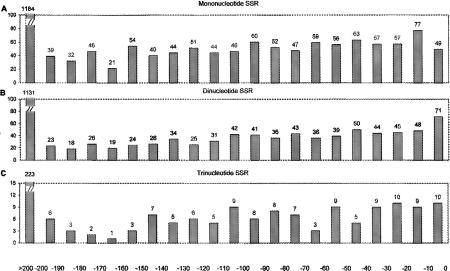
The presence and variation of SSRs in upstream regulatory elements might affect the expression of ORFs in either an on/off or a quantitative manner. We surveyed the fine positions of all SSRs in noncoding regions genome-wide relative to the ATG codon marking the start of translation of the adjacent gene (Fig. 2). There are 2178 mononucleotide SSR tracts >6 bp in length within 200 bp upstream of such ATG codons (Fig. 2A). The number of such SSRs in noncoding areas decreases with distance from the start of translation because the number of inter-ORF sequences of given length also decreases. Similar distributions, with decreasing numbers of SSRs at greater distances from the start of translation, were observed for di- and trinucleotide SSRs (Fig. 2B,C). Because this is a compact, prokaryotic genome, intergenic regions are usually short, and a subset of SSRs are in upstream areas where variation could affect gene expression (see Discussion below).
Figure 2.
Histograms showing frequencies of fine locations of SSR tracts in the entire E. coli genome relative to start of translation for particular ORFs downstream of the SSR tracts for mononucleotide SSRs >6 bp (A), dinucleotide SSRs >6 bp (B), and trinucleotide SSRs >9 bp (C). The horizontal axis shows position relative to the ATG codon marking the start of translation.
Polymorphism of SSRs
Screening for polymorphisms among strains of E. coli was conducted at 14 arbitrarily chosen loci containing SSR repeats (Table 4). DNA at chosen loci was amplified by PCR using primers flanking the particular SSR locus. Repeat number polymorphism at the ycgW locus was observed as differential mobility of radioactively labeled amplification products through polyacrylamide gels (Fig. 3), demonstrating hypervariable single-locus DNA fingerprint bands distinguishing among E. coli strains. DNA sequence alignments (Fig. 4) showed that a number of mononucleotide SSR arrays in noncoding regions were polymorphic, exhibiting two to four alleles for SSR repeat number. At three loci, additional polymorphisms observed in sequences flanking the targeted SSR tract proved to be due to different numbers of mononucleotides (Fig. 4A–C). Two SSR polymorphisms at the ycgW gene (Fig. 4A) were located upstream of the ORF at the −77 position and, depending on the strain, at the −84 to −89 position relative to the ATG codon at the start of translation. DNA from some of the pathogenic strains did not exhibit PCR amplification; presumably, one or both primers did not anneal because of sequence variation at the site.
Table 4.
Summary of Variation for 14 SSR Loci Screened Among Strains of E. coli
| Strain and substrain | No. of Repeats | Core | Coding or noncoding region | Genomic location, name of ORF, or downstream ORF |
|---|---|---|---|---|
| Mononucleotide SSRs | ||||
| K12:DH5α, K12:W3110 | 8 | G | noncoding | G1787407, ycgW |
| B SR9b | 4 | |||
| ETEC | 10 | |||
| EPEC | 8 | |||
| K12:DH5α,K12:W3110 | 10 | C | noncoding | G1786555, yaiN |
| B SR9c | 8 | |||
| E:1 | 10 | |||
| E:54, E:68 | 9 | |||
| K12:DHα,K12:W3110, BSR9b, BSR9c EHEC, E18 | 8 7 | A | noncoding | G1787106, serW |
| K12, B, EHEC, EPEC, ETEC | 9 | T | noncoding | G1790782, YjiD |
| K12:W3110, BSR9c, ETEC, E18 EHEC | 9 8 | T | noncoding | G1786218, caiF |
| K12:W3110, B SR9b, EHEC, EPEC, ETEC | 8 | C | coding | G1787051, b0829 |
| K12:W3110, B SR9b, EHEC, EPEC, ETEC | 9 | A | coding | G1790021, yibA |
| Dinucleotide SSRs | ||||
| K12, B, EHEC, EPEC, ETEC, E:1-69 | 4.5 | GT | noncoding | G1790630, aidB |
| K12, B, EHEC, EPEC, ETEC, E:1-69 | 4.5 | TC | noncoding | G1788430, molR_1 |
| K12:W3110, B SR9b, EHEC, EPEC, ETEC | 6 | GC | coding | G1786541, mhpR |
| Trinucleotide SSRs | ||||
| K12, B, EHEC, EPEC, ETEC, E:1-69 | 4 | GGT | noncoding | G1787973, b1688 |
| K12, B, EHEC, EPEC, ETEC, E:1-69 | 5 | CGG | coding | G1786284, ftsZ |
| Trinucleotide SSRs | ||||
| K12, B, EHEC, EPEC, ETEC, E:1-69 | 3 | ATTA | noncoding | G1789986, yiaB |
| K12, B, EHEC, EPEC, ETEC, E:1-69 | 4 | CTGG | coding | G1788332, hisC |
Figure 3.
