Abstract
Platelet derived growth factor (PDGF) regulates gene transcription by binding to specific receptors. PDGF plays a critical role in oncogenesis in brain and other tumors, regulates angiogenesis, and remodels the stroma in physiologic conditions. Here, we show by using microRNA (miR) arrays that PDGFs regulate the expression and function of miRs in glioblastoma and ovarian cancer cells. The two PDGF ligands AA and BB affect expression of several miRs in ligand-specific manner; the most robust changes consisting of let-7d repression by PDGF-AA and miR-146b induction by PDGF-BB. Induction of miR-146b by PDGF-BB is modulated via MAPK-dependent induction of c-fos. We demonstrate that PDGF regulates expression of some of its known targets (e.g. cyclin D1) through miR alterations and identify the epidermal growth factor receptor (EGFR) as a new PDGF-BB target. We show that its expression and function are repressed by PDGF-induced miR-146b and that mir-146b and EGFR correlate inversely in human glioblastomas. We propose that PDGF-regulated gene transcription involves alterations in non-coding RNAs and provide evidence for a miR-dependent feedback mechanism balancing growth factor receptor signaling in cancer cells.
INTRODUCTION
Two structurally similar receptor tyrosine kinases are activated by the platelet derived growth factor, PDGF (1,2). PDGF induces cell growth and survival (3), transformation (4), migration, vascular permeability and wound healing (5). PDGF is a dimeric molecule composed of two disulfide-bound chains (A and B) (6). It binds selectively to receptor subunits, based on specific epitopes (7), PDGF-AA binding PDGFR-α, PDGF-AB and -BB recognizing both PDGFR-α and PDGFR-β subunits (2). Two recently cloned PDGF chains (C and D) (8,9) bind the PDGFR-α and PDGFR-β, respectively. PDGFR activation occurs through dimerization and autophosphorylation of tyrosine residues in the intracellular domain (10). The two PDGFRs mediate similar, but not identical processes, PDGFR-β being more efficient in mediating chemotactic effects (11), transformation (12) and intracellular Ca++ traffic (13).
Experimental studies have implicated aberrant PDGFR signaling in oncogenesis, particularly in glioblastomas (GBM) (14), sarcomas (15) and selected epithelial cancers [breast and ovarian cancer (16,17)]. PDGFR activation in cancer occurs as a consequence of gene amplification [GBMs (14)], chromosomal rearrangements (18) or activating mutations (19). Activation of PDGFR in tumors can also occur through autocrine or paracrine stimulation (20–23) as both tumor and normal cells in the stroma secrete PDGF. Work in our laboratory demonstrated that PDGFR and its ligands are expressed in ovarian tumors, PDGF is detectable in malignant ascites, and the PDGFR is activated by tumor cell-secreted ligand (24). Autocrine PDGFR activation was also described in brain tumors as an important step to tumor progression (21,25). PDGF signaling in neural stem cells was linked to tumor initiation (26) and autocrine PDGF stimulation of de-differentiated astrocytes induced gliomagenesis (21,23). Autocrine activation of the receptor may contribute to adverse clinical outcome in ovarian (16) and other tumors (22,27,28). PDGFR stimulation leads to activation of intracellular signaling, particularly of Akt, Src and ras/MAPK1/2, with significant cross-talk documented between these pathways (17,24,29). The mechanisms accounting for differences between effects elicited by distinct PDGF ligands are not well understood, having been previously attributed to cell-specific contexts.
MicroRNAs (miR) are single stranded 16–24 nt regulatory RNAs that repress target gene expression by inhibiting translation or by promoting target mRNA degradation. Emerging evidence implicates miRs in human cancer, where they act either as oncogenic factors (through repression of tumor suppressor genes) or as tumor suppressors (30,31). As each miR targets multiple genes, it is predicted that ∼30–40% of genes deregulated in human cancer are under the control of cancer-associated microRNAs (32). Through their effects on gene expression in cancer cells, miRs regulate key cellular mechanisms such as cell differentiation, survival, proliferation and metabolism (33). miR dysregulation in tumor cells may occur constitutively (through deletion, amplification of chromosomal fragments or mutations) (34) or as consequence of micro-environmental factors (e.g. hypoxia, cytokines, estrogens) (35–37). The goal of this study was to identify miRs regulated by PDGFs and their functions in cancer cells.
Here we identify non-coding RNAs (ncRNAs) specifically regulated by each of the two PDGF ligands, AA and BB. We show that PDGF-BB, but not AA, induces miR-146b expression through a Fos-dependent mechanism and that in turn, this miR regulates the expression of the epidermal growth factor receptor, the EGFR. We provide evidence that a ncRNA modulates the balance between two growth factor receptor pathways in cancer cells (PDGFR and EGFR), suggesting the existence of a feedback loop that allows cancer cells to use one pathway selectively over the other. We propose that alterations in miR expression represent one potential mechanism by which PDGFs regulate target gene expression (e.g. cyclin D1 and EGFR). Other miRs induced or repressed by PDGFs and their effects on gene transcription are identified and discussed.
MATERIALS AND METHODS
Materials and cell lines
PDGF BB, PD98059 and LY294002 were from Sigma (St Louis, MO, USA), PDGF AA from PeproTech (Rocky Hill, NJ, USA) and EGF and VEGF from R&D (Minneapolis, MN). All cells were treated with PDGF BB and AA at 25 ng/ml unless otherwise specified and with EGF at 20 and 150 ng/ml. U118-MG, U87-MG glioblastoma cells, and human WI-38 fibroblasts (ATCC, Manassas, VA, USA) were grown according to ATCC instructions. The immortalized ovarian cells C272/hTert/E7 were previously described (38) and grown in a 1:1 mixture of MCDB 105 (Sigma) and M199 media (Cellgro, Herndon, VA, USA) supplemented with FBS. Neuroblastoma cells SHSY-5Y were from Dr Carol Thiele of the National Cancer Institute (39). Nineteen de-identified snap-frozen human glioblastoma specimens were acquired from the Indiana University Cancer Center Tissue Bank and from the Brain Tumor Tissue Bank (BTTB) of London Health Sciences Centre (London, ON, Canada). Their use was approved by the Institutional Review Board.
MicroRNA microarrays and statistical analysis
RNA extracted from cells stimulated with PDGF or control was biotin labeled and hybridized to custom made microRNA chips containing 900 probes for human mature and precursor miRs; 700 probes four mouse miRs, 900 probes for ultraconserved sequences and controls (array express accession, A-MEXP-1246). All probes on the array were duplicated, and average values were used for the analysis. Two biological replicates for each time-point (0, 2, 6, 12 and 24 h) were analyzed. Data were normalized using quantiles algorithm (www.bioconductor.org). Absent calls were assigned at a threshold of 4.5 (log2) before statistical analysis. miR probes were excluded from analysis if <20% of expression data values had at least a 1.5-fold change in either direction from the miR median value. Two-sample t-test (with random variance model) utilizing the Biometric Research Branch (BRB) array tool version 3.6.0 (https://linus.nci.nih.gov/BRB-ArrayTools.html) was used to perform the class comparison between unstimulated and PDGF-stimulated samples or between PDGF-AA and PDGF-BB treated samples. miRs significant at the 0.01 level (two-sided) of the univariate test are reported. Hierarchical clustering was performed by using the expression of the differentially regulated miRs across experiments. The mean was computed over all samples from the same class. MiRs and arrays were mean-centered and normalized by using GENE CLUSTER 3.0. Average linkage clustering was performed by using Euclidean distance and displayed using TreeView program (http://rana.lbl.gov/EisenSoftware.htm). Clustering trees display average expression values after log2 transformation. Principal component analysis (PCA) used unsupervised and supervised classification algorithms from Partek Genomics Suite (http://www.partek.com/) of normalized and filtered data.
