Abstract
Differential allelic expression (DAE) is a powerful tool to identify cis-regulatory elements for gene expression. The UDP-glucuronosyltransferase 2 family, polypeptide B15 (UGT2B15) is an important enzyme involved in the metabolism of multiple endobiotics and xenobiotics. In the present study, we measured the relative expression of two alleles at SNP c.1568C>A (rs4148269) in this gene, which causes an amino acid substitution (T523K). An excess of the C over the A allele was consistently observed in both liver (P=0.0021) and breast (P=0.012) samples, suggesting that SNP(s) in strong linkage disequilibrium (LD) with c.1568C>A can regulate UGT2B15 expression in both tissues. By resequencing, one such SNP, c.1761T>C (rs3100) in 3′untranslated region (UTR), was identified. Reporter gene assays showed that the 1761T allele results in a significantly higher gene expression level than the 1761C allele in HepG2, MCF-7, LNCaP, and Caco-2 cell lines (all P<0.001), thus indicating that this variation can regulate UGT2B15 gene expression in liver, breast, colon, and prostate tissues. Considering its location, we postulated that this SNP is within an unknown microRNA binding site and can influence microRNA targeting. Considering the importance of UGT2B15 in metabolism, we proposed that this SNP might contribute to multiple cancer risk and variability in drug response.
Keywords: UGT2B15, Differential allelic expression, cis-regulation, gene expression, 3′untranslated region
Introduction
Glucuronidation is an important clearance pathway for many endogenous and exogenous molecules, including steroid hormones, bile acid, carcinogens and clinical drugs (King et al., 2000; Tukey and Strassburg, 2000; Belanger et al., 2003). This reaction can transfer the glucuronic acid from UDP glucuronic acid to appropriate substrates, which can make them more water soluble and more easily excreted through the biliary and renal systems than their parent compound, and is catalyzed by UDP-glucuronosyltransferase (UGT) family in human body (King et al., 2000; Guillemette, 2003; Mackenzie et al., 2005). In this family, UGT2B15 (Chen et al., 1993; Turgeon et al., 2000) has particular importance due to its relatively high expression level (Ohno and Nakajin, 2009) and activity (Turgeon et al., 2001), especially on steroid hormones and multiple clinical agents (Tukey and Strassburg, 2000; Court et al., 2004). The UGT2B15 expression is mainly observed in liver, breast, prostate, and colon (Gardner-Stephen and Mackenzie, 2008; Nakamura et al., 2008; Ohno and Nakajin, 2009). Considering that steroid hormones play a central role in multiple cancers and that UGT2B15 is essential in the metabolism of steroid hormones, it has long been proposed that this gene is involved in breast (Sparks et al., 2004) and prostate (MacLeod et al., 2000; Hajdinjak and Zagradisnik, 2004; Park et al., 2004) cancer risk (Nagar and Remmel, 2006).
Differential allelic expression (DAE, or allelic imbalance, AI) has been shown to be a robust and accurate way to identify cis-regulatory elements (Pastinen and Hudson, 2004; Stamatoyannopoulos, 2004; Yan and Zhou, 2004; Bray and O’Donovan, 2006). Recently, by studying DAE at the c.253G>T (relative to translation start, rs1902023) site, two tissue-specific cis-regulatory elements for UGT2B15 were identified in the promoter region (Sun et al., 2010). Besides c.253G>T, there are other coding region variants in this gene, such as c.1568C>A (rs4148269) (Iida et al., 2002), which causes a T523K amino acid substitution but is not likely to influence enzyme activity, at least on oxazepam (Court et al., 2004). However, the DAE based on this variation has not been investigated so far.
MicroRNA (miRNA) is a group of endogenous, noncoding, and small (~22 nt in mature type) RNA molecule (Bartel, 2004). It has been established that miRNA is involved in a broad range of physiological processes, including cell proliferation, differentiation, apoptosis, signal transduction, viral infection, and tissue morphogenesis (including myogenesis, cardiogenesis, hematopoiesis, etc.), and in the development of various human diseases, especially cancer (Esquela-Kerscher and Slack, 2006; Kloosterman and Plasterk, 2006; Bushati and Cohen, 2007; Chang and Mendell, 2007). In most cases, miRNA exerts its function through binding to mRNA 3′ untranslated region (UTR) and negatively regulating gene expression by suppressing translation or cleaving mRNA (Bartel, 2004; Filipowicz et al., 2008; Flynt and Lai, 2008; Ghildiyal and Zamore, 2009). Therefore, some polymorphisms in miRNA target sites, i.e., 3′UTR in mRNA, can influence the miRNA-mRNA interactions and have been observed to associate with phenotype variability, such as muscularity in sheep (Clop et al., 2006), asthma (Tan et al., 2007), stroke (Chen et al., 2010), chondrodysplasia (Simon et al., 2010), and Tourette’s syndrome (Abelson et al., 2005) risk. However, no cis-regulatory element in 3′UTR and potential miRNA interaction has been reported in the UGT gene family so far.
