Abstract
Microsatellite enrichment is an excess of repetitive sequences characteristic to all studied eukaryotes. It is thought to result from the accumulated effects of replication slippage mutations. Enrichment is commonly measured as the ratio of the observed frequency of microsatellites to the frequency expected to result from random association of nucleotides. We have compared enrichment of specific types of microsatellites in coding sequences with those in noncoding sequences across seven eukaryotic clades. The results reveal consistent differences between coding and noncoding regions, in terms of both the quantity of repetitive DNA and the types present. In noncoding regions, all types of microsatellite (mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats) are found in excess, and in all cases, these excesses scale in a similar exponential fashion with the length of the microsatellite. This suggests that all types of noncoding repeats are subject to similar mutational and selective processes. Coding repeats, however, appear to be under much stronger and more specific constraints. Tri- and hexanucleotide repeats are found in consistent and significant excess over a wide range of lengths in both coding and noncoding sequences, but other repeat types are much less frequent in coding regions than in noncoding regions. These findings suggest that the differences between coding and noncoding microsatellite frequencies arise from specific selection against frameshift mutations in coding regions resulting from length changes in nontriplet repeats. Furthermore, the excesses of tri- and hexanucleotide coding repeats appear to be controlled primarily by mutation pressure.
Microsatellites are repetitive DNAs consisting of directly repeated short motifs of 1–6 nucleotides. These repeats are subject to a high rate of single-motif insertion and deletion mutations, through the process of replication slippage (Levinson and Gutman 1987; Tautz and Schlotterer 1994). Over time, the cumulative effect of these mutations generates highly significant excesses of repetitive tracts in the genomes of eukaryotes (Hancock 1995; Cox and Mirkin 1997). Because of their high rate of mutation, eukaryotic microsatellites have become a primary source of polymorphic markers for studies of population genetics and linkage analysis (Slatkin 1995). Microsatellites, and their associated mutational processes, have also been implicated in a variety of human genetic maladies, including myotonic dystrophy and Fragile X syndrome (Caskey et al. 1992).
A recent analysis of microsatellite distributions in whole genomic DNA of Saccharomyces cerevisiae has shown that enrichment of all types of microsatellites with motifs of from 1 to 5 nucleotides is primarily dependent on the number of base pairs contained in a repeat sequence, as opposed to the length or number of individual motifs (Pupko and Graur 1999). We observed similar patterns of enrichment in the noncoding sequences of all eukaryotic clades analyzed in this paper (for primates and plants, this relationship can be seen in Fig. 1). This suggests that microsatellites of a variety of motif types undergo similar rates and types of length mutation (or the fixation thereof) in both genomic DNA considered as a whole and in noncoding regions analyzed separately. This relationship between microsatellite length and genomic enrichment has been suggested to derive from the fact that replication slippage is dependent on secondary structure formation, which is itself dependent on sequence length (Cox and Mirkin 1997).
Figure 1.
Microsatellite enrichment in primate and plant sequences, with excess (observed number of repeats/expected number of repeats) shown as a function of repeat length in base pairs. The top two graphs show coding and noncoding enrichment of nontriplet microsatellites; and bottom two graphs show the same information for triplet repeats.
It has been noted for S. cerevisiae that noncoding regions have more long mono- and dinucleotide repeats (more than eight motifs in length) than coding regions, whereas trinucleotide repeat abundances are similar in coding and noncoding regions (Field and Wills 1998). Others have also recognized general differences in repeat frequencies between coding and noncoding regions (Tautz et al. 1986; Hancock 1996), and it has been suggested that these differences arise from increased selection in coding sequences. Because microsatellites are characterized by length mutations, we hypothesized that limitation of microsatellite expansion occurs in coding regions as a direct result of selection against frameshift mutations. To address this hypothesis, we have examined the differences between coding and noncoding microsatellite enrichment for all microsatellite motif lengths, in several widely diverged eukaryotic clades.
The hypothesis predicts that microsatellites with a motif length not divisible by three (that is, mono-, di-, tetra-, and pentanucleotide repeats) would be enriched less in coding regions than in noncoding regions. Length mutations in these nontriplet microsatellite types would lead to frameshift mutations when they occurred in coding DNA, and selection against these mutations would reduce their chances of fixation. Enrichment, being dependent on the rate of fixation, would likewise be reduced in coding regions. In contrast, microsatellites with a repeat length divisible by three (tri- and hexanucleotide repeats) should not generate frameshift mutations. Fixation of mutants (and further expansion) of these microsatellites should not be prevented by selection against frameshift events.