Immunoblotting
Actively growing cells were lysed into RIPA buffer containing leupeptin (1 µg/ml), aprotinin (1 µg/ml), PMSF (400 µM) and Na3VO4 (1 mM). Cell lysates were sonicated and incubated on ice for 30 min. Cellular debris was removed by centrifugation for 15 min. Equal amounts of protein were fractionated by SDS–PAGE, transferred to a nitrocellulose membrane and treated with primary antibodies against phospho-PDGFRα (Santa Cruz, sc-12911-R), PDGFRα (Cell Signaling, #3164), phospho-PDGFRβ (Cell Signaling, #3166S), PDGFRβ (Santa Cruz, sc-339), phospho-AKT (Cell Signaling, #4060X), phospho-MAPK-P42/44 (Cell Signaling, #9101S), phospho-EGFR (Cell Signaling, #2236S), EGFR (Santa Cruz, sc-03), c-fos (Santa Cruz, sc-52), cyclin D1 (Cell Signaling, #2922) and GAPDH (Santa Cruz) during an overnight incubation at 4°C. After incubation with corresponding HRP-labeled secondary antibodies, enhanced chemiluminescence was used for protein visualization. Western blots were repeated in independent conditions at least twice.
Quantitative real-time reverse transcription–PCR
Mature miR expression was quantified by qPCR from total RNA extracted from cell cultures or tumors with RNA STAT-60 reagent (Tel-Test Inc.). Total RNA (200–500 ng) was reverse transcribed with TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). miR expression levels were quantified by using probes from TaqMan® MicroRNA Assays (Applied Biosystems) on an ABI Prism 7900 platform (Applied Biosystems) according to the manufacturer’s protocol. At the end of the PCR reaction a melting curve was generated and the cycle threshold (Ct) was recorded. Relative expression of microRNA of interest was calculated as ΔCt, measured by subtracting the Ct of the reference microRNA from that of the RNU49 control (for cancer cell lines) and snRNA U6 (for human glioblastoma samples). qPCR for EGFR used the FastStart Taqman Probe Master (Roche) and results were normalized to GAPDH. Primers are included in Supplementary Table S1. Results are presented as means ± standard deviation (SD) of replicates. Each measurement was performed in duplicate and repeated. Student’s t-test compared mean miR expression levels between groups and Pearson coefficient measured the correlation between EGFR and miR-146b. A P < 0.05 was deemed significant.
Transfection
To knockdown let-7d and miR-146b, we used locked nucleic acids (LNA) MiRCURY LNA™ and scrambled LNA from Exiqon (Denmark). To overexpress let-7d and miR-146b, we used pre-miR™ miRNA Precursor and negative control #1 from Ambion (Austin, TX, USA). Transfection was facilitated by Dreamfect (Oz Biosciences) according to manufacturer’s instructions. SiRNA against c-fos and scrambled siRNA (control) were purchased from Dharmacon (Lafayette, CO, USA). Typically miR or gene knock-down or overexpression were achieved within 24–48 h of transfection. After transfection and/or treatments, cells were harvested for RNA or protein analyses at specified time-points.
Reporter assay
A Dual-Luciferase Assay (Promega) was performed to quantify EGFR 3′-UTR activities. EGFR 3′-UTR reporter clone pMirTarget-EGFR was from Origene (SC216236). The predicted miR-146b target site between 1064 and 1085 bp was deleted by using the QuickChange II Site-directied Mutagenesis Kit (Agilent Technologies #200523-5) to generate the pMirTarget-Mut clone and verified by sequencing. U118 cells were co-transfected with pMirTarget-EGFR or pMirTarget-Mut and control reporter plasmid (renilla) at a ratio of 10:1 by using FuGENE HD transfection Reagent (Roche). Luminescence was measured by using a TD-20/20 luminometer (Turner Biosystems). Experiments were performed in triplicate and repeated in independent conditions two times. To control for transfection efficiency, values for luminescence were normalized to renilla activity.
BrdU assay
An ELISA based assay (Roche Applied Science) measured cell proliferation of C272-hTert/E7 cells transfected with let-7d precursor and control and stimulated with PDGF-AA, according to manufacturer’s instructions.
Chromatin immunoprecipitation
To detect the interaction between c-fos and the miR-146b promoter, we used chromatin immunoprecipitation (ChiP). In brief, C272-hTert/E7 cells were treated with PDGF-BB for 3 h, then fixed in 1% formaldehyde, lysed into 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl (pH 8.1), 167 mM NaCl and sonicated. The soluble chromatin was pre-cleared by incubation with sheared salmon sperm DNA–protein. One portion of the supernatant was used as a DNA input control, and the remainder was incubated with rabbit IgG or a Fos antibody (overnight, 4°C). Immunoprecipitated complexes were incubated with protein A slurry, washed successively with low-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl (pH 8.1), 150 mM NaCl], high-salt buffer (500 mM NaCl), LiCl buffer [0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl (pH 8.1)] and Tris–EDTA (pH 8.0), and then eluted with 1% SDS, 100 mM NaHCO3 buffer. Cross-linking of protein–DNA complexes was reversed by incubation with 5 M NaCl at 65°C for 4 h, and DNA was digested with 10 mg/ml of proteinase K (1 h, 45°C). The DNA was extracted with phenol–chloroform and subjected to PCR amplification using primers designed for the predicted c-fos binding domain in the miR-146b promoter: CAGTGGCTCACACCTGTAATTC (f) and GTAGCACAATCACGGCTTACTG (r). The 242-bp PCR products were resolved by 2% agarose–ethidium bromide gel electrophoresis, visualized by UV and analyzed with the Kodak gel analysis system. As another negative control, DNA immunoprecipitated with c-fos antibody was amplified with primers designed for a fragment located 1000 bp downstream of miR-146b (F: 5′-GCCTACCAAAGTGCTGGGATTAC-3′ and R: 5′-ATATGCTGGACGCTGGACTAGAC-3′).
Statistical analysis
The Student's t-test compared quantitative data between groups; P < 0.05 being significant.
RESULTS
MiRNAs regulated by PDGF
We hypothesized that the response to PDGF involves alterations in miR expression and employed miR microarrays to identify such changes. U118 glioblastoma cells expressing both subunits of the PDGFR, α and β, were treated with PDGF-AA or PDGF-BB over a 24 h time-course. RNA extracted at 0, 2, 6, 16 and 24 h was used for miR expression profiling. A two-sample t-test with random variance was used to perform the class comparison between un-stimulated and PDGF-stimulated samples and miRs significant at the P < 0.01 level of the univariate test are reported. Twenty miRs were induced or repressed by PDGF, with 7 being altered by PDGFAA (four up-regulated and three down-regulated) and 13 affected by PDGF BB (11 up-regulated and two down-regulated). Tables 1 and 2 summarize miRs whose expression is altered by PDGFs compared to un-stimulated cells. Surprisingly no miR was altered in a similar manner by both PDGF-AA and PDGF-BB at P < 0.01. Comparison of miR profiles combining all time-points after PDGF-AA or PDGF-BB stimulation revealed two miRs (let-7g and miR-556) most differentially altered by the two PDGFs (P < 0.01, Supplementary Table S1).