In the present study, we investigated the allele specific expression marked by the c.1568C>A coding variant. A relative excess of the C allele were observed in both liver and breast samples, thus suggesting the presence of cis-regulatory variation in linkage disequilibrium (LD) with c.1568C>A. Re-sequencing of the UGT2B15 exon 6 identified one such SNP in nearby region. By comparing luciferase activity for different plasmid constructs, we verified that this SNP could affect UGT2B15 gene expression. Considering the position of this SNP, we proposed that the regulation might result from alteration of the affinity of an unknown miRNA.
Materials and methods
Tissue samples, RNA and DNA extraction, and genotyping
Thirty-one normal liver (3 European American [CA] and 1 African American [AA], 27 unknown) and 81 normal breast (4 CA and 8 AA, 69 unknown) tissue samples were retrieved from the University of Chicago Tissue Core Facility. RNA and DNA were extracted by RNeasy Lipid Tissue and QIAamp DNA Mini Kit (Qiagen, Valencia, CA), respectively. cDNA was synthesized by High Capacity Reverse Transcription Kit (Applied biosystems, Foster City, CA). Genotype of c.1568C>A polymorphism was determined by a Taqman genotyping assay C_9440184_20 (Applied Biosystems) according to the manufacturer’s protocol. Among all tissues, 13 liver (6 female and 7 male) and 30 breast (28 female, 1 male and 1 unknown) samples were heterozygous for the c.1568C>A variant.
DAE
Allele specific real time PCR was performed with the same abovementioned Taqman assay in cDNA sample from c.1568C>A heterozygous individuals. Each Taqman probe, which is specific for one allele, was labeled by a different dye and fluorescence was detected on a StepOne Plus Realtime PCR System (Applied Biosystems). For normalization, a heterozygous genomic DNA sample was serially diluted as standard. The real time PCR was performed in triplicate for each sample and the AI ratio was expressed as Aquantity/Cquantity. The robustness and sensitivity of this method has been fully described in our recent study (Sun et al., 2010).
Resequencing
Fifty-six unrelated Hapmap samples (24 Yoruba in Ibadan [YRI], 22 CEPH Collection [CEU] and 10 Han Chinese in Beijing or Japanese in Tokyo [ASN]) were chosen for resequencing. Amplification of UGT2B15 exon 6 was performed by using the primer pair 5′-TGGCTAAAGTAAAACAAAAAT-3′ and 5′-CTTACTTATAGCACTTAGAA-3′. After exonuclease I and Shrimp Alkaline Phosphatase (United States Biochemicals, Cleveland, OH) treatment, sequencing was performed by using internal primers 5′-TGCATCCAGTAACTCGTCATT-3′ and 5′-TTTTCAAAGACCATCCATAG-3′ and BigDye Terminator v3.1 (Applied Biosystems). Polymorphisms were scored by PolyPhred (Stephens et al., 2006) and confirmed visually. Visual genotype and LD were determined by using the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). FST (Wright, 1950; Weir and Cockerham, 1984) were calculated by Slider (http://genapps.uchicago.edu/labweb/index.html).
Plasmid construction
The full 3′UTR region of UGT2B15 (from positions 1594 to 2065 relative to the translation start site) was amplified by nested PCR from individuals with specific haplotypes. The first round of PCR was performed by using the above PCR primers and the second round by using primers 5′-CAGTC-TCTAGA-TTATATCAAAAGCCTGAAGTG-3′ and 5′-CAGTC-GGATCC-TTTTTATGGCTTGGATGACA-3′, which introduced restriction sites for XbaI and BamHI (New England Biolabs, Ipswich, MA), respectively. PCR was performed by iProof High-Fidelity DNA Polymerase (Bio-Rad, Hercules, CA) to avoid artificial mutations. After digestion, the segment was fused with pGL3-Promoter vector (the original 3′UTR of the vector was removed by the same enzymes, Promega, Madison, WI). Both plasmids were sequenced to rule out PCR errors and to verify the haplotype orientation before transfection.