Mobility differences in PCR products harboring specific SSR tracts among strains of E. coli following electrophoresis in a 5% acrylamide TBE denaturing sequencing gel. PCR was performed using primer pairs, one radiolabeled, flanking the poly(G) tract at a genomic site ∼77 bp upstream from the ATG site of the ycgW locus. The dried gel was exposed to a PhosphorImager. The expected size of the E. coli K12 amplification product was 200 bp. Shown are amplification products for the following strains: (lane 1) K12:DH5α; (lane 2) B:SR9c; (lane 3) B:SR9b; (lane 4) ETEC:O78:H [E10407]; (lane 5) EPEC: O111[E639616]; (lane 6) E:1; (lane 7) E:7; (lane 8) E:18; (lane 9) E:47.
Figure 4.
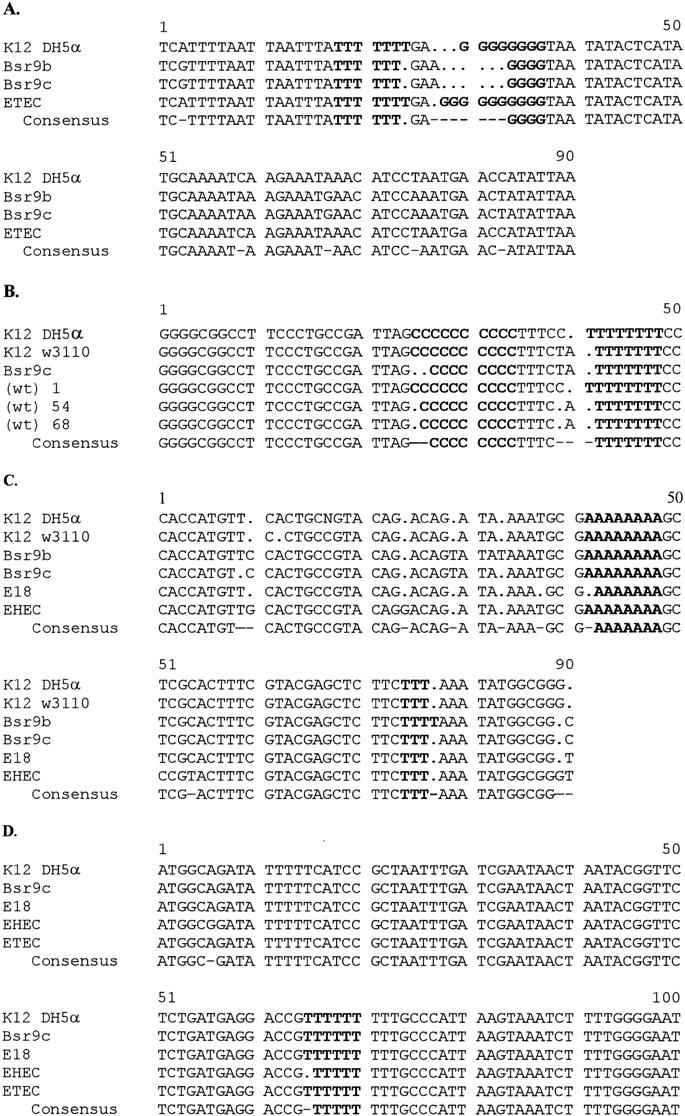
DNA sequence alignments for complementary DNA strands for four loci bearing mononucleotide repeat polymorphisms among strains of E. coli. PCR products were sequenced using the dideoxy-chain termination method and aligned using the Pile-up GCG program. Polymorphic SSR tracts are shown in boldface letters. (A) Poly(G) tract 77 bp and poly(T) tract 84–89 bp upstream of the ATG element of ycgW; (B) poly(C) tract ∼90 bp and poly(T) tract 76 bp upstream of the ATG element of yaiN; (C) poly(A) tract 35 bp and poly(T) tract 67 bp downstream of the tRNA gene serW; (D) poly(T) tract 38 bp upstream of the ATG element of caiF.
Overall, four SSR tracts exhibited length polymorphism among strains of E. coli (Table 4). All four polymorphic sites shared three characteristics. Namely, they involved mononucleotide SSRs in noncoding regions. This is particularly striking because, in all, only five sites meeting these criteria were examined. In contrast, length polymorphism was not shown by two mononucleotide SSRs in coding regions or by seven higher-order SSRs in either coding or noncoding regions. The numbers examined in these categories, however, were too small to determine which of the two defining characteristics (mononucleotide motif or noncoding location) was more important for the presence of polymorphism. All SSRs examined had a tract length of at least 8 nucleotides in the sequenced E. coli K12 genome. Thus, mononucleotide SSRs of this length appear to have a high likelihood of being polymorphic among E. coli strains. In all, there are 240 mononucleotide tracts of this length in the E. coli K12 genome. Polymorphism among tracts of lesser length was not examined.