Table 1.
Specific PDGF AA inducible or repressible microRNAs
| miR | Chromosome | Fold-changes | P-value | Expression in cancer | Function |
|---|---|---|---|---|---|
| mir-126-5p | 9q24 | −19.1 | 4.54E–05 | Lost in breast cancer | Suppresses cell proliferation |
| miR-149 | 2q37 | 4.8 | 0.0003 | Down in glioblastoma, melanoma | Not known |
| miR-185 | 22q11.21 | 3.9 | 0.0089 | Up in hepatocellular and bladder cancer | Not known |
| miR-663 | 20p11.21 | 5.5 | 0.0011 | Up in ovarian, lost in breast cancer | Epigenetic silencing in breast cancer |
| miR-769-5p | 19q13.33 | 5.2 | 0.0087 | Not known | Not known |
| Let-7d | 9q22 | −9.5 | 0.0048 | Down in hepatocellular, ovarian, lung cancer | Suppresses cell proliferation |
| miR-770P | 14q42 | −6.0 | 0.002 | Not known | Not known |
Table 2.
Specific PDGF BB inducible or repressible microRNAs
| miR | Chromosome | Fold-changes | P-value | Expression in cancer | Function |
|---|---|---|---|---|---|
| miR-106b | 7q22.1 | 4.6 | 0.0433 | Up in prostate cancer | Suppresses E2F1 and p21 |
| miR-30b | 8q24 | −9.2 | 0.005 | Down in cancer stem cells | Self renewal and survival of cancer stem cells |
| miR-30c | 1p34 | −5.6 | 1.16E−05 | Down in cancer stem cells | Self renewal and survival of cancer stem cells |
| miR-146b | 10q24.32 | 9.3 | 1.96E−05 | Up in thyroid, breast, pancreatic cancer | Suppresses KIT Inducible by TNF |
| miR-198 | 3q13 | 3.2 | 0.003 | Down in hepatocellular carcinoma up in prostate cancer | Not known |
| miR-181a1 | 1q31.32 | 6.2 | 0.006 | Up in pancreatic, thyroid and colon cancer | Target of p53 |
| miR-631 | 15.q24 | 4.7 | 0.0006 | Not known | Not known |
| miR-1 | 18q11 | 3.1 | 0.004 | Up in prostate cancer, down-regulated in other cancers | Suppresses E2F1 Synaptic signaling |
| miR-613 | 12p13 | 3.4 | 0.004 | Not known | Not known |
| miR-129-1 | 7q32.1 | 5.3 | 0.039 | Up in retinoblastoma, down-regulated in other cancers | Not known |
| miR-216 | 2p18.1 | 2.9 | 0.0017 | Down-regulated in pancreatic cancer | Not known |
| miR-499 | 20q11.22 | 3.7 | 0.0062 | Not know | Expressed in myocytes, involved in cell senescence |
| miR-564 | 3p21.31 | 9.4 | 1.84E−05 | Not known | Not known |
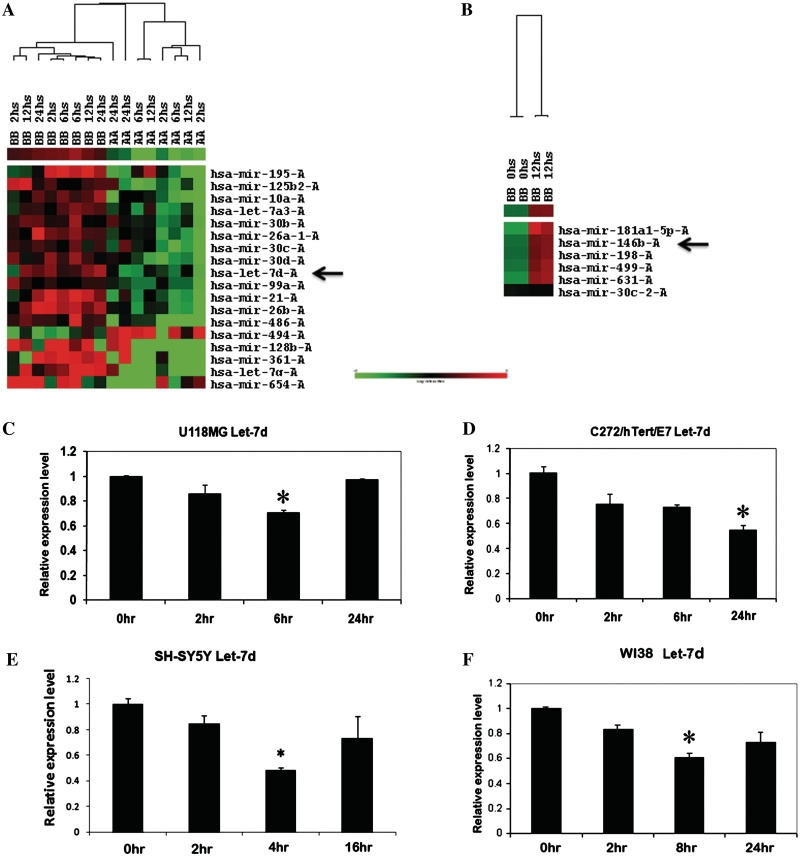
Cluster analysis shows global differences between miRs altered by PDGF-AA and PDGF-BB, let-7d being one of the miRs repressed by PDGF-AA (Figure 1A) and mir-146b being one of the miRs induced by PDGF-BB (Figure 1B). These two miRs were studied in further detail in subsequent analyses. Supervised and unsupervised PCA miR classification was performed for samples stimulated with PDGF-AA or PDGF-BB at different timepoints versus un-stimulated samples. Analyses summarized in Supplementary Figure S1A and B demonstrate significant differences in miR expression patterns induced by the two PDGFs versus un-stimulated cells. Likewise PCA was used to classify miRs altered by PDGF-AA versus PDGF-BB (Supplementary Figure S1C–F) demonstrating discriminatory miR profiles elicited by the two PDGF ligands. Selected miRs emerging from the microarray analysis were confirmed by qPCR. The U118MG, C272/hTert/E7 and WI38 cells express both PDGF receptor subunits, while U87MG cells express predominantly PDGFR-β (Supplementary Figure S2A). Supplementary Figure S2 includes selected validation studies confirming by qPCR miR alterations induced by PDGF, such as miR-106b up-regulation by PDGF-BB.
Figure 1.