Tissue culture
Human hepatocellular carcinoma cell line HepG2 was cultured in minimum essential medium (MEM, ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA). Human breast adenocarcinoma cell line MCF-7 was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen) with 10% FBS and 0.1% insulin (Sigma, St. Louis, MO). Human prostate carcinoma cell line LNCaP and colon adenocarcinoma cell line Caco-2 were maintained in DMEM with 10% FBS.
Transient transfection
Cells (105) were seeded into a 24-well plate 24 hours before transfection. Plasmid constructs (1.9 μg DNA) were transfected by using FuGene HD (Roche, Indianapolis, IN) according to the manufacturer’s recommendations. Plasmid pRL-TK (0.1 μg DNA; Promega) was co-transfected as an internal control. Thirty-six hours after transfection, cells were harvested and luciferase activity was read by Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. The result was expressed as the ratio between Firefly and Renilla luciferase. Six replicates were performed for each experiment.
Statistical Analyses
To investigate whether DAE deviated from the null expectation, one-sample t-test was performed. Shapiro-Wilk test was utilized to determine whether the allele intensity ratio follows normal distribution. The DAE difference in c.253G>T genotype and gender groups, and luciferase result for different alleles was compared by independent two-tailed t-test. The correlation between DAE and UGT2B17 copy number variation (CNV) or age was determined by linear regression, in which age of donors (coded as a continuous integer) or CNV (coded as 0, 1, and 2 for homozygous of deletion, heterozygous, and homozygous of non-deletion, respectively) was set as an independent variable and the DAE value as dependent. All analyses were performed by SPSS 15.0 (SPSS Inc., Chicago, IL) and the null hypothesis was rejected when P< 0.05.
Results and Discussion
As shown in Fig 1a, except for one individual (7.7%), all liver samples showed an excess of C allele over the A allele and consequently a significant deviation from 1:1 ratio was observed (minimum, 0.25; median, 0.75; maximum, 1.15; mean ± standard deviation [SD], 0.73 ± 0.25; 95% confidence interval [CI], 0.32–0.85, t-test, P=0.0021). In breast, most individuals (76.7%) also showed the same pattern (minimum, 0.33; median, 0.85; maximum, 1.56; mean ±SD, 0.88 ± 0.25; 95% CI, 0.66–0.91, t-test, P=0.012, Fig 1b). The observed allele intensity ratio did not violate normal distribution in either liver (P=0.325) or breast (P=0.611). On average, the proportion of C allele was 17.6–212.5% and 9.9–51.5% higher than that of A allele in liver and breast samples, respectively. These results indicated that c.1568C>A variant, or other SNPs in LD with it, influence UGT2B15 expression levels in both tissues.
Fig 1.