To test these predictions, we compared the excess of observed over expected frequencies of microsatellites in coding and noncoding regions. This comparison was carried out with DNA libraries representing a variety of widely diverged eukaryotes, including S. cerevisiae, Caenorhabditis elegans, Schizosaccharomyces pombe, Mus, Drosophila, plants, and primates. Observed and expected frequencies of microsatellites in each library were computed independently for each motif size (1–6 bp) and all possible lengths from 1 to 30 bp. Expected frequencies of repeats were primarily derived from nucleotide frequencies, but we controlled for di- and trinucleotide (codon) biases by recalculating statistics for di- and trinucleotide repeats on the basis of motif frequencies. Exponential regression analysis was used to define the relationship between microsatellite excess and length, and nonparametric statistics were used to determine the overall significance of the effect of functional status (coding vs. noncoding) on the excess of each type of repeat.
RESULTS
Table 1 shows the raw data from the primate libraries. Raw data for other organisms is not shown, but demonstrated similar patterns. Figure 1 summarizes the data for triplet and nontriplet repeats with motifs of 1–6 nucleotides in primates and plants, depicting the proportional excess of microsatellite DNA as a function of length for both coding and noncoding sequences. In Figure 1, gaps in the distribution are treated as missing data.
Table 1.
Observed and Expected Frequencies of Microsatellites of Varying Motif Length in Coding and Noncoding DNA from the Primate Library
| Mononucleotidesa | Dinucleotidesa | Tetranucleotidesa | ||||||||||||
| (noncoding) | (coding) | (noncoding) | (coding) | (noncoding) | (coding) | |||||||||
| bp | obs | exp | obs | exp | bp | obs | exp | obs | exp | bp | obs | exp | obs | exp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.3 e6 | 5.7 e6 | 4.6 e6 | 4.7 e6 | 2 | 6.2 e6 | 6.7 e6 | 5.5 e6 | 5.5 e6 | 4 | 7.7 e6 | 8.4 e6 | 6.3 e6 | 7.0 e6 |
| 2 | 1.4 e6 | 1.5 e6 | 1.3 e6 | 1.1 e6 | 4 | 4.5 e5 | 4.3 e5 | 3.5 e5 | 3.6 e5 | 8 | 54078 | 36729 | 24996 | 31426 |
| 3 | 4.6 e5 | 3.8 e5 | 3.1 e5 | 3.1 e5 | 6 | 38775 | 29695 | 20658 | 24232 | 12 | 3200 | 191 | 193 | 166 |
| 4 | 1.5 e5 | 1.0 e5 | 90432 | 85196 | 8 | 5553 | 2144 | 1817 | 1732 | 16 | 1272 | 1.25 | 3 | 1.08 |
| 5 | 51597 | 29419 | 24827 | 23894 | 10 | 1345 | 164 | 160 | 130 | 20 | 465 | 0.0114 | 0 | 9.4 e-3 |
| 6 | 14092 | 8540 | 5526 | 6902 | 12 | 587 | 13.3 | 13 | 10.4 | 24 | 235 | 1.7 e-4 | 0 | 1.1 e-4 |
| 7 | 5426 | 2546 | 1110 | 2050 | 14 | 329 | 1.15 | 8 | 0.880 | 28 | 95 | 4.0 e-6 | 0 | 1.9 e-6 |
| 8 | 1961 | 778 | 227 | 625 | 16 | 219 | 0.105 | 0 | 0.0788 | Pentanucleotidesa | ||||
| 9 | 1121 | 243 | 49 | 195 | 18 | 151 | 0.0102 | 0 | 7.5 e-3 | (noncoding)(coding) | ||||
| 10 | 750 | 77.9 | 9 | 62.5 | 20 | 93 | 1.0 e-3 | 0 | 7.5 e-4 | bp | Obs | Exp | Obs | Exp |
| 11 | 489 | 25.6 | 1 | 20.5 | 22 | 83 | 1.2 e-4 | 0 | 7.9 2-5 | |||||
| 12 | 408 | 8.60 | 0 | 6.86 | 24 | 61 | 1.4 e-5 | 0 | 8.8 e-6 | 5 | 8.3 e6 | 1.0 e7 | 6.9 e6 | 8.3 e6 |
| 13 | 325 | 2.98 | 1 | 2.36 | 26 | 49 | 1.9 e-6 | 0 | 1.0 e-6 | 10 | 21135 | 11845 | 9624 | 9850 |
| 14 | 295 | 1.06 | 0 | 0.829 | 28 | 64 | 2.8 e-7 | 0 | 1.2 e-7 | 15 | 1369 | 18.6 | 27 | 15.4 |
| 15 | 285 | 0.394 | 0 | 0.300 | 30 | 47 | 4.4 e-8 | 0 | 1.6 e-8 | 20 | 442 | 0.0586 | 6 | 0.0405 |
| 16 | 226 | 0.152 | 2 | 0.112 | Trinucleotidesa | 25 30 | 116 31 | 9.6 e-4 4.4 e-5 | 2 3 | 4.0 e-4 1.7 e-5 | ||||
| 17 | 183 | 0.0611 | 0 | 0.0425 | (noncoding) | (coding) | Hexanucleotidesa | |||||||
| 18 | 141 | 0.0258 | 0 | 0.0168 | (noncoding) | (coding) | ||||||||
| 19 | 156 | 0.0114 | 0 | 6.9 e-3 | bp | obs | exp | obs | exp | |||||