Hierarchical clustering demonstrates specific effects of PDGF-AA and PDGF–BB on miR expression in U118 cells. (A) Differential expression of miRs is induced by PDGF-AA and PDGF-BB at different timepoints. Among other miRs, let-7d is suppressed by PDGF-AA (see arrow). (B) Effects of PDGF-BB on miR expression (0 versus 12 h). Among other miRs, miR-146b is induced by PDGF-BB (see arrow). qPCR quantifies let-7d expression levels in U118 MG (C), C272/hTert/E7 (D), SH-SY5Y (E) and WI38 cells (F) after PDGF-AA treatment. Statistical significance is marked by asterisks.
PDGF-AA represses let-7d in cancer cells
Further confirmation of the microarray generated data focused on let-7d as a specific target of PDGF-AA and on miR-146b as a PDGF-BB inducible miR. Up to 50% let-7d repression by PDGF-AA was evident U118MG, C272hTert/E7, SH-SY5Y and WI38 cells within 6 h of PDGF-AA stimulation. The effect of PDGF-AA on let-7d occurred rapidly (2–6 h, Figure 1C–F), although some variability in the kinetic response of let-7d to PDGF-AA was evident in the different cell lines tested. For instance in C272/hTert/E7 cells the repression of let-7d persisted for 24 h, whereas in U118MG cells the level of let-7d returned to normal 24 h after PDGF-AA stimulation. While some of the PDGFR-expressing cancer cells tested here secrete PDGF, the neuroblastoma cells SH-SY5Y cells express both PDGF receptors but do not secrete PDGFs (40). Thus the effects of PDGF-AA on let-7d expression are independent of autocrine secretion of this growth factor (Figure 1E).
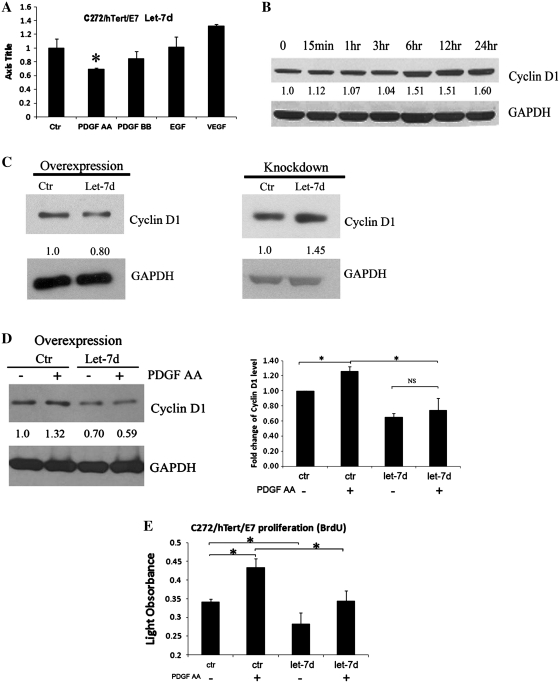
To test the specificity of the miR response to individual growth factors, let-7d expression was quantified in C272hTert/E7 cells treated with PDGF-AA, PDGF-BB, EGF and VEGF. A ∼40% decrease in Let-7d level was observed in response to PDGF-AA, with no measurable changes observed in PDGF-BB, EGF or VEGF-treated cells (Figure 2A).
Figure 2.
PDGF-AA down-regulates let-7d expression. (A) qPCR quantifies let-7d expression in C272/hTert/E7 cells in response to PDGF-AA, PDGF-BB, VEGF and EGF. RNU49 was used as control. (B) Western blotting for cyclin-D1 in C272/hTert/E7 cells treated with PDGF-AA. (C) Western blotting for cyclin D1 in U118MG cells transfected with let-7d precursor or control (left panel). Western blotting for cyclin D1 in U118MG cells transfected with LNA targeting let-7d or scrambled LNA (right panel). (D) Western blotting assessed cyclin D1 in C272/hTERT/E7 cells treated with PDGF-AA for 6 h after transfection of let-7d precursor or control (left panel). Densitometry quantifies cyclin D1 expression levels relative to GAPDH in independent experiments. Statistical significance is indicated by asterisks (NS, not significant). (E) BRDU assay quantifies cell proliferation of C272/hTert/E7 cells transfected with let-7d precursor and control and stimulated with PDGF-AA or vehicle Statistical significance is marked by asterisks.
To test the functional significance of let-7d repression by PDGF-AA, we measured its effects on one of its best documented targets, cyclin D1 (33). Cyclin D1 is induced by PDGF-AA (Figure 2B) and its up-regulation is known to be implicated in PDGF-stimulated cell proliferation (41). To test its role in the context of PDGF-regulated miR alterations, Let-7d was overexpressed and knocked down and cyclin D1 expression levels were measured. Overexpression in U118MG cells was achieved by transfection of the let-7d pre-miR and confirmed by qPCR (not shown). Overexpression of let-7d repressed cyclin D1 expression (Figure 2C, left panel). Likewise let-7d knockdown was achieved by transient transfection of an LNA targeting this miR and confirmed by qPCR (not shown). Let-7d knockdown induced cyclin D1 (Figure 2C, left panel). To show that repression of let-7d by PDGF-AA was significant to the regulation of cyclin D1 by PDGF, this miR was overexpressed. Subsequent treatment with PDGF-AA in cells overexpressing let-7d did not significantly affect cyclin D1 expression (P = 0.3) as compared to the effect of PDGF-AA on cyclin-D1 in cells transfected with a control sequence (P = 0.02, Figure 2D). Further, this effect was implicated in PDGF-AA-induced cell proliferation, as let-7d overexpression inhibited PDGF-AA induced BRDU incorporation in proliferating cells (Figure 2E, P = 0.03). Collectively, these data indicate that PDGF-AA regulates critical cell cycle components and S-phase entry in part via down-regulation of let-7d.
PDGF-BB induces miR-146b in cancer cells
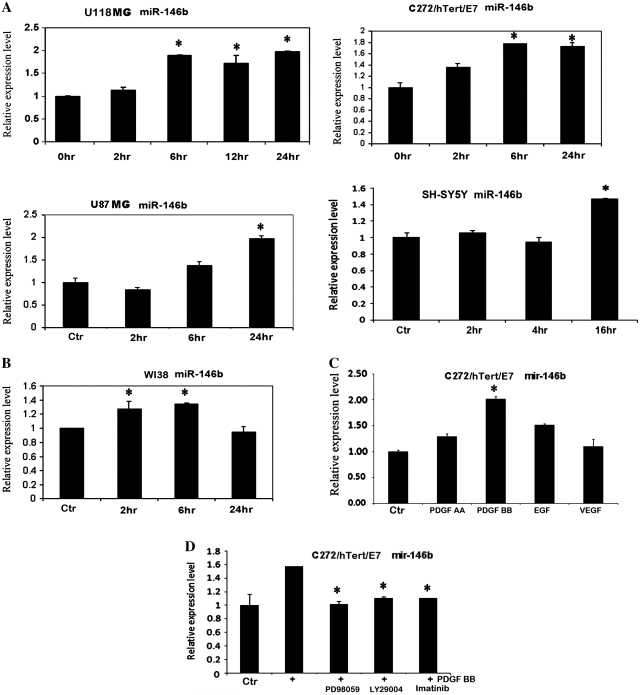
Next, we focused on the effects of PDGF-BB and showed by qPCR that miR-146b was induced by up to 2-fold in U118MG, U87MG, C272hTert/E7 and SH-SY5Y cells within 6–16 h (Figure 3A). The induction in miR-146b expression was sustained (up to 24 h) in the cancer cells tested here. Lesser induction of miR-146b (∼30%) was observed in non-transformed WI38 fibroblasts stimulated with PDGF-BB (Figure 3B). To test the specificity of this miR’s response to individual growth factors, C272/hTert/E7 cells were treated with PDGF-AA, PDGF-BB, EGF and VEGF and miR-146b was quantified. A 2-fold increase in miR-146b expression level was noted in response to PDGF-BB, while PDGF-AA, EGF and VEGF did not induce significant variation of miR-146b levels (Figure 3C).