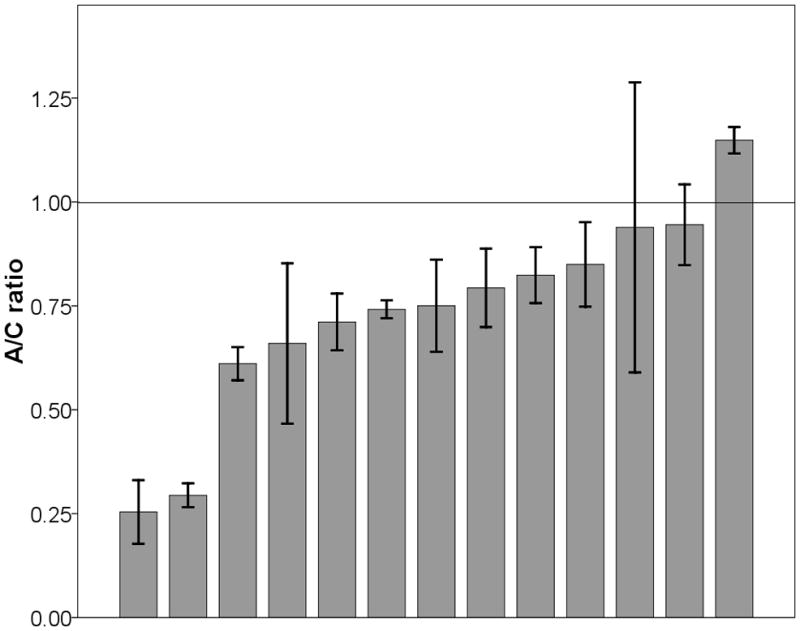
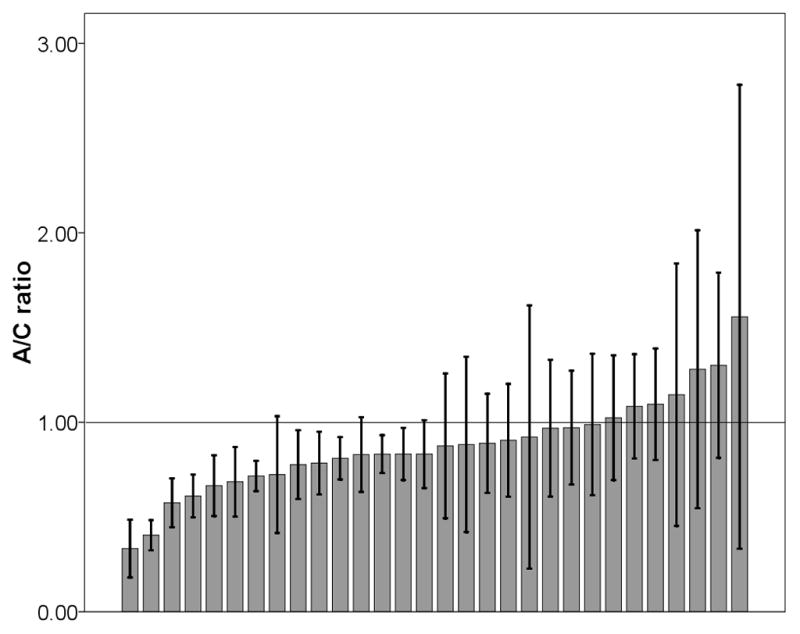
The ratio of A/C alleles at C1568A in liver (a) and breast (b) samples. Each bar represents one individual and data is expressed as mean ± standard error (SE).
A recent study found that one or more promoter SNPs in LD with c.253G>T can modify UGT2B15 expression (Sun et al., 2010). To investigate whether these SNPs can influence DAE at c.1568C>A, we classified the individuals in our sample into heterozygotes (n=8 in liver and 16 in breast) and homozygotes of either allele for c.253G>T (n=5 in liver and 14 in breast) (Sun et al., 2010) and compared the mean of the ratio of the two alleles at c.1568C>A by an independent t-test. No significant difference was observed between these two groups in both liver (P=0.67) and breast (P=0.46), thus suggesting that DAE at c.1568C>A is likely to be independent on c.253G>T status or that our test does not have sufficient power.
It has been proposed that UGT2B17 CNV can influence UGT2B15 expression (Jakobsson et al., 2008). To test whether UGT2B17 CNV can influence UGT2B15 DAE at c.1568C>A, we retrieved the UGT2B17 CNV genotype data for the current samples from our recent study (Sun et al., 2010; Sun et al., 2011). We found 0, 7, and 6 liver and 2, 16, and 12 breast samples that are homozygous of deletion, heterozygous, and homozygous of non-deletion, respectively. When linear regression was performed, no correlation was observed in either liver (r=0.103, P=0.737) or breast (r=0.126, P=0.506), thus indicating that UGT2B17 CNV does not influence UGT2B15 c.1568C>A DAE. However, this conclusion should be interpreted with caution since multiple groups have small sample size.
A recent study has also showed that UGT2B15 is differentially expressed in female and male in liver (Sun et al., 2011). To test whether gender contributes to c.1568C>A DAE, we compared the DAE between gender groups. The mean±SD for DAE was 0.81±0.27 in male while 0.64±0.21 in female. Consequently, no significant difference was observed in liver (t-test, P=0.24).
It has been observed that UGT2B15 expression in liver increases with age (Sun et al., 2011). In our liver sample group, the age varies from 32–79 years, with mean±SD 58.4±13.4 and median 57. For our breast cohort, the minimum, median, maximum age, and mean±SD was 15, 42, 62, and 38.3±13.1 years, respectively. To test whether age influences c.1568C>A DAE, we performed linear regression and no significant result was obtained in liver (r=0.215, P=0.481) and breast (r=0.098, P=0.613).