| 20 | 106 | 5.2 e-3 | 0 | 2.9 e-3 | 3 | 9.0 e6 | 9.4 e6 | 7.5 e6 | 7.7 e6 | bp | Obs | Exp | Obs | Exp |
| 21 | 125 | 2.5 e-3 | 1 | 1.3 e-3 | 6 | 1.8 e5 | 1.6 e5 | 2.1 e5 | 1.3 e5 | |||||
| 22 | 103 | 1.2 e-3 | 1 | 5.9 e-4 | 9 | 7080 | 3074 | 11189 | 2484 | 6 | 8.7 e6 | 9.2 e6 | 7.0 e6 | 7.6 e6 |
| 23 | 69 | 6.4 e-4 | 0 | 2.8 e-4 | 12 | 867 | 66.4 | 1694 | 52.9 | 12 | 10308 | 2797 | 8000 | 2335 |
| 24 | 67 | 3.3 e-4 | 0 | 1.4 e-4 | 15 | 295 | 1.64 | 548 | 1.28 | 18 | 608 | 1.21 | 155 | 1.00 |
| 25 | 46 | 1.7 e-4 | 0 | 7.0 e-5 | 18 | 148 | 0.0472 | 226 | 0.0360 | 24 | 114 | 1.2 e-3 | 29 | 8.3 e-4 |
| 26 | 39 | 9.6 e-5 | 0 | 3.7 e-5 | 21 | 55 | 1.6 e-3 | 97 | 1.2 e-3 | 30 | 16 | 5.9 e-6 | 2 | 2.2 e-6 |
| 27 | 32 | 5.3 e-5 | 0 | 2.0 e-5 | 24 | 44 | 7.0 e-5 | 22 | 4.4 e-5 | |||||
| 28 | 24 | 2.9 e-5 | 0 | 1.1 e-5 | 27 | 12 | 3.8 e-6 | 28 | 1.9 e-6 | |||||
| 29 | 23 | 1.6 e-5 | 0 | 6.0 e-6 | 30 | 4 | 2.5 e-7 | 13 | 9.0 e-8 | |||||
| 30 | 15 | 9.1 e-6 | 0 | 3.4 e-6 | ||||||||||
(bp) Length in base pairs. (Obs and Exp) Observed and expected frequencies of occurrence in library. The expected frequencies are derived from the nucleotide frequencies of each sequence in the library. Decimal scientific notation is represented by eX, where X represents the exponent. Negative decimal scientific notation is represented by e-X, where X represents the negative exponent.
All nontriplet microsatellite types (mono-, di-, tetra-, and pentanucleotide repeats) were found at least once in the noncoding primate sequence library for all possible lengths up to 30 bp. The same was true for plants, with the exception of the longest di- and tetranucleotides (30 and 28 bp, respectively). In contrast, most types of nontriplets were not consistently represented at all lengths in the coding libraries (e.g., we found no primate coding mononucleotide runs with lengths of 17–20 bp, nor are there any of 23–30 bp). It is clear from the data shown in Table 1 that nontriplet microsatellites are far less common and less consistently represented across the range of lengths in coding regions than in noncoding regions.
There are no gaps in the length distributions of either coding or noncoding triplet repeats from primates and plants, and there appears to be very little difference in the length distributions of these repeats between coding and noncoding regions. Similar patterns for both triplet and nontriplet repeats were obtained for the other organisms.
For each organism and each type of repeat, the slope (growth coefficient) of the exponential regression of excess as a function of length is shown in Tables 2 and 3. These slopes represent the regression lines fitted to the data in the form shown in Figure 1. To avoid the use of zero values in exponential regressions, these analyses were carried out by treating gaps in the distribution (zero frequencies) as missing data. This treatment of gaps should be conservative with respect to our hypothesis, because zero frequencies occur primarily in coding nontriplet microsatellites, and if they could be included, they would lower the value of the growth coefficient. The fit of the equation was excellent in all cases. The mean fit and its standard deviation for all 84 exponential regressions was R = 0.9621 ± 0.09518.
The regression slope for each type of nontriplet microsatellite, averaged across all organisms, was lower in coding regions than in noncoding regions. This relationship is independently significant for all polynucleotide nontriplet repeats (di-, tetra-, and pentanucleotides) and strongly significant for nontriplet repeats as a whole, although not for mononucleotide repeats considered independently. Wilcoxon P values for each type of microsatellite, as well as for sets of all triplet and nontriplet microsatellites, are shown in Tables 2 and 3.