Figure 3.
PDGF BB up-regulates miR-146b. (A) qPCR quantifies miR-146b in U118, U87-MG, C272/hTert/E7 and SH-SY5Y after PDGF-BB treatment. (B) qPCR quantifies miR-146b in WI38 fibroblasts after PDGF-BB treatment. (C) qPCR quantifies miR-146b in C272/hTert/E7 cells in response to PDGF-AA, PDGF-BB, VEGF and EGF. (D) qPCR quantifies miR-146b in C272hTert/E7 cells treated with PDGF-BB and PD98059, LY294002 and imatinib mesylate. RNU49 was used as control for all qPCR reactions.
Akt and MAPK (42) are the two major oncogenic pathways activated by PDGF stimulation, and we sought to determine their contribution to miR-146b up-regulation. Therefore we measured the effects of a MEK (PD98059), PI3K (LY294002) and PDGFR (imatinib mesylate) inhibitors on miR-146b. Pre-treatment with these inhibitors prevented miR-146b induction by PDGF-BB (Figure 3D), suggesting that both the ERK (inhibited by PD98059) and the AKT (inhibited by LY294002) signaling pathways are involved in miR-146b induction.
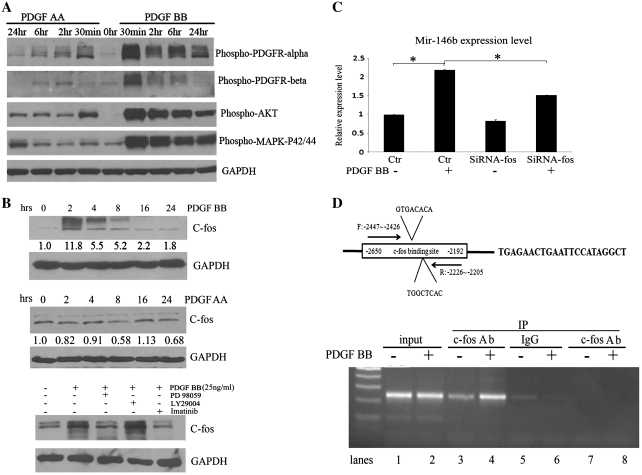
To generate mechanistic insight into PDGF chains specific effects on miR expression, we assessed the differential impact of PDGF-AA versus PDGF-BB on the major downstream signaling pathways. In C272/hTert/E7 cells, PDGF-AA induced phosphorylation of PDGFRα, but not of PDGFRβ within 30 min (Figure 4A). In contrast, PDGF-BB led to phosphorylation of both receptor subunits and its effects were more prominent than those of PDGF-AA. Akt was rapidly phosphorylated after stimulation with each of the two PDGFs. At equivalent doses of PDGF (25 ng/ml), PDGF-BB induced Akt phosphorylation more prominently compared to PDGF-AA. Likewise, MAPK p42/p44 was and more rapidly and robustly activated by PDGF-BB compared to PDGF-AA (Figure 4A), suggesting that the former is a more potent stimulator of this pathway. An important transcription factor induced by PDGF is c-Fos(43). In C272HTERT/E7 cells, c-fos expression was markedly induced by PDGF-BB within 1–3 h, but not measurably by PDGF-AA (Figure 4B, upper and middle panels). As the main mechanism implicated in c-fos engagement by PDGF is the activation of MAPK pathway (44), we measured the effects of its inhibition. The MAPK inhibitor, PD98059, blocked c-fos induction by PDGF-BB, while LY294002 did not affect it (Figure 4B, lower panel), indicating that PDGF-BB engages c-fos more effectively than PDGF-AA, and does so by activating the MAPK pathway.
Figure 4.
Mechanism of miR-146b regulation by PDGF BB. (A) Phosphorylation of PDGFRα and β, Akt and MAPKp42/p44 was measured by western blotting in C272hTert/E7 cells stimulated with PDGF-AA (25 ng/ml) or PDGF-BB (25 ng/ml) for 30 min to 24 h. (B) Western blotting for c-Fos in C272/hTert/E7 cells treated with PDGF-BB for 2–24 h (upper panel). Western blotting for c-Fos in C272/hTert/E7 cells treated with PDGF-AA for 2–24 h (middle panel). Western blotting quantifies c-fos in C272hTert/E7 cells stimulated with PDGF-BB in the presence or not of PD98059 and LY294002 inhibitors (lower panel). (C) C272Htert/E7 cells were transfected with c-fos targeted siRNA or scrambled siRNA (control) and treated with PDGF BB. MiR-146b was quantified by qPCR (P = 0.007). Statistical significance is marked by asterisks. (D) Schematic representation of the c-fos binding site at position −2300 upstream of the miR-146b and the position of primers chosen for amplification in the ChIP assay. ChIP assay tests binding of c-Fos to the miR-146b promoter region after PDGF-BB stimulation. A 242-bp PCR product was detected by gel agarose electrophoresis from chromatin immunoprecipitated with an antibody against c-fos (lanes 3 and 4) using primers flanking the c-fos binding region of the miR-146b promoter (−2447 to −2205 bp). Positive controls are amplicons from input chromatin (lanes 1 and 2). Negative controls consist of chromatin immunoprecipitated with non-specific IgG and amplified with primers specific to the miR-146b promoter (lanes 5 and 6) and chromatin immunoprecipitated with c-fos antibody and amplified with primers away from the predicted c-fos binding site (lanes 7 and 8).
To further understand the relevance of PDGF to the induction of miR-146b, we performed an in silico search upstream of the miR-146b sequence for known transcription factors activated by PDGF (http://microrna.sanger.ac.uk/sequences/ftp/genomes/hsa.gff). This led to the identification of a putative binding site for c-fos at position −2300 upstream of miR-146b (Figure 4D). c-fos knockdown by siRNA repressed the expression level of miR-146b induced by PDGF-BB (Figure 4C, P = 0.007). Binding of c-fos to the miR-146b promoter was shown by ChIP utilizing primers encompassing from −2447 to −2205 region upstream of the transcription starting site of miR-146b. c-fos interaction with this miR-146b promoter region was enhanced by PDGF-BB (Figure 4D, lower panel), further supporting the significance of c-fos to PDGF-BB induced up-regulation of miR-146b.