Although our sample size was modest, which might limit the power of our test, all above evidence indicated that c.1568C>A DAE is likely to be independent of c.253G>T genotype, UGT2B17 CNV, gender, and age. To investigate whether DAE is affected by the combination of all these factors, we further performed multiple regression and failed to observed any significant correlation (P>0.39 and >0.18 for all variables in liver and breast, respectively). These analysis suggests that c.1568C>A or SNP(s) in LD with it can alter UGT2B15 expression liver and breast. Because c.1568C>A lies in the coding region, it could influence UGT2B15 expression by altering the mRNA secondary structure and stability, as previously reported for other genes (Nackley et al., 2006). To explore this possibility, we used the software RNA Mfold (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=mfold#) (Mathews et al., 1999; Zuker, 2003) to predict and compare the secondary structure induced by these two alleles. For all predictions for each haplotype, the Gibbs free energy (dG) fluctuated around 570kcal/mol, thus indicating there was not much difference between their predicted mRNA structures. Therefore, it was more likely that the observed DAE should be attributed to some other nearby SNP(s).
To uncover additional SNPs in LD with c.1568C>A, we resequenced the UGT2B15 exon 6 (Fig 2). The derived allele, 1568A, occurs at lower frequency in CEU (27%) compared to ASN and YRI (90%, Table S1). Besides c.1568C>A, our resequencing survey identified additional 10 SNPs (see Fig 2 and Table S1). Only one SNP, c.1761T>C (rs3100) in 3′UTR region, showed nearly complete LD with c.1568C>A (r2=0.814, 1, and 1 in YRI, CEU, and ASN, respectively; result not shown). Considering our DAE result, it could be postulated that c.1761T>C can affect UGT2B15 expression and that the T allele was correlated with a higher expression level than C allele. The remaining 9 SNPs showed a relative low minor allele frequency (<15%; see Fig 2 and Table S1) and low LD (all r2<0.27; result not shown) with c.1568C>A.
Fig 2.
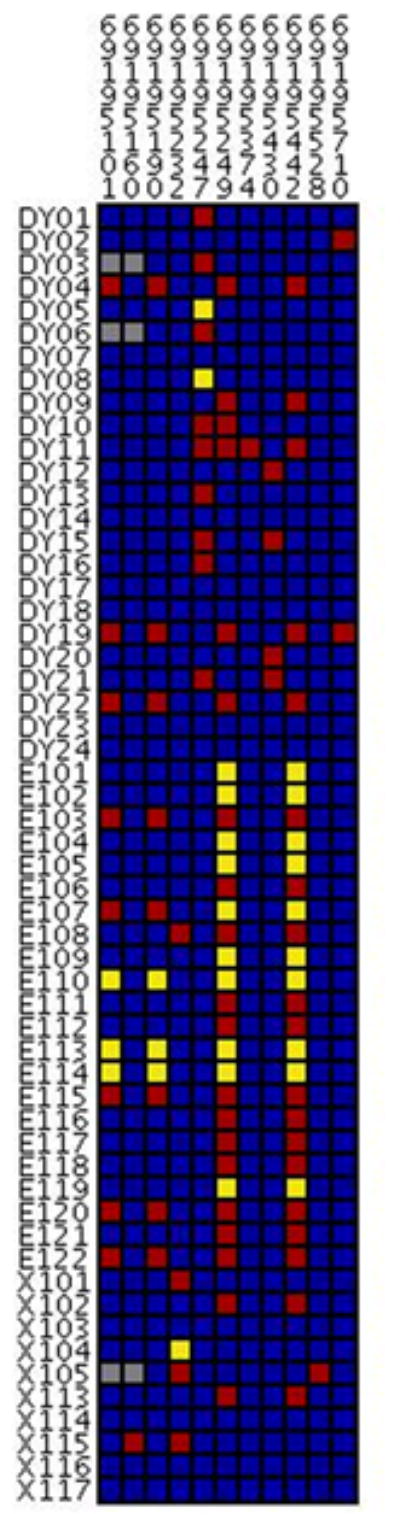
Visual genotype of the UGT2B15 exon 6 region. Each column indicates one SNP while each row denotes one individual. Blue, red, yellow, and grey represent homozygous of common allele, heterozygous, homozygous of rare allele, and missing data, respectively. DY, E, and X indicate YRI, CEU, and ASN HapMap populations, respectively. All positions refer to the genome sequence (build 36) for chromosome 4.