DISCUSSION
Until recently, SSR regions in E. coli were thought to be rare and limited to dinucleotide SSRs with a maximum of five repeat units per locus (van Belkum et al. 1998). However, Field and Wills (1998) presented data reporting tens of thousands of mononucleotide SSRs in E. coli and showing the existence of SSRs with longer motifs. Our results confirm that SSR tracts in E. coli are numerous and diverse in terms of motif and repeat number and show that they are widely distributed throughout the genome. We show that mononucleotide SSRs occur more frequently than expected in noncoding areas. SSRs of many motif lengths differ in composition from the genomic regions in which they occur, with mononucleotide SSRs with poly(A) or poly(T) strongly over-represented in both coding and noncoding regions. We show polymorphism of mononucleotide SSRs in noncoding regions.
Distribution of SSR Tract Length and Structure
Mutation at SSR loci is believed to be the consequence of slipped-strand mispairing during DNA replication (Strand et al. 1993). This is because the tertiary structure of repetitive DNA allows mismatching of neighboring repeats, and depending on the strand orientation, repeats can be inserted or deleted during DNA polymerase-mediated DNA duplication (Coggins and O'Prey 1989; Hauge and Litt 1993; Chiurrazzi et al. 1994). The resulting mutations are not always repaired by DNA mismatch-repair mechanisms (Strand et al. 1993; Modrich and Lahue 1996). Our observation of upper limits for SSR array lengths in E. coli (i.e., 9 bp for mononucleotides and 12 bp for SSRs with longer motifs; Fig. 1) suggests that the tendency for repeat length at a locus to rise via mutation is counteracted by selection. Such selection might operate through an uncharacterized mechanism on the length of the SSR sequence itself or on gene expression as affected by the SSR sequence at issue.
Interacting processes of mutation and selection can be invoked to explain observations regarding motif length and repeat number at SSR tracts. Slipped-strand mispairing is more likely for mononucleotide SSRs than for higher-order SSRs, because both strand separation and slippage are more likely. This is particularly important for poly(A) and poly(T), as strand separation is easier than for poly(C) and poly(G). For higher-order SSRs having small repeat number, there is very little mutability in repeat number. Thus, selection would have considerable opportunity to operate only against larger repeat numbers. In coding regions, variation in mononucleotide SSR repeat number causes frame-shift, nonsense mutations and, hence, will be selected against strongly. Thus, there will be a balance between production of SSRs and selection against them. In noncoding regions, any effects of mononucleotide SSR repeat number variation are less obvious. The tremendous lack of poly(C) and poly(G) SSR tracts is remarkable and requires explanation.
Expectations of SSR frequencies were calculated on the basis of the genome-wide nucleotide composition of ORFs and noncoding regions. Within ORFs, this approach does not reflect differential nucleotide composition at the first, second, and third codon positions (Andachi et al. 1987); differential nucleotide compositions among different classes of genes (Hirosawa et al. 1997); and the effect of codon usage bias on the distribution of polynucleotides within ORFs (Sharp 1991; Sharp et al. 1995). The importance of these effects remains to be evaluated in future study.
Locations of SSRs Relative to ORFs
To assess the likelihood that SSRs might affect gene expression, we examined the positions of all SSRs in the genome with regard to ORFs. In Figure 2, we show the distribution of SSRs upstream of ORFs in relation to the first ATG codon of ORFs. Substantial numbers of SSR tracts are localized up to 200 bp from the ATG. The DNA sequence immediately upstream of an ORF contains proximal regulatory elements that play an important role in controlling expression of the gene. In E. coli, mononucleotide SSRs occurred in noncoding regions more frequently than expected by chance. Given the compact nature of the E. coli genome, almost any genetic variation might affect gene function; however, variation in SSR arrays at regulatory regions of genes must affect gene expression in a way that can be tolerated by the host (Kashi et al. 1997; King et al. 1997). Variation at or near regulatory elements can influence gene expression by affecting binding of regulatory elements (Bewley et al. 1998), distance between regulatory elements, bending of DNA (Perez-Martin et al. 1994), blocking of DNA replication elongation (Krasilnikov et al. 1999), phasing on the DNA helix, formation of unusual DNA structures (Williamson 1994; Soyfer and Potaman 1995), DNA coiling, DNA packaging (Pettijohn 1988), or other mechanisms (Kashi 1998). Some of these variations affect gene expression in a gross on-off manner (Rosenberg et al. 1994; Moxon and Wills 1999), whereas others affect fine-tuning of the level of gene expression (Kashi et al. 1997; King et al. 1997). Hypervariable SSR loci serve as transcriptional or translational switches in a variety of pathogenic (Himmelreich et al. 1996; Karlin et al. 1996; Henaut et al. 1998) and nonpathogenic (Field and Wills 1998) microbes. Our computer-based screening showed that large repeat tracts with motifs of 2 or more bp do not occur in the E. coli K12 genome. It has been shown in eukaryotes that tracts of certain types of repetitive DNA are localized to the 5′ or 3′ flanking regions of genes, where they may affect nucleosome organization, recombination, or regulation of gene expression or gene product activity (Tripathi and Brahamachari 1977; Kashi et al. 1997; King et al. 1997; Kashi and Soller 1998), suggesting the need for further study in prokaryotes.