MiR-146b modulates the balance between PDGF and EGF signaling
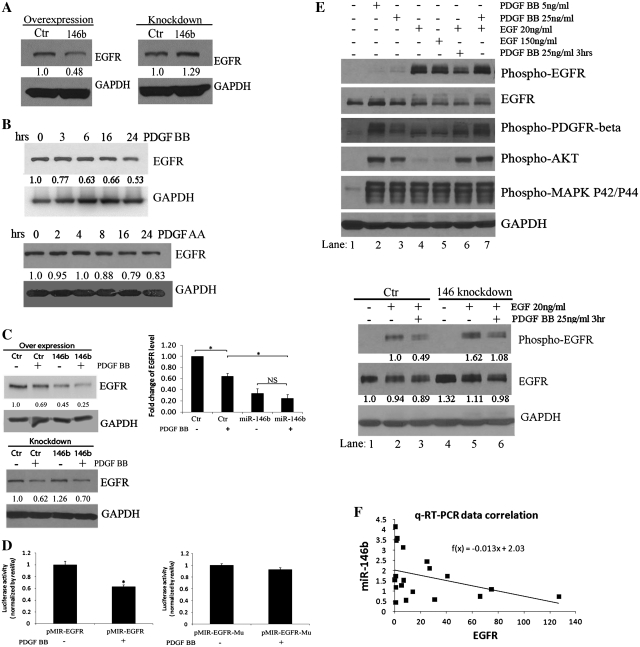
We next searched for possible targets of miR-146b by using bioinformatic predictive tools (http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi). Having learned that in other systems EGFR is a miR-146b target (45), we tested whether changes in miR-146b expression alter EGFR expression. Transfection of a commercial miR-146b mimic led to repression of EGFR protein expression level (Figure 5A, left panel). Conversely, transfection of a LNA targeting miR-146b caused its knock down (not shown) and subsequent up-regulation of EGFR protein expression level (Figure 5A, right panel). However, as expected, the variation in miR-146b expression levels resulting from ectopic overexpression or LNA-mediated knock down of miR-146b achieved in these experiments exceeded the range of PDGF-BB induced changes in cancer cells. Therefore, to test whether by altering miR-146b’s expression, PDGF-BB regulates the expression level of EGFR; we measured the receptor’s expression after PDGF-BB treatment and noted down-regulation of EGFR protein level 6–24 h after PDGF-BB stimulation (Figure 5B, upper panel). In contrast, PDGF-AA did not decrease measurably EGFR expression, in concordance with the known lack of induction of miR-146b by PDGF-AA (Figure 5B, lower panel).
Figure 5.
MiR-146b targets the EGFR. (A) Western blotting for EGFR in U118MG cells transfected with miR-146b precursor or control (left) or with LNA targeting miR-146b or scrambled LNA (right). (B) Western blotting for EGFR in C272/hTert/E7 cells treated with PDGF-BB for 3–24 h (upper panel). Western blotting for EGFR in C272/hTert/E7 cells treated with PDGF-AA for 2–24 h (lower panel). (C) Western blotting assessed EGFR expression in C272/hTERT/E7 cells treated with PDGF-BB for 6 h after transfection of miR-146b precursor or control leading to miR-146b over-expression. Densitometry quantifies EGFR expression levels relative to GAPDH in independent experiments. Statistical significance is indicated by asterisks (upper panels, NS, not significant). Reporter assay measured the activity of an EGFR 3′-UTR-luciferase construct before and after stimulation with 50 ng/ml of PDGF-BB (lower panels). Reporter activity of wild-type EGFR 3′-UTR (left) and mutated 3′-UTR (right) were measured and normalized to renilla activity. Statistical significance is marked by asterisks; NS, non-significant. (D) Phosphorylation of PDGFRβ, EGFR, Akt and MAPK p42/p44 were measured by western blotting (upper panel) in C272hTert/E7 cells stimulated for 30 min with PDGF BB (5 ng/ml, lane 2 and 25 ng/ml, lane 3), EGF (20 ng/ml, lane 4 and 150 ng/ml, lane 5) or a combination of PDGF and EGF (pre-treatment with PDGF for 3 h, followed by EGF; lane 6; or concomitant stimulation with PDGF and EGF for 30 min; lane 7). Phosphorylation of EGFR and total expression of EGFR were measured by Western blotting in C272hTert/E7 cells (lower panel) transfected with LNA targeting miR-146b or control. Cells were then treated with EGF (20 ng/ml, lanes 2 and 5) or with PDGF for 3 h prior to stimulation with EGF (lanes 3 and 6). Densitometry quantifies EGFR phosphorylation and total expression relative to GAPDH. (E) EGFR and miR-146b relative expression plot obtained by using relative expression data after standardization of EGFR relative expression values and miR-146b. F(x) is the estimated linear trend function.
To investigate whether miR-146b has a significant contribution to the effect of PDGF-BB on EGFR, this miR was overexpressed and cells were treated with vehicle or PDGF-BB. Overexpression of miR-146b in C272hTERT/E7 cells down-regulated EGFR expression as compared to cells transfected with control miR. EGFR expression was further suppressed in cells overexpressing miR-146b after treatment with PDGF-BB (Figure 5C, upper panels). To further validate that the regulation of EGFR is mediated by miR-146b through binding to its 3′-UTR region, we measured the effects of PDGF-BB on the activity of an EGFR’s 3′-UTR-luciferase reporter. PDGF-BB repressed the reporter activity of the wild-type 3′-UTR fragment (P = 0.04; Figure 5C, lower left panel), but not that of a 3′-UTR fragment lacking the miR-146b seed sequence (P = 0.56, Figure 5C, lower right panel). These data suggest that one possible mechanism through which PDGF-BB regulates EGFR expression occurs via modulation of miR-146b expression levels.
To gain further insight into the functional significance of the effects of PDGF on EGF-EGFR signaling, C272/hTert/E7 cells that express both PDGFRs and the EGFR, were stimulated with PDGF-BB and EGF, either alone, in combination, or in succession. Two doses of PDGF (5 and 25 ng/ml) and of EGF (20 and 150 ng/ml) were used (Figure 5D, upper panel). As anticipated, PDGF-BB phosphorylated the PDGFR-β, but not the EGFR; while EGF potently phosphorylated the EGFR. MAPK42/44 activation was induced by PDGF-BB as well as by EGF at comparable levels, but Akt phosphorylation was more potently induced by PDGF-BB compared to EGF (Figure 5C, top panel). Interestingly, pre-stimulation with PDGF-BB for 3 h prior to EGF, attenuated EGFR phosphorylation as compared to EGF-induced receptor activation (Figure 5D, top panel, lane 6 versus 4 and 5). This suggested that PDGF-BB may induce a factor which inhibits the effects of EGF. This effect was not observed when the two growth factors were administered simultaneously (Figure 5D, top panel, lane 7 versus 6). Activation of Akt and MAPK42/44 downstream of the EGFR was not affected by the sequence of growth factor administration, likely because of the overlapping effects of PDGF and EGF on these pathways.
To demonstrate that PDGF-induced inhibition of EGFR phosphorylation occurred at least in part via induction of miR-146b, we knocked-down miR-146b prior to stimulation with EGF or with PDGF followed by EGF. miR-146b knockdown accentuated EGFR activation in response to EGF when compared to control-LNA transfected cells (Figure 5D, lower panel, lanes 2 and 5) and this occurred in parallel with an increase in the expression level of EGFR. Further, miR-146b knockdown prevented partially the attenuation of EGFR phosphorylation induced by pre-treatment with PDGF prior to EGF (Figure 5D, lower panel, lanes 3 and 6). These data demonstrate how a miR fine tunes the cancer cells’ response to different growth signals.