The regression slopes of triplet microsatellites, in contrast, were not significantly different between coding and noncoding regions. No differences were found across the range of organisms for tri- or hexanucleotide repeats considered independently or for both types of triplet repeat considered together.
In Table 4, slope data are shown for analyses carried out on di- and trinucleotide microsatellite frequencies with the motif frequencies instead of the nucleotide frequencies to generate expected values. This data, and the associated Wilcoxon P value statistics, demonstrate that dinucleotide enrichment is still significantly decreased in coding regions compared with noncoding regions when dinucleotide biases are controlled for, and trinucleotide enrichment still shows no significant difference between coding and noncoding regions when trinucleotide biases are controlled for.
Table 4.
Growth Coefficients of the Regression Line of Microsatellite Excess as a Function of Length
| Database | Dinucleotides | Trinucleotides | ||||
|---|---|---|---|---|---|---|
| noncoding | coding | Δ(slope) | noncoding | coding | Δ(slope) | |
| S. cerevisiae | 0.69 | 0.61 | −0.08 | 0.47 | 0.60 | 0.13 |
| C. elegans | 0.54 | 0.34 | −0.20 | 0.36 | 0.22 | −0.14 |
| Plants | 0.57 | 0.070 | −0.50 | 0.37 | 0.50 | 0.13 |
| Primates | 0.58 | 0.073 | −0.51 | 0.37 | 0.59 | 0.22 |
| S. pombe | 0.69 | 0.15 | −0.54 | 0.61 | 0.59 | −0.02 |
| Drosophila | 0.54 | 0.58 | 0.04 | 0.32 | 0.55 | 0.23 |
| Mus | 0.62 | 0.32 | −0.30 | 0.41 | 0.44 | 0.03 |
| Mean | 0.60 | 0.31 | −0.29 | 0.41 | 0.50 | 0.09 |
| Wilcoxon paired sample P (two-tailed) | 0.05 > P > 0.02 | 0.5 > P > 0.2 | ||||
Expectations calculated on the basis of motif frequencies (dinucleotide frequencies in the case of dinucleotide repeats and trinucleotide frequencies in the case of trinucleotide repeats).
Table 5 shows growth coefficient data for analyses carried out with only in-frame trinucleotide frequencies to generate observed and expected frequencies. This procedure, which controls for the effects of amino acid and codon biases, still fails to reveal any significant difference between coding and noncoding trinucleotide microsatellite frequencies.
Table 5.
Growth Coefficients of the Regression Line of Microsatellite Excess as a Function of Length
| Database | Trinucleotides | ||
|---|---|---|---|
| noncoding | coding | Δ(slope) | |
| S. cerevisiae | 0.34 | 0.41 | 0.07 |
| C. elegans | 0.29 | 0.17 | −0.12 |
| Plants | 0.36 | 0.17 | −0.19 |
| Primates | 0.48 | 0.31 | −0.17 |
| S. pombe | 0.42 | 0.34 | −0.08 |
| Drosophila | 0.28 | 0.29 | 0.01 |
| Mus | 0.38 | 0.20 | −0.18 |
| mean | 0.36 | 0.27 | −0.09 |
| Wilcoxon paired sample P (two-tailed) | 0.1 > P > 0.05 | ||
Expectations calculated on the basis of motif frequencies (codons in the case of trinucleotide coding repeats and arbitrary codons assigned starting with the first base in the case of noncoding trinucleotide repeats. Observed values used in these calculations were made only for in-frame repeats (codon repeats).
In Figure 1, it can be seen that the excesses of primate noncoding mono- and pentanucleotide repeats appear to be somewhat depressed relative to noncoding di-, tri, tetra, and hexanucleotide repeats at lengths above 20 bp. This relationship appeared consistently for mononucleotide repeats across all libraries analyzed here, but is not apparent for pentanucleotides in several libraries.
The only observed deficiencies of repetitive DNA occurred among primates, Mus, and Drosophila, and involved mononucleotide repeats between the lengths of 5 and 15 bp. An example of this can be seen in the Primate Nontriplet graph in Figure 1.
It can also be seen that noncoding dinucleotide repeat excesses consistently give greater growth coefficients than other types of noncoding repeats. This effect is significant for all comparisons of dinucleotides to other repeat types on the basis of two-tailed Wilcoxon-paired sample tests (P ≤ 0.02 in all cases).