Practical and Evolutionary Implications of SSR Polymorphisms
Observation of repeat number variation at SSR loci in E. coli suggests that SSRs may prove a ready source of polymorphisms for marking its genome. SSR loci in other prokaryotes also have been shown to exhibit length polymorphisms (for review, see Moxon et al. 1994; van Belkum et al. 1998). For example, variation for specific trinucleotide repeats of very large tract size was shown for Neisseria meningitidis, Mycoplasma genitalium, and Mycobacterium leprae (Field and Wills 1996), and SSR variation has been observed in Staphylococcus aureus and Hemophilus influenzae (van Belkum et al. 1996, 1997a,b). Polymorphism of SSR tracts in prokaryotes poses both practical and evolutionary implications.
Although E. coli is part of the normal human microflora, there are pathogenic strains for which rapid detection and strain identification are important. Present-day approaches for typing of prokaryotes (Vanderzant and Splittstoesser 1992) have limited ability to distinguish among E. coli strains and are time consuming (Padhye and Doyle 1992; Yu and Kaper 1992; Witham et al. 1996). Screenings of SSR variation may provide the basis for rapid and sensitive identification of pathogenic and nonpathogenic E. coli strains. Polymorphic mononucleotide sites found in E. coli exhibited 1–4 bp size differences. The small numbers of repeats are well suited for development of SSR allele-specific oligonucleotides (ASOs). Such ASOs may be used, for example, as PCR primers, or as hybridization probes that can be spotted on DNA microarrays (Southern 1996; Marshall and Hodgson 1998; Ramsey 1998) for rapid, automated characterization of variation at a given set of loci for purposes of DNA fingerprinting of E. coli strains. Similarly, knowledge of SSR variation in other pathogenic microbes, such as H. influenzae (van Belkum et al. 1997a), Candida albicans (Field et al. 1996; Bretagne et al. 1997), Bacteroides fragilis and Bacteroides thetaiotaomicron (Claros et al. 1997), Helicobacter pylori (Marshall et al. 1996), and N. meningitidis (Tonjum et al. 1998), has been or could be applied for rapid detection and strain characterization. A DNA-fingerprinting approach based on SSR polymorphism also can be used for epidemiological purposes, for example, to determine whether a pathogenic E. coli strain detected in a patient matches a known or suspected source of a given disease outbreak. SSRs have been used as markers for such purposes for several pathogenic microbes (for review, see van Belkum 1999), including Mycobacterium tuberculosis (van Soolingen et al. 1993), H. pylori (Marshall et al. 1996), and H. influenzae (van Belkum et al. 1997a). To demonstrate a similar approach in E. coli, a collection of allelic SSR markers distinguishing relevant strains will have to be developed. Recent work with hypervariable markers in pathogenic microbes (van Belkum et al. 1996; Moxon and Wills 1999) shows that the variability at particular markers will have to be evaluated to determine that it reflects the overall rate of evolution of the E. coli genome.
SSRs can be screened to determine whether such molecular variation gives rise to phenotypic variation. For example, SSR variability poses clear implications for virulence in pathogenic microbes. Tracts of SSRs have been found within confirmed or potential virulence genes of H. influenzae (Karlin et al. 1997); Neisseria sp., Hemophilus parainfluenzae, and Moraxella catarrhalis (Peak et al. 1996), and repeat number variation seems to be related to modulation of expression of virulence factors. Contingency genes containing SSRs exhibit high mutation rates, allowing the bacterium to respond rapidly to challenging environmental conditions (Moxon et al. 1994). Locating SSR repeat arrays by computerized search of the genomic sequence and localization of such arrays with regard to expressed genes, as we report here, could provide a basis for discovering new virulence- or other key phenotype-determining loci in bacteria.
All of the SSR polymorphisms observed at the arbitrarily chosen sites screened in this study were in noncoding regions. Over an evolutionary time frame, E. coli has allowed these polymorphisms to persist. Allelic variation mostly was conserved within each E. coli strain that we screened. These SSR sites were not hypervariable, as were SSRs at contingency genes in pathogens such as H. influenzae (Karlin et al. 1997; van Belkum et al. 1997a; Moxon and Wills 1999). These observations may support the hypotheses (Moxon et al. 1994; Kashi et al. 1997; King et al. 1997; Moxon and Wills 1999) that mutation rates are higher in genes whose products interact with the environment in unpredictable ways and that SSRs affect mutability so that different classes of genes have adaptively appropriate mutation rates. Mutability rate may be mediated by SSR motif length and overall tract length. We hypothesize that certain SSR variation drives fine-tuning of gene expression as well as variation of key phenotypes, providing an important target for natural selection, thereby affecting evolution of both pathogenic and nonpathogenic E. coli strains. It is possible that some portion of between-strain functional variation in E. coli results from differences in SSR repeat number in gene regulatory regions. Further DNA sequencing in genomes of pathogenic E. coli strains could yield insights into relative rates of mutability among SSR loci and into the phenotypic consequences of SSR variation.