Lastly, to illustrate the functional significance of miR-146b expression to EGFR’s expression in vivo, we measured by qPCR the expression levels of EGFR and miR-146b in 19 human glioblastoma specimens. mir-146b and EGFR expression levels were inversely correlated (Pearson coefficient −0.38, P = 0.05, one tail, Figure 5E), demonstrating that this miR-receptor interaction can still be detected in tumors despite the complexity of in vivo systems.
DISCUSSION
Significant literature, including our previous studies, demonstrates that autocrine PDGFR activation plays a critical role in cancer progression via regulation of specific molecular networks (21,22,24). In the current study, we provide new evidence that PDGF-initiated signaling alters the human miRnome, with significant repercussions on target gene expression and function. This report implicates alterations of ncRNAs in the effects of PDGF signaling on gene regulation in cancer cells and demonstrates that distinct PDGF ligands have specific effects on miR expression.
The original observations that a significant proportion of miRs exhibit altered expression patterns in tumors, with or without corresponding genomic alterations, established the foundation for the ongoing ‘ncRNAs revolution’ in oncology (34,46). Mechanistically, cancer-associated miRs directly or indirectly regulate genes with critical roles in tumor formation, including oncogenes or tumor supressors (46). Additionally, miRs respond to cytokines, estrogen, and a variety of stresses, which are relevant to our understanding of the cancer microenvironment (47), as well as of other non-malignant disorders or of normal physiological responses.
The impact of our studies is at least 3-fold. First, the two PDGF ligands (AA and BB) exhibit compelling differences in their impact on miRs expression. Repression of let-7d is specifically induced by PDGF-AA, whereas induction of miR-146b is specific to PDGF-BB. To our knowledge, this represents the first evidence for PDGF ligand-specific miRs response, and may help explain some of the observed different effects elicited by the two forms (12,48). PDGFs’ effects on miR expression have been described in non-transformed mesenchymal cells in rapport to cell proliferation and migration induced by these growth factors (49–51). However, miRs previously described to be inducible by PDGF in smooth muscle cells (e.g. miR-24, miR-221) were not identified by the global approach (miR arrays) utilized in our study. These differences may be attributable to the cancer phenotype studied here or to the threshold for detection of differences in gene expression levels imposed by the use of miR arrays.
Second, transcription factors regulate miR expression in a fashion similar to the regulation of classic genes, well documented examples including HIF-miR-210 (47) and p53-miR-34a (52). Here we show that PDGF induces miR-146b through a c-fos dependent mechanism. These findings expand the accepted classical pathway of gene expression modulated by PDGF through recruitment of factors that activate gene transcription to include a new mechanism dependent on ncRNAs. This model may help to improve our understanding of how different downstream responses are derived from relatively similar upstream growth factors.
Third, EGFR expression is regulated by miR-146b in response to PDGF-BB. While miR-146b has been shown to target EGFR (45), we provide evidence for a feedback mechanism stemming from PDGFR signaling, potentially providing proof of principle for a shift between growth factor signaling in cancer cells. We also demonstrate that the EGFR and miR-146b expression are inversely correlated in vivo. These findings suggest that ncRNAs participate in the fine tuning of cellular responses to external stimuli, balancing one growth factor pathway versus another. Given that the two growth factors’ effects on cell proliferation and survival are to some extent redundant, this mechanism may explain how cells adapt to stimuli from the micro-environment by turning off un-necessary growth or survival pathways. It is also possible that the ncRNA-modulated shift between response to PDGF (a mesenchymal factor) and EGF (an epithelial factor) may have implications for maintaining the balance between epithelial and mesenchymal features of cancer cells, and perhaps influence the transition between the two states. For this study, we dissected this mechanism in the context of cancer cells, however we do not exclude the possibility that a similar feedback loop may operate in non-transformed cells during physiologic responses.
The above findings also point to potential clinical applications of this regulatory mechanism. When PDGF signaling is turned off by inhibitors, PDGF inducible miRs may be silenced and subsequently their targets, including the alternative growth pathway (e.g. EGF-EGFR), may become de-repressed. If operative in tumors in vivo, this link may contribute to resistance to PDGFR inhibitors. Understanding this mechanism may help to develop strategies that take into account the interconnected arrays of growth factors in the tumor environment.
In summary, our findings expand the classical view of PDGF-initiated signaling and provide insight into the regulation of known and/or new targets of PDGF. The involvement of PDGF-inducible ncRNAs may explain differences between biological effects of specific PDGF ligands and feedback regulatory mechanisms between growth pathways in cancer cells. These findings have implications for development of therapies targeting the PDGFR in cancer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Cancer Society (ACS), Mentored Research Scholar Grant (MRSG#107613 to D.M.); research funds (Fellow of the UTM to G.A.C.); D. Anderson Research Trust and Research Scholar of the UT System Regents; Ladjevardian Regents Research Scholar Fund. Funding for open access charge: American Concern Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr David Donner for review of the article and Chang-gong Liu for printing the arrays.