DISCUSSION
The data support the hypothesis that selection against frameshift mutations limits expansion of nontriplet microsatellites in coding regions. The rate of generation of excess microsatellites, as represented by the slope of the exponential regression line fit to proportional microsatellite excess as a function of length, is significantly lower for nontriplet microsatellites in coding regions than it is in noncoding regions. The most probable explanation for this difference is that coding nontriplet microsatellites generate negatively selected frameshifts when they undergo length mutations involving single motifs, and that this type of mutation is the most common variety contributing to changes in microsatellite length, and therefore to enrichment.
In contrast, triplet microsatellites do not generate frameshifts through single-motif length mutations. The results show that the rate of generation of excess triplet microsatellites is not significantly affected by coding status. This suggests that both coding and noncoding triplet microsatellites are subject to similar rates of repeat expansion. More specifically, expansion of triplet microsatellites is not subject to differential selective pressures in coding and noncoding regions, whereas nontriplet microsatellites are subject to greater purifying selection in coding regions. An important inference from this observation is that the excess of triplet microsatellites in both coding and noncoding regions is primarily the result of mutation pressure.
Among noncoding repeats, dinucleotides showed significantly higher growth coefficients than other types of microsatellites. This finding appears primarily to be the result of pervasive noncoding-region dinucleotide biases, which increase the likelihood of dinucleotide runs above the frequencies expected on the basis of nucleotide frequencies alone.
Among primates, Mus, and Drosophila, a deficiency of mid-length (5–15 bp) coding mononucleotide runs was seen. This effect cannot be explained by selection against the products of replication slippage, which in theory should act only to prevent enrichment. Such deficiencies might represent selection against the targets rather than the products of replication slippage, or selection against mononucleotide runs resulting from pressures unrelated to length mutability. Alternatively, these mononucleotide repeats could be subject to different patterns of mutation than other repeats.
METHODS
Library Construction
We obtained sequence data directly from nonredundant and whole-genome collections in GenBank (release 105.0, February 15, 1998). Sequences were collected for each library (see Table 6) by downloading files that contained, in the associated annotation, the name of the appropriate clade and functional status indicator (coding or noncoding). Library construction was accomplished with AWK programs (The AWK Programming Language; A.V. Aho, P.J. Weinberger, B.W. Kernighan, Prentice Hall, Englewood Cliffs, NJ) developed by J. Bytof. For each library, we randomly collected up to 10,000 sequences. Library sizes and other parameters are shown in Table 6. In some cases, sequence availability limited library size. In cases for which specific-species data was limited, we combined species to form higher-level libraries (primate and plant libraries). Because we are not comparing groups to one another, but rather using groups as independent samples to address a phylogeny-independent hypothesis, any evolutionarily defensible grouping strategy would suffice, so long as none of the groups overlap. The types of sequence used to construct the noncoding libraries also varied by nature of their availability. For S. cerevisiae, which has very few introns, the noncoding library consisted primarily of intergenic sequence, whereas for the other clades it consisted entirely of intronic sequences. Previous work has shown that both varieties of noncoding DNA tend to be more repetitive than nonspecific coding DNA in eukaryotic genomes (Hancock 1995), and they are equivalent in terms of our hypothesis.
Table 6.
Size Parameters of Sequence Libraries
| Coding libraries | Noncoding librariesa | |||
|---|---|---|---|---|
| sequences | bases | sequences | bases | |
| S. cerevisiae | 6145 | 8664450 | 5453 | 3289903 |
| C. elegans | 10000 | 12914199 | 10000 | 10285584 |
| Plants | 10000 | 9705006 | 10000 | 5927449 |
| Primates | 10000 | 8629770 | 10000 | 19933942 |
| S. pombe | 2752 | 3765135 | 3471 | 1786100 |
| Drosophila | 4310 | 5785722 | 4856 | 1822840 |
| Mus | 10000 | 9070887 | 5439 | 4178459 |
Noncoding libraries represent introns in all cases except S. cerevisiae, for which the noncoding library is primarily composed of intergenic sequences.
The assignment of coding or noncoding status relies on the accuracy of the annotation in GenBank. This annotation is partially based on predictive statistics, as opposed to known functionality. Any errors in assignment, however, should be conservative with regard to the hypothesis because such errors would tend to reduce the statistical significance of observed differences between coding and noncoding regions.
It has been shown that individual regions of high repetitivity are not conserved, even in otherwise highly conserved coding regions, at evolutionary distances as short as those between chickens and rats (Tautz et al. 1986). In this work the two most closely related clades are Mus and primates, and we did not find significant evidence of microsatellite conservation between these even more closely related groups. We therefore consider that the frequencies of repetitive DNA in the widely diverged clades that are compared in this work are effectively independent.
Generation of Observed and Expected Values
The libraries were surveyed to determine the observed and expected numbers of all mono- through hexanucleotide repeats. Expected numbers of these repeats were calculated according to the method of deWachter (1981).
To control for local variation in base content, sequences were analyzed individually to obtain observed and expected repeat frequencies. The values obtained for individual sequences were summed for each library.