METHODS
DNA Sequence Analysis Software
We developed DNA sequence analysis software in the programming language C that screens entire genomes for SSRs and reports motif, number of repeats, and genomic position. It is available for downloading from our university's ftp site at ftp://ftp.technion.ac.il/pub/supported/biotech/ssr.exe. It searches for all of the SSRs with motif lengths up to 10 bp; records motif, repeat number, and genomic location; and reports the results in an output file. The complete genomic sequence of E. coli was obtained from http://mol.genes.nig.ac.jp/ecoli/ and screened for SSRs, their motif sequence, number of repeats, and genomic locations.
A second program in the programming language C characterizes the locations of SSR arrays in relation to ORFs in genomic sequence data sets. It reports the numbers of occurrences of SSRs of specified motif length and repeat number in both ORFs and noncoding sequences. For SSRs occurring upstream of ORFs, it reports the number of nucleotides between the SSR tract and the ATG codon marking the start of translation.
Statistical Testing of SSR Frequencies
To determine whether frequencies of SSRs of given motif length and repeat number occurred as expected by chance, ten simulated genomes were constructed by randomly choosing nucleotides at the frequencies characterizing the E. coli genome. The simulated genomes then were analyzed using the genome scanning software described above to determine the number of SSRs of given motif length and repeat number. Results of the ten runs were summarized in terms of means and standard errors, yielding expected numbers of tracts of given motif length and repeat number. Departures of observed numbers of SSRs of given motif length and repeat number from expectations were tested using parametric statistics.
Were all nucleotides equally frequent in the genome, the relative frequencies of the six possible combinations of nucleotides in dinucleotide SSRs all would equal 0.167. However, because frequencies of the respective nucleotides were not equal, expectations for the relative frequencies of particular dinucleotides (E) were adjusted, as E = (fN1 + fN2) × 2 × 0.167, where fN1 and fN2 are the frequencies of nucleotides 1 and 2, respectively. For example, the frequencies of both C and G in ORFs are 0.26, and we seek the frequencies of CG and GC dinucleotides. Hence, E = (0.26 + 0.26) × 2 × 0.167 = 0.173.
Screening for Variability of SSRs Among E. coli Strains
Nonpathogenic and pathogenic strains of E. coli screened for variation at SSR loci included K12 (DH5α, W3110), B (SR9b, SR9c), E (1, 7, 11, 18, 47, 52, 54, 63, 68, 69); EHEC O157:H7 (FEB, Rowe no. E304810, HER 1057, 1058, 1261, 1265, 1266), EPEC [serotype O111ac (Rowe no. E639616)], ETEC [serotype O78:H (Rowe no. E10407)]. The K and B strains were obtained from the microbiology laboratory collection of our department. The E strains were isolated by and obtained from Ochman and Selander (1984). The EHEC O157:H7 HER strains were isolated by and obtained from Ahmed et al. (1987). Cultures for DNA extraction were grown on Luria broth agar plates for 24 hr at 37°C. A large loop of colonies from the plate was transferred to a microcentrifuge tube containing 500 μl of TE buffer (pH 7.5) and vortexed thoroughly. Bacterial cells were lysed at 80°C for 10 min and centrifuged for 10 min at 14,000 rpm (20,800g). The pellet was suspended in 100 μl of TE, boiled for 5 min, and centrifuged at 14,000 rpm for 2 min (Kirschner and Bottger 1996). The supernatant was held at −20°C until used for PCR.
Fourteen SSR loci of E. coli were selected for detailed analysis. The forward (F) and reverse (R) PCR primer sequences for the loci examined were as follows: ycgW (F = 5′-GATTTTGCATATGAGTATATTAC-3′, R = 5′-TTAATTACAGGATGTTCAGTC-3′); yaiN (F = 5′-AATTTATCCGGTGAATGTGGT-3′, R = 5′-CAACTTAATCTCGGGCTGAC-3′); serW (F = 5′-TTCCACAGGTAACATACTCCAC-3′, R = 5′-TTTGGTGAGGTCTCCGAG-3′); YjiD (F = 5′-TACATGGCTGATTATGCGG-3′, R = 5′-TCGCTATGAATATCTACTGAC-3′); aidB (F = 5′-GTCAGAGCAGATCCAGAATG-3′, R = 5′-TCTACAGCAAATGAACAATG-3′); molR_1 (F = 5′-GGTCATCAGGTGAAATAATC-3′, R = 5′-CGTCCTGATAGATAAAGTGTC-3′); ftsZ (F = 5′-CAATGGAACTTACCAATGAC-3′, R = 5′-TACCGCGAAGAATTCAACAC-3′); b1668 (F = 5′-AGCATCAGCGCACAATGCAC-3′, R = 5′-TGTATGCAGGCTGGCACAAC-3′); yiaB (F = 5′-ATAACGATCTCCATATCTAC-3′, R = 5′-CTCTATCAGCAACTTCTGCC-3′); hisC (F = 5′-ATCCGCAGGATTTTCGCACC-3′, R = 5′-TGCCAGCGTAAATCCGCAAC-3′); MhpR (F = 5′-AATCACCCGTTGTTCACT-3′, R = 5′-CGGAACAAGACCGCAAGGA-3′); b0829 (F = 5′-ACCGCAACATCCTTACAC-3′, R = 5′-TGACAAGATTACGCACTC-3′); yibA (F = 5′-AATCGGACTTTCCTACAGA-3′, R = 5′-AACTCACGCTATGAACGC-3′); and caiF (F = 5′-TGAATGCCGATGCGACTG-3′, R = 5′-GTATGCAACTTCACCGTC-3′).
