Abstract
Proteins of the Argonaute family are small RNA carriers that guide regulatory complexes to their targets. The family comprises two major subclades. Members of the Ago subclade, which are present in most eukaryotic phyla, bind different classes of small RNAs and regulate gene expression at both transcriptional and post-transcriptional levels. Piwi subclade members appear to have been lost in plants and fungi and were mostly studied in metazoa, where they bind piRNAs and have essential roles in sexual reproduction. Their presence in ciliates, unicellular organisms harbouring both germline micronuclei and somatic macronuclei, offers an interesting perspective on the evolution of their functions. Here, we report phylogenetic and functional analyses of the 15 Piwi genes from Paramecium tetraurelia. We show that four constitutively expressed proteins are involved in siRNA pathways that mediate gene silencing throughout the life cycle. Two other proteins, specifically expressed during meiosis, are required for accumulation of scnRNAs during sexual reproduction and for programmed genome rearrangements during development of the somatic macronucleus. Our results indicate that Paramecium Piwi proteins have evolved to perform both vegetative and sexual functions through mechanisms ranging from post-transcriptional mRNA cleavage to epigenetic regulation of genome rearrangements.
INTRODUCTION
RNA interference (RNAi) is one of several related regulatory mechanisms that can be defined by the use of small RNAs (sRNAs) as specificity factors, recognizing target sequences by base-pairing interactions. A variety of effector complexes allows regulation of gene expression at the levels of transcription, mRNA stability or translation, but the core component which binds the sRNA and guides the complex is invariably a member of the Argonaute family, also known as PPD [PIWI Argonaute Zwille (PAZ)-P-element Induced Wimpy Testis (PIWI) domain] proteins. In recent years, functional and structural studies have led to a detailed understanding of the roles of the three conserved domains of Argonaute proteins (1,2). The PAZ domain binds the 3′-end of the sRNA, while the 5′-end is anchored in a conserved pocket at the junction of the Middle (MID) and PIWI domains. The PIWI domain has an RNase H-like fold and contains the catalytic DDH triad responsible for slicer activity, the endonucleolytic cleavage of a target RNA molecule paired with the sRNA. Genes encoding Argonaute proteins have undergone a high degree of duplication in some eukaryotic phyla and their numbers vary greatly between species, ranging from one in Schizosaccharomyces pombe to 27 in Caenorhabditis elegans. Phylogenetic analyses have shown that they can be classified into three subclades: the Ago and Piwi subclades, and a third group only found in C. elegans, the worm-specific Agos (Wagos) (3). The distribution of Ago and Piwi proteins in extant species suggests that both were present in the last common ancestor of eukaryotes, where it has been proposed that the Ago type was already specialized for post-transcriptional gene silencing (PTGS) in the cytoplasm and the Piwi type for transcriptional gene silencing (TGS) in the nucleus, possibly through the targeting of histone modifications (4).
This view was inspired by the first studies of metazoan Ago proteins, which revealed their roles in siRNA-mediated mRNA cleavage and miRNA-mediated translation inhibition. Both classes of sRNAs are produced by Dicer-like ribonucleases, but from different precursors: siRNAs are cut from long double-stranded RNAs (dsRNAs), whereas miRNAs are processed from non-coding endogenous transcripts forming stem–loop structures (5). In C. elegans, secondary siRNAs are synthesized in a Dicer-independent manner by an RNA-dependent RNA polymerase (RdRP) using the targeted mRNA as a template. These siRNAs are loaded onto Wago proteins and appear to be responsible for the potency of RNAi-mediated PTGS (3,6–9). The mechanisms of action of Ago proteins, however, are not limited to PTGS. Indeed, mammalian Agos have also been implicated in siRNA-mediated TGS (10,11); furthermore, a large body of experimental evidence has established that Ago proteins are used for both TGS and PTGS in eukaryotes that lack Piwi-subclade members, such as Arabidopsis thaliana (12,13) or S. pombe (14).
Our current understanding of the functions of Piwi proteins derives almost exclusively from studies of metazoan species, where they play essential roles in several aspects of sexual reproduction, from germline stem cell maintenance to gametogenesis (15,16). Their expression is mostly restricted to the germline and gonadal somatic cells and they were found to bind a new class of sRNAs typically longer than siRNAs and miRNAs (17). The so-called piRNAs are unique in that they are amplified by a Dicer-independent mechanism, which may involve the Piwi slicer activity and, at least in vertebrates, they are massively produced during meiosis or just before. Metazoan Piwis are required for repression of transposable elements through the targeting of histone and DNA methylation (18–22) and/or PTGS mechanisms (23–25), and they may also play positive roles on chromatin structure and mRNA translation (26,27). Very little is known about Piwi proteins in other eukaryotic phyla, and it is unclear whether specialized functions in sexual reproduction are a conserved feature of the subclade.