Observed numbers were generated as follows: A repeat motif M is defined as a specific sequence of bases. If a repeat motif appears in a sequence, but the bases following it do not consist of the same motif, the repeat has length one. If the sequence of bases that follows M is the same as that found in the motif, but the repeat ends at this point, the repeat has length two, and so on. There is redundancy in this measure; for example, if a triplet repeat CATCATCATCAT occurs in the sequence, the algorithm will also count repeats ATCATCATC and TCATCATCA. However, this does not effect the direct comparison of observed and expected numbers of repeats, as the same redundancy applies to both observed and expected figures.
We define the length of a repeat as the total number of nucleotides in the repeat sequence. Recent work has shown that the excess of observed over expected frequencies of repeats is a function of the absolute length of the repeat (in nucleotides) rather than the number of times the motif is repeated (Pupko and Graur 1999). In the present analysis, both observed and expected numbers of all repeats up to a length of 30 were determined. That is, we examined mononucleotide runs of up to 30 repeats, dinucleotide runs of up to 15 repeats, trinucleotide runs of up to 10 repeats, and so on.
A correction factor was applied to the data to prevent autocorrelation between repeat types. An (AA)3 repeat, for example, could also be counted as an (A)6 repeat. Therefore, runs in which the first base of the repeat motif is the same as the second were excluded from the dinucleotide totals. Corrections to remove similar ambiguities were applied to tri- through hexanucleotide repeats.
To obtain the expected numbers, all possible motifs of 1 to 6 bases were examined in turn. For each DNA segment made up of N bases, the expected numbers of a repeat of motif M and repeat number t is given by
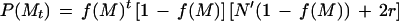 |
1 |
where f(M) is the probability of a motif, t is the number of motifs in the repeat, r is the length of the motif in nucleotides, and
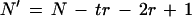 |
2 |
where N is the number of nucleotides in the analyzed sequence (deWachter 1981).
As with the observed numbers of repeats, a correction factor was applied to the data to prevent autocorrelation between repeat types. For example, probabilities for dinucleotide repeats that were also mononucleotide repeats were counted only in mononucleotide totals, not dinucleotide totals.
Dinucleotide biases can generate apparent microsatellite excesses that disappear when dinucleotide rather than mononucleotide frequencies are used to generate the expected frequencies, and this false-positive effect primarily influences apparent excesses of dinucleotide repeats (Hancock and Armstrong 1994). The frequency of trinucleotide repeats in coding regions is likely to be influenced by a similar effect, due to amino acid and codon biases that generate nonrandom trinucleotide frequencies with respect to the underlying nucleotide frequencies.
To control for apparent excesses of di- and trinucleotide repeats that result from biased di- and trinucleotide frequencies, as opposed to replication slippage, similar calculations to those described above were performed to generate repeat frequency expectations on the basis of motif usage instead of nucleotide usage. Motif frequencies were based on the averaged frequencies of all possible motifs (16 in the case of dinucleotides, 64 in the case of tris) in each of the possible frames for that motif (two for dinucleotides and three for tris). Observed values were then compared with these adjusted expected values (see Table 4).
To control specifically for codon biases in the reading frame, a third set of calculations were carried out for trinucleotide repeats, with in-frame codon frequencies to generate f(M) for coding trinucleotides, and one-frame-only trinucleotide frequencies (arbitrarily starting at the first base) for noncoding trinucleotides. In this case, observed microsatellites were limited to those occurring in frame (codon repeats), but otherwise the analysis was performed in an identical manner to that described above with nucleotide frequencies (see Table 5).
The above protocols for calculation of both observed and expected frequencies of repeats were combined in a TrueBasic for the Macintosh program available from C. Wills (cwills@ucsd.edu).
Analysis of Repeat Frequencies
The proportional excess of a particular type of microsatellite, generated by dividing the observed number of repeats of a particular length by the number expected to occur by chance, increases exponentially with length in eukaryotic genomes (Cox and Mirkin 1997; Pupko and Graur 1999). This relationship is predicted under a Markov chain model invoking the combined effects of base-pair mutation and replication slippage (Kruglyak et al. 1998). We fit our data to an exponential model:
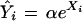 |
3 |
where Ŷi is the expected value of the ratio of the observed over expected microsatellite frequencies, for repeats of length i, and Xi is the growth coefficient. The slope of this relationship on a semilog plot, defined as the exponent (Xi) of the exponential regression of excess against length, is used here as a proxy for the rate of fixation of mutations. A description of the logic behind this statistic is as follows:
Let us call microsatellites that are generated by recursive slippage processes (as opposed to those expected to arise by chance association of nucleotides) “excess microsatellites”. Microsatellite mutations have been shown to be primarily insertions and deletions of single motifs in both empirical studies (Henderson and Petes 1992) and distributional analyses (Bell and Jurka 1997). Therefore, excess microsatellites consisting of a particular number of repeats (X) will generally arise through either (1) an insertion event affecting a microsatellite of X − 1 repeats or (2) a deletion event affecting a microsatellite of X + 1 repeats. In addition, the original microsatellite in case 2 must have, at one time, arisen from the growth of a microsatellite of X repeats and is therefore replacing itself in this length category. That is, the first time it reached length X, it did so by mechanism 1. Hence, excess microsatellites are assumed to have arisen at some point in the past from a microsatellite of the next shortest category. The rate of fixation of mutations determines how many microsatellites of length X will evolve to length X + 1, and so on.