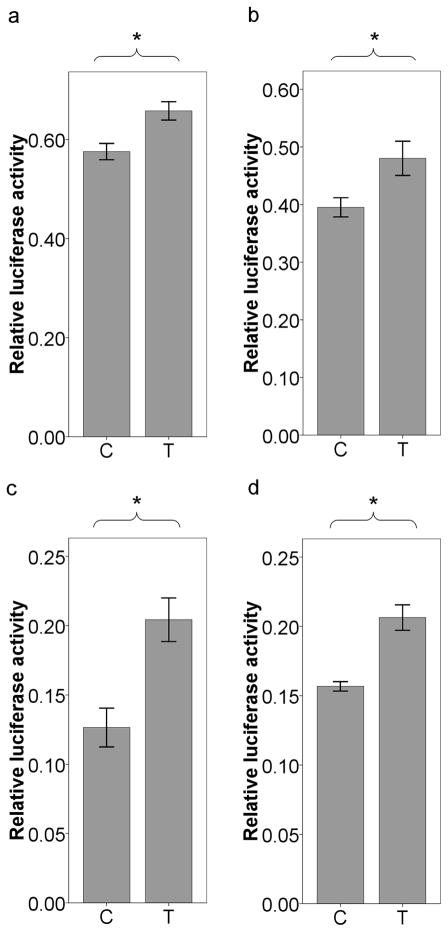
To investigate the cis-regulatory potential of c.1761T>C, we constructed a luciferase plasmid containing UGT2B15 3′UTR with two different alleles and transfected them into HepG2 and MCF-7 cell lines, which have been frequently used to examine the function of 3′UTR variants (Tan et al., 2007; Huang and Li, 2009; Chen et al., 2010; Simon et al., 2010). The relative luciferase activity of the plasmid with UGT2B15 3′UTR was only ~4.5% and ~12.3% (result not shown) of that of pGL3-promoter (empty) vector in HepG2 and MCF-7 cells, respectively, thus confirming that UGT2B15 3′UTR can dramatically reduce gene expression. On the other hand, the luciferase activity of the plasmid with UGT2B15 3′UTR was 3–150 thousand times higher than background (empty tube, result not shown) and 3–16 times higher than pGL3-basic plasmid (result not shown), thus indicating that our assay could be used to distinguish the effect of two alleles. When the two alleles were compared, the T allele showed ~14.3% (t-test, P<10−4, Fig 3a) and ~21.5% (t-test, P<0.001, Fig 3b) higher activity than C allele in HepG2 and MCF-7, respectively, consistent with the hypothesis that 1761T allele up-regulates UGT2B15 expression in both tissues.
Fig 3.
Transient transfection of plasmid constructs with different rs3100 alleles in HepG2 (a), MCF-7 (b), LNCaP (c), and Caco-2 (d). Data is expressed as mean ± SE. *P<0.001 (t-test).
We also transfected the plasmid into LNCaP and Caco-2 cell lines in order to examine whether this SNP can modify UGT2B15 expression in prostate and colon, respectively. Similar to the results in HepG2 and MCF-7, the relative luciferase activity of the plasmid with UGT2B15 3′UTR accounted for only ~11.1% and ~18.4% of that of pGL3-promoter vector in LNCaP and Caco-2 cell lines, respectively. When the two alleles were compared, the T allele showed ~61.5% (t-test, P<10−4, Fig 3c) and ~31.6% (t-test, P<10−5, Fig 3d) higher luciferase activity than C allele in LNCaP and Caco-2 cell lines, respectively. These results strongly suggest that c.1761T>C can alter UGT2B15 expression in all four tissues.