REFERENCES
- 1.Matsui T, Heidaran M, Miki T, Popescu N, La Rochelle W, Kraus M, Pierce J, Aaronson S. Isolation of a novel receptor cDNA establishes the existence of two PDGF receptor genes. Science. 1989;243:800–804. doi: 10.1126/science.2536956. [DOI] [PubMed] [Google Scholar]
- 2.Hart CE, Forstrom JW, Kelly JD, Seifert RA, Smith RA, Ross R, Murray MJ, Bowen-Pope DF. Two classes of PDGF receptor recognize different isoforms of PDGF. Science. 1988;240:1529–1531. doi: 10.1126/science.2836952. [DOI] [PubMed] [Google Scholar]
- 3.Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 4.Huang JS, Huang SS, Deuel TF. Transforming protein of simian sarcoma virus stimulates autocrine growth of SSV-transformed cells through PDGF cell-surface receptors. Cell. 1984;39:79–87. doi: 10.1016/0092-8674(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 5.Greenhalgh DG, Sprugel KH, Murray MJ, Ross R. PDGF and FGF stimulate wound healing in the genetically diabetic mouse. Am. J. Pathol. 1990;136:1235–1246. [PMC free article] [PubMed] [Google Scholar]
- 6.Antoniades HN, Hunkapiller MW. Human platelet-derived growth factor (PDGF): amino-terminal amino acid sequence. Science. 1983;220:963–965. doi: 10.1126/science.6844921. [DOI] [PubMed] [Google Scholar]
- 7.Yu JC, Mahadevan D, LaRochelle WJ, Pierce JH, Heidaran MA. Structural coincidence of alpha PDGFR epitopes binding to platelet-derived growth factor-AA and a potent neutralizing monoclonal antibody. J. Biol. Chem. 1994;269:10668–10674. [PubMed] [Google Scholar]
- 8.Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, Li H, et al. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nat. Cell Biol. 2000;2:302–309. doi: 10.1038/35010579. [DOI] [PubMed] [Google Scholar]
- 9.LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, et al. PDGF-D, a new protease-activated growth factor. Nat. Cell Biol. 2001;3:517–521. doi: 10.1038/35074593. [DOI] [PubMed] [Google Scholar]
- 10.Kazlauskas A, Cooper JA. Autophosphorylation of the PDGF receptor in the kinase insert region regulates interactions with cell proteins. Cell. 1989;58:1121–1133. doi: 10.1016/0092-8674(89)90510-2. [DOI] [PubMed] [Google Scholar]
- 11.Westermark B, Siegbahn A, Heldin CH, Claesson-Welsh L. B-type receptor for platelet-derived growth factor mediates a chemotactic response by means of ligand-induced activation of the receptor protein-tyrosine kinase. Proc. Natl Acad. Sci. USA. 1990;87:128–132. doi: 10.1073/pnas.87.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidaran MA, Beeler JF, Yu JC, Ishibashi T, LaRochelle WJ, Pierce JH, Aaronson SA. Differences in substrate specificities of alpha and beta platelet-derived growth factor (PDGF) receptors. Correlation with their ability to mediate PDGF transforming functions. J. Biol. Chem. 1993;268:9287–9295. [PubMed] [Google Scholar]
- 13.Fatatis A, Miller RJ. Platelet-derived growth factor (PDGF)-induced Ca2+ signaling in the CG4 oligodendroglial cell line and in transformed oligodendrocytes expressing the beta-PDGF receptor. J. Biol. Chem. 1997;272:4351–4358. doi: 10.1074/jbc.272.7.4351. [DOI] [PubMed] [Google Scholar]
- 14.Mao X, Hamoudi RA. Molecular and cytogenetic analysis of glioblastoma multiforme. Cancer Genet. Cytogenet. 2000;122:87–92. doi: 10.1016/s0165-4608(00)00278-8. [DOI] [PubMed] [Google Scholar]
- 15.Oda Y, Wehrmann B, Radig K, Walter H, Rose I, Neumann W, Roessner A. Expression of growth factors and their receptors in human osteosarcomas. Immunohistochemical detection of epidermal growth factor, platelet-derived growth factor and their receptors: its correlation with proliferating activities and p53 expression. Gen. Diagn. Pathol. 1995;141:97–103. [PubMed] [Google Scholar]
- 16.Henriksen R, Funa K, Wilander E, Backstrom T, Ridderheim M, Oberg K. Expression and prognostic significance of platelet-derived growth factor and its receptors in epithelial ovarian neoplasms. Cancer Res. 1993;53:4550–4554. [PubMed] [Google Scholar]
- 17.Matei D, Chang DD, Jeng MH. Imatinib mesylate (Gleevec) inhibits ovarian cancer cell growth through a mechanism dependent on platelet-derived growth factor receptor alpha and Akt inactivation. Clin. Cancer Res. 2004;10:681–690. doi: 10.1158/1078-0432.ccr-0754-03. [DOI] [PubMed] [Google Scholar]
- 18.Carroll M, Tomasson MH, Barker GF, Golub TR, Gilliland DG. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc. Natl Acad. Sci. USA. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 20.Betsholtz C, Westermark B, Ek B, Heldin CH. Coexpression of a PDGF-like growth factor and PDGF receptors in a human osteosarcoma cell line: implications for autocrine receptor activation. Cell. 1984;39:447–457. doi: 10.1016/0092-8674(84)90452-5. [DOI] [PubMed] [Google Scholar]
- 21.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D, Nutt CL, Shanehsaz P, Peng X, Louis DN, Kaetzel DM. Autocrine platelet-derived growth factor-dependent gene expression in glioblastoma cells is mediated largely by activation of the transcription factor sterol regulatory element binding protein and is associated with altered genotype and patient survival in human brain tumors. Cancer Res. 2005;65:5523–5534. doi: 10.1158/0008-5472.CAN-04-2582. [DOI] [PubMed] [Google Scholar]
- 23.Westermark B, Heldin CH, Nister M. Platelet-derived growth factor in human glioma. Glia. 1995;15:257–263. doi: 10.1002/glia.440150307. [DOI] [PubMed] [Google Scholar]
- 24.Matei D, Emerson RE, Lai YC, Baldridge LA, Rao J, Yiannoutsos C, Donner DD. Autocrine activation of PDGFRalpha promotes the progression of ovarian cancer. Oncogene. 2006;25:2060–2069. doi: 10.1038/sj.onc.1209232. [DOI] [PubMed] [Google Scholar]
- 25.Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res. 2002;62:3729–3735. [PubMed] [Google Scholar]
- 26.Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Dolloff NG, Russell MR, Loizos N, Fatatis A. Human bone marrow activates the Akt pathway in metastatic prostate cells through transactivation of the alpha-platelet-derived growth factor receptor. Cancer Res. 2007;67:555–562. doi: 10.1158/0008-5472.CAN-06-2593. [DOI] [PubMed] [Google Scholar]
- 28.Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nister M. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52:3213–3219. [PubMed] [Google Scholar]
- 29.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 31.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl Acad. Sci. USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 33.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 34.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, Benes V, Roche H, Dalenc F, Auboeuf D, Millevoi S, et al. Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 2009;69:8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- 37.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl Acad. Sci. USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 39.Beppu K, Jaboine J, Merchant MS, Mackall CL, Thiele CJ. Effect of imatinib mesylate on neuroblastoma tumorigenesis and vascular endothelial growth factor expression. J. Natl Cancer Inst. 2004;96:46–55. doi: 10.1093/jnci/djh004. [DOI] [PubMed] [Google Scholar]
- 40.Pahlman S, Johansson I, Westermark B, Nister M. Platelet-derived growth factor potentiates phorbol ester-induced neuronal differentiation of human neuroblastoma cells. Cell Growth Differ. 1992;3:783–790. [PubMed] [Google Scholar]
- 41.Reneker LW, Overbeek PA. Lens-specific expression of PDGF-A alters lens growth and development. Dev. Biol. 1996;180:554–565. doi: 10.1006/dbio.1996.0328. [DOI] [PubMed] [Google Scholar]
- 42.Heldin CH, Ostman A, Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim. Biophys. Acta. 1998;1378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 43.Barone MV, Courtneidge SA. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature. 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu T, Kinugawa K, Yao A, Sugishita Y, Sugishita K, Harada K, Matsui H, Kohmoto O, Serizawa T, Takahashi T. Platelet-derived growth factor induces cellular growth in cultured chick ventricular myocytes. Cardiovasc Res. 1999;41:641–653. doi: 10.1016/s0008-6363(98)00261-2. [DOI] [PubMed] [Google Scholar]
- 45.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 47.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15:667–671. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Deuel TF, Kim HR. Platelet-derived growth factor (PDGF) receptor-alpha activates c-Jun NH2-terminal kinase-1 and antagonizes PDGF receptor-beta -induced phenotypic transformation. J. Biol. Chem. 2000;275:19076–19082. doi: 10.1074/jbc.M910329199. [DOI] [PubMed] [Google Scholar]
- 49.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J. Biol. Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan MC, Hilyard AC, Wu C, Davis BN, Hill NS, Lal A, Lieberman J, Lagna G, Hata A. Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 2010;29:559–573. doi: 10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quintavalle M, Elia L, Condorelli G, Courtneidge SA. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J. Cell Biol. 2010;189:13–22. doi: 10.1083/jcb.200912096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.