The general exponential nature of the relationship between microsatellite excess and length is primarily driven by the exponential decrease in expected values with increasing length in nucleotides (Cox and Mirkin 1997) (e.g., given unbiased nucleotide frequencies, any sequence is expected to occur by chance at a rate of 0.25 raised to the power of its length in nucleotides). For any organism, the expected value for a particular length (in base pairs) is the same for microsatellites of all motif sizes. We controlled for this similarity by representing all excess values as a function of length, in the form of an exponential regression equation of excess against length. Differences between the slopes (growth coefficients) of these regression lines are assumed to represent differences in the rate at which microsatellites with different motif lengths evolve (through mutation and fixation) to generate excess microsatellites.
If comparable mutation pressures are affecting microsatellite expansion, then growth coefficients should be the same in coding and noncoding regions, at least for a particular type of microsatellite in a specific organism or clade. However, coding microsatellites with a motif length not divisible by three are expected to generate frameshifts, and length variants of these microsatellites should be subject to purifying selection. Hence, the model predicts that the rate of fixation of mutations in nontriplet microsatellites should be higher in noncoding regions than in coding regions, and therefore the related growth coefficients should be significantly higher for noncoding, as opposed to coding, sequences. In contrast, for both coding and noncoding triplet microsatellites, the growth coefficients should be the same.
To test this hypothesis, we used the two-tailed Wilcoxon paired-sample test (Zar 1984) (H0: Coding slope = Noncoding slope) to analyze the effect of functional status (coding vs. noncoding) on the growth coefficients of triplet and nontriplet microsatellites. A one-tailed test might have been appropriate for the nontriplet microsatellites, because the hypothesis predicts a specific directional effect of coding status, but would not have been appropriate for triplets, for which the hypothesis predicts no difference between coding and noncoding regions. Two-tailed tests were used throughout to maintain comparability of statistical parameters. We also analyzed each type of microsatellite independently for effect of functional status on microsatellite enrichment.
Table 2a.
Slope of the Exponential Regression of Microsatellite Excess as a Function of Length, for Nontriplet Repeats
| Database | Mononucleotides | Dinucleotides | ||||
|---|---|---|---|---|---|---|
| noncoding | coding | Δ(slope)a | noncoding | coding | Δ(slope)a | |
| S. cerevisiae | 0.45 | 0.48 | 0.03 | 0.76 | 0.70 | −0.06 |
| C. elegans | 0.42 | 0.45 | 0.03 | 0.70 | 0.64 | −0.06 |
| Plants | 0.43 | 0.28 | −0.15 | 0.70 | 0.094 | −0.61 |
| Primates | 0.59 | 0.32 | −0.27 | 0.81 | 0.13 | −0.68 |
| S. pombe | 0.50 | 0.099 | −0.40 | 0.67 | 0.13 | −0.54 |
| Drosophila | 0.46 | 0.47 | 0.01 | 0.78 | 0.70 | −0.08 |
| Mus | 0.58 | 0.23 | −0.35 | 0.80 | 0.55 | −0.25 |
| mean | 0.49 | 0.33 | −0.16 | 0.75 | 0.42 | −0.33 |
| Wilcoxon paired sample P (two-tailed) | 0.5 > P > 0.2 | P ≤ 0.02 | ||||
| Database | Tetranucleotides | Pentanucleotides | ||||
| noncoding | coding | Δ(slope)a | noncoding | coding | Δ(slope)a | |
| S. cerevisiae | 0.57 | 0.042 | −0.53 | 0.52 | 0.51 | −0.01 |
| C. elegans | 0.51 | 0.33 | −0.18 | 0.46 | 0.089 | −0.37 |
| Plants | 0.50 | −0.031 | −0.53 | 0.45 | 0.051 | −0.40 |
| Primates | 0.78 | 0.095 | −0.69 | 0.61 | 0.57 | −0.04 |
| S. pombe | 0.51 | 0.0050 | −0.50 | 0.55 | 0.37 | −0.18 |
| Drosophila | 0.62 | −0.079 | −0.70 | 0.58 | 0.26 | −0.32 |
| Mus | 0.76 | 0.40 | −0.36 | 0.68 | 0.24 | −0.44 |
| mean | 0.61 | 0.11 | −0.50 | 0.55 | 0.30 | −0.25 |
| Wilcoxon paired sample P (two-tailed) | P ≤ 0.02 | P ≤ 0.02 | ||||
Δ(slope) is defined as the difference between the coding slope and the noncoding slope.