Since this SNP is located within the 3′UTR, the most plausible mechanism is that it alters miRNA binding affinity. To identify the putative miRNA, we used TargetScan (http://www.targetscan.org/) (Lewis et al., 2005) and MicroInspector (http://bioinfo.uni-plovdiv.bg/microinspector/) (Rusinov et al., 2005) to predict the interaction between UGT2B15 mRNA and known miRNAs. However, no miRNA target site was obtained in vicinity of c.1761T>C. Considering the fact that the full miRNA profile is unlikely to be known, we hypothesized that c.1761T>C exerts its function through an unknown miRNA. Since c.1761T>C is functional in all four tissues, this miRNA is hypothesized to be ubiquitously expressed. Moreover, the luciferase activity difference between T and C allele varied markedly, from 14% to 61%, among four tissues. This phenomenon, if it is not due to noise in the experimental measurement, could be attributed to differences in abundance of this putative miRNA among tissues.
Owing to the importance of UGT2B15, the associations between the genetic polymorphisms in this gene and drug response or cancer risk have been repeatedly investigated (as reviewed by (Desai et al., 2003; Guillemette, 2003; Maruo et al., 2005; Nagar and Remmel, 2006)). However, the role of T1761C in these phenotypes has never been tested. Our results point to the c.1761T>C variant as a strong candidate risk factor for hormone-dependent diseases such as breast and prostate cancer and for inter-individual variability in drug metabolism and response, especially in populations of European ancestry due to the high minor allele frequency.
Supplementary Material
Acknowledgments
We thank Ms. Maria Tretiakova, David B. Witonsky, Dr. Weihua Huang (University of Virginia), and Dr. Zheng Tan (Mount Sinai School of Medicine) for technical advice. We also thank two anonymous reviewers for their helpful comments. This research was supported by the University of Chicago Breast SPORE NCI Grant CA125183 and by grant U01 GM61393. The sponsor had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- UGT
UDP-glucuronosyltransferase
- DAE
differential allelic expression
- AI
allelic imbalance
- miRNA
microRNA
- 3′UTR
3′ untranslated region
- LD
linkage disequilibrium
- CNV
copy number variation
Footnotes
Conflict of interest statement
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, Davis NR, Ercan-Sencicek AG, Guez DH, Spertus JA, Leckman JF, Dure LSt, Kurlan R, Singer HS, Gilbert DL, Farhi A, Louvi A, Lifton RP, Sestan N, State MW. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Belanger A, Pelletier G, Labrie F, Barbier O, Chouinard S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab. 2003;14:473–479. doi: 10.1016/j.tem.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Bray NJ, O’Donovan MC. Investigating cis-acting regulatory variation using assays of relative allelic expression. Psychiatr Genet. 2006;16:173–177. doi: 10.1097/01.ypg.0000218612.35139.84. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Chen F, Ritter JK, Wang MG, McBride OW, Lubet RA, Owens IS. Characterization of a cloned human dihydrotestosterone/androstanediol UDP-glucuronosyltransferase and its comparison to other steroid isoforms. Biochemistry. 1993;32:10648–10657. doi: 10.1021/bi00091a015. [DOI] [PubMed] [Google Scholar]
- Chen J, Yang T, Yu H, Sun K, Shi Y, Song W, Bai Y, Wang X, Lou K, Song Y, Zhang Y, Hui R. A functional variant in the 3′-UTR of angiopoietin-1 might reduce stroke risk by interfering with the binding efficiency of microRNA 211. Hum Mol Genet. 2010;19:2524–2533. doi: 10.1093/hmg/ddq131. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, von Moltke LL, Greenblatt DJ. UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther. 2004;310:656–665. doi: 10.1124/jpet.104.067660. [DOI] [PubMed] [Google Scholar]
- Desai AA, Innocenti F, Ratain MJ. UGT pharmacogenomics: implications for cancer risk and cancer therapeutics. Pharmacogenetics. 2003;13:517–523. doi: 10.1097/01.fpc.0000054116.14659.e5. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner-Stephen DA, Mackenzie PI. Liver-enriched transcription factors and their role in regulating UDP glucuronosyltransferase gene expression. Curr Drug Metab. 2008;9:439–452. doi: 10.2174/138920008784746409. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 2003;3:136–158. doi: 10.1038/sj.tpj.6500171. [DOI] [PubMed] [Google Scholar]