Five microliters of DNA extract (∼50 ng), 2.5 μl of 10× PCR buffer (ProMega, 25 mm Mg2+ added), 0.2 μl of 25mm dNTPs, 1.0 units of Taq polymerase (Promega), and l0 pmoles each of F and R primers were brought to a final volume of 25 μl with sterile ddH2O. Mineral oil (15–20 μl) was added for PCR in a MJ Research thermocycler without a heating cover. The cycling conditions for PCR consisted of denaturation at 95°C for 5 min, followed by 5 cycles (1 min at 95°C, 1 min at Tm, and 1 min at 72°C), 20 cycles (1 min at 95°C, 1 min at Tm − 5°C, and 1 min at 72°C), a final step of 7 min at 72°C, and cooling to room temperature.
Methods for radioactive PCR were as follows: To label primers, 2 μl (1 ng) of primer DNA, 2 μl of 10× T4 kinase buffer (NEB), 4 μl of [γ-35S] ATP (250 mCi, NEN), and 1 μl (10 units) of T4 DNA kinase (NEB) were brought to a final volume of 20 μl with sterile ddH2O. The contents were mixed and incubated at 37°C for 1 hr. The reaction was stopped by incubation at 70°C for 10 min. For the radioactive PCR reaction, 0.5 μl of nonradioactive and 0.5 μl of radioactive primer (together, 10 pmoles) were used following the PCR protocol described above. To observe small size differences among PCR products, electrophoresis of radioactive products was carried out in a 5% denaturing TBE acrylamide gel. The gels were dried (80°C for 1.5 hr) and exposed to a PhosphorImager cassette, and the results were read using a PhosphorImager (Bas reader 100, Fuji).
PCR products were eluted from electrophoretic gels using Jetsorb (Genomed) and sequenced by the dideoxy-chain termination method using an ABI automated sequencing machine (Biological Services, Weizmann Institute, Rehovot, Israel).
Acknowledgments
This research was supported in part by the Technion Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation, Germany. R.G.-A. was supported by the Food Control Administration in the Israel Ministry of Health. E.H. was supported by Virginia Polytechnic Institute and State University and by the U.S. Fulbright Senior Scholars Program. We are grateful to A. Korol, T. Haran, M. Soller, N. Ulitzur, and two anonymous reviewers for constructive comments on drafts of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kashi@tx.technion.ac.il; FAX 972-4-8320742.
REFERENCES
- Ahmed R, Bopp C, Bonczyk A, Kasatiya S. Phage typing scheme for Escherichia coli 0157:H7. J Infect Dis. 1987;155:806–809. doi: 10.1093/infdis/155.4.806. [DOI] [PubMed] [Google Scholar]
- American Public Health Association (APHA); American Water Works Association; Water Pollution Control Association. Standard methods for the examination of water and wastewater. 16th ed. Washington, D.C.: APHA; 1985. p. 878. [Google Scholar]
- Andachi Y, Yamao F, Iwami M, Muto A, Osawa S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNA in Mycoplasma capriculum. Proc Natl Acad Sci. 1987;84:7398–7402. doi: 10.1073/pnas.84.21.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: Structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998;27:105. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AS, McClelland M. DNA mismatch correction by Very Short Patch repair may have altered the abundance of oligonucleotides in the E. coli genome. Nucleic Acids Res. 1992;20:1663–1668. doi: 10.1093/nar/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FM, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bretagne S, Costa JM, Besmond C, Carsique R, Calderone R. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as a basis for a typing system. J Clin Microbiol. 1997;35:1777–1780. doi: 10.1128/jcm.35.7.1777-1780.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi P, Kozak L, Neri G. Unstable triplets and their mutational mechanism: Size reduction of the CGG repeat versus germline mosaicism in the fragile X syndrome. Am J Med Genet. 1994;51:517–521. doi: 10.1002/ajmg.1320510446. [DOI] [PubMed] [Google Scholar]
- Claros MC, Gerardo SH, Citron DM, Goldstein EJ, Schonian G, Rodloff AC. Use of the polymerase chain reaction fingerprinting to compare clinical isolates of Bacteroides fragilis and Bacteroides thetaiotaomicron from Germany and the United States. Clin Infect Dis (Suppl. 2) 1997;25:S295–S298. doi: 10.1086/516200. [DOI] [PubMed] [Google Scholar]
- Coggins LW, O'Prey M. DNA tertiary structures formed in vitro by misaligned hybridization of multiple tandem repeat sequences. Nucleic Acids Res. 1989;17:7417–7426. doi: 10.1093/nar/17.18.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Economic Community (EEC) Council directive 80/777/EEC on the laws of member states relating to the exploitation and marketing of natural mineral water. Off J Eur Commun. 1980;23(L229):1–10. [Google Scholar]