Table 2b.
Summary Statistics for Nontriplet Microsatellites in All Clades
| mean slope for all noncoding nontriplets | 0.60 |
| mean slope for all coding nontriplets | 0.29 |
| mean Δ(slope) for all nontriplets | −0.31 |
| Variance (s2) in Δ(slope) for all nontriplets | 0.055 |
| Wilcoxon P for all nontriplets (coding vs. noncoding) | P << 0.001 |
Table 3a.
Growth Coefficients of the Regression Line of Microsatellite Excess as a Function of Length for Triplet Repeats
| Database | Trinucleotides | Hexanucleotides | ||||
|---|---|---|---|---|---|---|
| noncoding | coding | Δ(slope) | noncoding | coding | Δ(slope) | |
| S. cerevisiae | 0.64 | 0.78 | 0.14 | 0.60 | 0.73 | 0.13 |
| C. elegans | 0.56 | 0.65 | 0.09 | 0.64 | 0.53 | −0.11 |
| Plants | 0.66 | 0.62 | −0.04 | 0.54 | 0.59 | 0.05 |
| Primates | 0.70 | 0.74 | 0.04 | 0.66 | 0.61 | −0.05 |
| S. pombe | 0.63 | 0.63 | 0 | 0.47 | 0.32 | −0.15 |
| Drosophila | 0.65 | 0.79 | 0.14 | 0.65 | 0.70 | 0.05 |
| Mus | 0.72 | 0.78 | 0.06 | 0.70 | 0.69 | −0.01 |
| mean | 0.65 | 0.71 | 0.06 | 0.61 | 0.60 | −0.01 |
| Wilcoxon paired sample P (two-tailed) | 0.2 > P > 0.1 | P > 0.5 | ||||
Table 3b.
Summary Statistics for All Triplet Microsatellites in All Clades
| mean slope for all noncoding triplets | 0.63 |
| mean slope for all coding triplets | 0.65 |
| mean Δ(slope) for all triplets | 0.02 |
| Variance (s2) in Δ(slope) for all triplets | 0.0079 |
| Wilcoxon P for all triplets (coding vs. noncoding) | 0.5 > P > 0.2 |
Acknowledgments
D.M. was supported by a fellowship from the NASA NSCORT program. This work was personally supported by C.W. We thank Christian Hansen for help with statistical analyses.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cwills@ucsd.edu; FAX (619) 534-7108.
REFERENCES
- Bell GI, Jurka J. The length distribution of perfect dimer repetitive DNA is consistent with its evolution by an unbiased single-step mutation process. J Mol Evol. 1997;44:414–421. doi: 10.1007/pl00006161. [DOI] [PubMed] [Google Scholar]
- Caskey CT, Pizzuti A, Fu YH, Fenwick RG, Nelson DL. Triplet repeat mutations in human disease. Science. 1992;256:784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- Cox R, Mirkin SM. Characteristic enrichment of DNA repeats in different genomes. Proc Natl Acad Sci. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deWachter R. The number of repeats expected in random nucleic acid sequences and found in genes. J Theor Biol. 1981;91:71–98. doi: 10.1016/0022-5193(81)90375-1. [DOI] [PubMed] [Google Scholar]
- Field D, Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Natl Acad Sci. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JM. The contribution of slippage-like processes to genome evolution. J Mol Evol. 1995;41:1038–1047. doi: 10.1007/BF00173185. [DOI] [PubMed] [Google Scholar]
- ————— Simple sequences and the expanding genome. BioEssays. 1996;18:421–425. doi: 10.1002/bies.950180512. [DOI] [PubMed] [Google Scholar]
- Hancock JM, Armstrong JS. SIMPLE34: An improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput Appl Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Petes TD. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S, Durrett RT, Schug MD, Aquadro CF. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc Natl Acad Sci. 1998;95:10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Pupko T, Graur D. Evolution of microsatellites in the yeast Saccharomyces cerevisiae: Role of length and number of repeated units. J Mol Evol. 1999;48:313–316. doi: 10.1007/pl00006474. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Schlotterer C. Simple sequences. Curr Opin Genet Dev. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Tautz D, Trick M, Dover GA. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986;322:652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical analysis. (ed.) B. Kurtz pp. 153-156. Prentice-Hall, Englewood Cliffs, NJ. 1984. Paired-sample testing by ranks (10.4) [Google Scholar]