- Hajdinjak T, Zagradisnik B. Prostate cancer and polymorphism D85Y in gene for dihydrotestosterone degrading enzyme UGT2B15: Frequency of DD homozygotes increases with Gleason Score. Prostate. 2004;59:436–439. doi: 10.1002/pros.20024. [DOI] [PubMed] [Google Scholar]
- Huang W, Li MD. Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol Psychiatry. 2009;65:702–705. doi: 10.1016/j.biopsych.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Saito S, Sekine A, Mishima C, Kitamura Y, Kondo K, Harigae S, Osawa S, Nakamura Y. Catalog of 86 single-nucleotide polymorphisms (SNPs) in three uridine diphosphate glycosyltransferase genes: UGT2A1, UGT2B15, and UGT8. J Hum Genet. 2002;47:505–510. doi: 10.1007/s100380200075. [DOI] [PubMed] [Google Scholar]
- Jakobsson J, Lorentzon M, Ohlsson C, Lundmark J, Roh HK, Rane A, Ekstrom L. Genetic aspects of epitestosterone formation and androgen disposition: influence of polymorphisms in CYP17 and UGT2B enzymes. Pharmacogenet Genomics. 2008;18:477–485. doi: 10.1097/FPC.0b013e3282fad38a. [DOI] [PubMed] [Google Scholar]
- King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr Drug Metab. 2000;1:143–161. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- MacLeod SL, Nowell S, Plaxco J, Lang NP. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann Surg Oncol. 2000;7:777–782. doi: 10.1007/s10434-000-0777-3. [DOI] [PubMed] [Google Scholar]
- Maruo Y, Iwai M, Mori A, Sato H, Takeuchi Y. Polymorphism of UDP-glucuronosyltransferase and drug metabolism. Curr Drug Metab. 2005;6:91–99. doi: 10.2174/1389200053586064. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene. 2006;25:1659–1672. doi: 10.1038/sj.onc.1209375. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos. 2008;36:1461–1464. doi: 10.1124/dmd.108.021428. [DOI] [PubMed] [Google Scholar]
- Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- Park J, Chen L, Shade K, Lazarus P, Seigne J, Patterson S, Helal M, Pow-Sang J. Asp85tyr polymorphism in the udp-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J Urol. 2004;171:2484–2488. doi: 10.1097/01.ju.0000117748.44313.43. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D, Laloo B, Barillot M, Barnetche T, Blanchard C, Rooryck C, Marche M, Burgelin I, Coupry I, Chassaing N, Gilbert-Dussardier B, Lacombe D, Grosset C, Arveiler B. A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum Mol Genet. 2010;19:2015–2027. doi: 10.1093/hmg/ddq083. [DOI] [PubMed] [Google Scholar]
- Sparks R, Ulrich CM, Bigler J, Tworoger SS, Yasui Y, Rajan KB, Porter P, Stanczyk FZ, Ballard-Barbash R, Yuan X, Lin MG, McVarish L, Aiello EJ, McTiernan A. UDP-glucuronosyltransferase and sulfotransferase polymorphisms, sex hormone concentrations, and tumor receptor status in breast cancer patients. Breast Cancer Res. 2004;6:R488–498. doi: 10.1186/bcr818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA. The genomics of gene expression. Genomics. 2004;84:449–457. doi: 10.1016/j.ygeno.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- Sun C, Southard C, Huo D, Hernandez RD, Witonsky DB, Olopade OI, Di Rienzo A. SNP discovery, expression, and cis-regulatory variation in the UGT2B genes. Pharmacogenomics J. 2011 doi: 10.1038/tpj.2011.2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Southard C, Witonsky DB, Olopade OI, Di Rienzo A. Allelic imbalance (AI) identifies novel tissue-specific cis-regulatory variation for human UGT2B15. Hum Mutat. 2010;31:99–107. doi: 10.1002/humu.21145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- Turgeon D, Carrier JS, Levesque E, Beatty BG, Belanger A, Hum DW. Isolation and characterization of the human UGT2B15 gene, localized within a cluster of UGT2B genes and pseudogenes on chromosome 4. J Mol Biol. 2000;295:489–504. doi: 10.1006/jmbi.1999.3374. [DOI] [PubMed] [Google Scholar]
- Turgeon D, Carrier JS, Levesque E, Hum DW, Belanger A. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787. doi: 10.1210/endo.142.2.7958. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Genetical structure of populations. Nature. 1950;166:247–249. doi: 10.1038/166247a0. [DOI] [PubMed] [Google Scholar]
- Yan H, Zhou W. Allelic variations in gene expression. Curr Opin Oncol. 2004;16:39–43. doi: 10.1097/00001622-200401000-00008. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.