- Field D, Wills C. Long, polymorphic microsatellites in simple organisms. Proc Royal Acad London B. 1996;263:209–251. doi: 10.1098/rspb.1996.0033. [DOI] [PubMed] [Google Scholar]
- ————— Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Natl Acad Sci. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Eggert L, Metzgar D, Rose R, Wills C. Use of polymorphic short and clustered coding-region microsatellites to distinguish strains of Candida albicans. FEMS Immunol Med Microbiol. 1996;15:73–79. doi: 10.1111/j.1574-695X.1996.tb00056.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez G, Casadesus J, Oliver JL, Marin A. Compositional heterogeneity of the E. coli genome: A role for VSP repair? J Mol Evol. 1994;39:340–346. doi: 10.1007/BF00160266. [DOI] [PubMed] [Google Scholar]
- Hauge XY, Litt M. A study of the origin of “shadow bands” seen when typing dinucleotide repeat polymorphisms by the PCR. Nucleic Acids Res. 1993;2:411–415. doi: 10.1093/hmg/2.4.411. [DOI] [PubMed] [Google Scholar]
- Hays PR. Food microbiology and hygiene. 2nd ed. 1992. p. 8. , 70. Elsevier Applied Science, Amsterdam, The Netherlands. [Google Scholar]
- Henaut A, Lisacek F, Nitschke P, Moszer I, Danchin A. Global analysis of genomic texts: The distribution of ACGT tetranucleotides in the Escherichia coli and Bacillus subtilis genomes predict translational frameshifting and ribosomal hopping in several genes. Electrophoresis. 1998;19:515–527. doi: 10.1002/elps.1150190411. [DOI] [PubMed] [Google Scholar]
- Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4421–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa M, Isono K, Hayes W, Borodovsky M. Gene identification and classification in Synechocystis genomic sequence by recursive gene mark analysis. DNA Sequencing. 1997;8:17–19. doi: 10.3109/10425179709020881. [DOI] [PubMed] [Google Scholar]
- Jay JM. Modern food microbiology. 5th ed. New York, NY: Chapman and Hall; 1996. p. 195. [Google Scholar]
- Johnson JR. Virulence factors in Escherichia coli UTI. Clin Microbiol Rev. 1991;4:82–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Mrazek J, Campbell AM. Frequent oligonucleotides and peptides of the Hemophilus influenzae genome. Nucleic Acids Res. 1996;21:4263–4272. doi: 10.1093/nar/24.21.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Compositional biases of bacterial genomes and evolutionary implications. J Bacteriol. 1997;179:3899–3913. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi Y, Soller M. Functional roles of microsatellites and minisatellites. In: Goldstein DD, Schlotterer C, editors. Microsatellite evolution and application. Oxford, U.K: Oxford University Press; 1998. pp. 10–23. [Google Scholar]
- Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- King DG, Soller M, Kashi Y. Evolutionary tuning knobs. Endeavor. 1997;21:36–40. [Google Scholar]
- Kirschner P, Bottger EC. Detection of mycobacterium resistance to streptomycin and clarithromycin. In: Pershing DH, editor. PCR protocols for emerging infectious diseases. Washington, D.C.: ASM Press; 1996. pp. 130–137. [Google Scholar]
- Krasilnikov MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through simple DNA repeats blocks replication elongation. EMBO J. 1999;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzler P, Matsuo K, Schaffner W. Pathological, physiological, and evolutionary aspects of short unstable DNA repeats in the human genome. Biol Chem. 1995;376:201–211. doi: 10.1515/bchm3.1995.376.4.201. [DOI] [PubMed] [Google Scholar]
- Marshall A, Hodgson J. DNA chips: An array of possibilities. Nat Biotechaol. 1998;16:27–31. doi: 10.1038/nbt0198-27. [DOI] [PubMed] [Google Scholar]
- Marshall DG, Coleman DC, Sullivan DJ, Xia H, Morain CA, Smyth CJ. Genomic DNA fingerprinting of clinical isolates of Helicobacter pylori using short oligonucleotide probes containing repetitive sequences. J Appl Bacteriol. 1996;81:509–517. doi: 10.1111/j.1365-2672.1996.tb03540.x. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Moxon ER, Wills C. DNA microsatellites: Agents of evolution? Sci Am. 1999;280:94–99. doi: 10.1038/scientificamerican0199-94. [DOI] [PubMed] [Google Scholar]
- Moxon ER, Rainey PR, Nowak MA, Lenski RE. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- Murata-Kamiya N, Kamiya H, Kaji H, Kasai H. Mutational specificity of glyoxal, a product of DNA oxidation, the lacI gene of wild-type Escherichia coli W3110. Mut Res. 1997;377:255–262. doi: 10.1016/s0027-5107(97)00083-3. [DOI] [PubMed] [Google Scholar]
- Niedhardt FC. Escherichia coli and Salmonella. In: Niedhardt FC, et al., editors. Cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1–3. [Google Scholar]
- Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsivik O, Wastenson Y, Lund A, Hornes E. Pathogenic Escherichia coli found in food. Int J Microbiol. 1992;12:103–113. doi: 10.1016/0168-1605(91)90051-p. [DOI] [PubMed] [Google Scholar]
- Padhye NV, Doyle MP. Escherichia coli O57:H7: Epidemiology, pathogenesis, and methods for detection in food. J Food Protect. 1992;55:555–565. doi: 10.4315/0362-028X-55.7.555. [DOI] [PubMed] [Google Scholar]
- Peak IRA, Jennings MP, Hood DW, Bisercic M, Moxon ER. Tetrameric repeat units associated with virulence factor phase variation in Hemophilus also occur in Neiserria spp. and Moraxella catarrhalis. FEMS Microbiol Lett. 1996;137:109–114. doi: 10.1111/j.1574-6968.1996.tb08091.x. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: A common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn DE. Histone-like proteins and bacterial chromosome structure. J Biol Chem. 1988;263:12793–12796. [PubMed] [Google Scholar]
- Ramsey G. DNA chips: State of the art. Nat Biotechnol. 1998;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Sedgwick WD, Brown OE, Glickman BW. Deoxyuridine misincorporation causes site-specific mutational lesions in the lacI gene of Escherichia coli. Mutat Res. 1986;162:7–20. doi: 10.1016/0027-5107(86)90066-7. [DOI] [PubMed] [Google Scholar]
- Sharp PM. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurum: Codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Avrof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution: The sound of silence. Phil Trans Royal Soc Lond B Biol Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- Southern EM. DNA chips: Analysing sequence by hybridization to oligonucleotides on a large scale. Trends Genet. 1996;12:110–115. doi: 10.1016/0168-9525(96)81422-3. [DOI] [PubMed] [Google Scholar]
- Soyfer VN, Potaman VN. Triple helical nucleic acids. New York, NY: Springer-Verlag; 1995. [Google Scholar]
- Strand M, Prolla T, Liskay R, Petes T. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989;17:6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjum T, Caugant DA, Dunham SA, Koomy M. Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol Microbiol. 1998;29:111–124. doi: 10.1046/j.1365-2958.1998.00910.x. [DOI] [PubMed] [Google Scholar]
- Tripathi J, Brahamachari SK. Synthesis of hybrid bacterial plasmids containing highly repeated satellite DNA. Cell. 1977;10:509–518. doi: 10.1016/0092-8674(77)90038-1. [DOI] [PubMed] [Google Scholar]
- Van Belkum A. The role of short sequence repeats in epidemiologic typing. Curr Opin Microbiol. 1999;2:306–311. doi: 10.1016/S1369-5274(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Van Belkum A, Riewerts Eriksen N, Sijmons M, van Leeuwin W, VandenBergh M, Kluytmans J, Espersen F, Verbrugh H. Are variable repeats in the spa gene suitable targets for epidemiological studies of methicillin-resistant Staphylococcus strains? Eur J Clin Microbiol Infect Dis. 1996;15:768–769. doi: 10.1007/BF01691972. [DOI] [PubMed] [Google Scholar]
- Van Belkum A, Melchers WJG, Ijsseldijk C, Nohlmans L, Verbrugh HA, Meis JFGM. Outbreak of amoxycillin-resistant Haemophilus influenzae type b: Variable number of tandem repeats as novel molecular markers. J Clin Microbiol. 1997a;35:1517–1520. doi: 10.1128/jcm.35.6.1517-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum A, Scherer S, van Leeuwen W, Willemse D, van Alphen L, Verbrugh H A. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect Immunol. 1997b;65:5017–5027. doi: 10.1128/iai.65.12.5017-5027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, de Haas PEW, Hermans PWM, Groenen PMA, van Embden JDA. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderzant C, Splittstoesser DF. Compendium of methods for microbiological examination of foods. 3rd ed. Ann Arbor, MI: Edward Brothers; 1992. [Google Scholar]
- Vogt P. Potential genetic functions of tandemly repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code:” Hum. Genet. 1990;84:301–336. doi: 10.1007/BF00196228. [DOI] [PubMed] [Google Scholar]
- Weber JL. Informativeness of human poly(GT)n polymorphisms. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Williamson JR. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomolec Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- Witham PK, Yamashiro CT, Livak KJ, Batt CA. A PCR-based assay for the detection of Escherichia coli Shiga-like toxin genes in ground beef. Appl Environ Microbiol. 1996;62:1347–1353. doi: 10.1128/aem.62.4.1347-1353.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Kaper JB. Cloning and characterization of the eae gene of enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]