Ciliates are a monophyletic group of eukaryotes belonging to the Chromalveolata (28); being about equally distant from plants, fungi and animals, they offer an interesting perspective on the evolutionary diversification of Argonaute proteins and their functions. Although they are unicellular, ciliates have evolved a unique system for germline/soma differentiation, based on the coexistence of two different kinds of nuclei in the cytoplasm. The diploid micronucleus (MIC) is a germline nucleus: its genome is not expressed and its only role is to undergo meiosis and transmit genetic information to the next sexual generation. The highly polyploid macronucleus (MAC), on the other hand, is a somatic nucleus: it is responsible for all gene expression, but is lost during sexual reproduction and replaced by a new one that develops from a copy of the zygotic nucleus. MAC development involves extensive rearrangements of the germline (MIC) genome, which occur during its amplification to the final ploidy level (29). Rearrangements include the elimination of repeated sequences such as transposons and minisatellites, as well as the excision of many short, single-copy Internal Eliminated Sequences (IESs). In Paramecium tetraurelia, it has been estimated that ≥50 000 IESs are precisely excised from coding and non-coding sequences of the haploid MIC genome, a process that is required for the reconstitution of functional genes in the MAC (30).
Paramecium tetraurelia possesses at least two distinct sRNA pathways. One is constitutively active and mediates homology-dependent gene silencing, which can be experimentally induced throughout the life cycle. This can be achieved either by transformation of the MAC with high-copy, non-expressible transgenes (31,32), or by feeding cells with bacteria producing dsRNA (33). In both cases, silencing of the endogenous gene correlates with the accumulation of ∼23-nt siRNAs (34,35) that appear to depend on the Dicer gene DCR1 (36). DsRNA-induced silencing results, at least in part, from cleavage of the endogenous mRNA in the region targeted by the dsRNA (37). The sequencing of dsRNA-induced siRNAs (36), confirmed by northern blot data (38), revealed a distinct subset, which appears to represent secondary siRNAs; because they appear to be produced by RdRPs from the targeted mRNA, their continuous production implies continuous transcription of the target gene. Transgene-induced silencing, on the other hand, does not result in the production of secondary siRNAs, and whether it involves PTGS or affects transcription of the target gene in the MAC remains an open question (38).
The second sRNA pathway documented in P. tetraurelia is active only during sexual reproduction and is essential for the development of functional zygotic MACs. During early meiosis of the MIC, a highly complex population of ∼25-nt scnRNAs with a distinctive 5′-UNG signature is produced from much, if not all, of the germline genome by the meiosis-specific Dicer-like proteins Dcl2 and Dcl3, which are also required for genome rearrangements in the developing MAC (36). Microinjection of synthetic RNA duplexes mimicking scnRNAs into conjugating cells was further shown to target the deletion of homologous sequences in the zygotic MAC (39). The scnRNA pathway is also likely involved in the maternal inheritance of alternative genome rearrangements during conjugation and autogamy, a self-fertilization sexual process (40,41). For a subset of IESs called mcIESs (maternally controlled IESs), introduction of the IES sequence into the maternal MAC specifically inhibits excision of the homologous IES in sexual progeny, during development of the zygotic MAC (42,43). Similarly, experimental deletions of non-essential genes in the maternal MAC are spontaneously reproduced in the new MAC of sexual progeny (34,44,45). Recent evidence indicates that these trans-nuclear effects depend on non-coding transcripts of the maternal MAC which appear to antagonize the action of homologous scnRNAs (39), likely by sequestering them or inducing their degradation before the development of zygotic MACs. This genomic subtraction would select the MIC-specific scnRNAs that target DNA elimination in the developing MACs.
Both sRNA pathways appear to be conserved in the related ciliate Tetrahymena thermophila, which contains 12 Piwi genes (46–49). Only the Twi1 protein has been extensively studied and shown to be involved in the scnRNA pathway (50). Together with the RNA helicase Ema1 (51), it is required for the targeting of H3K27 and H3K9 methylation on MIC-specific sequences in the developing MAC, which in turns promotes their elimination (52,53). Although deep sequencing of associated sRNAs revealed that at least five other Twi proteins are loaded with ∼23–24-nt endogenous siRNAs of diverse types (49), their functions have not been experimentally determined. Here, we report phylogenetic and functional analyses of the 15 P. tetraurelia genes encoding Argonaute proteins, all of which belong to the Piwi subclade. Using RNAi to inactivate each of these genes, we have been able to assign functions to six of them. Distinct but overlapping sets of genes appear to be involved in transgene-induced versus dsRNA-induced silencing, suggesting mechanistic differences between these two processes. Our results further identify the meiosis-specific PTIWI01 and PTIWI09 paralogues as key players in genome scanning and in the programming of developmental genome rearrangements. Interestingly, joint inactivation of these two genes is required to impair IES excision while the single silencing of any of them is sufficient to inhibit macronuclear deletions of cellular genes, suggesting an intrinsic difference between IESs and genes.
MATERIALS AND METHODS
Paramecium strains and cultivation
All experiments were carried out with the entirely homozygous strain 51. The cell line carrying a macronuclear deletion of the A surface antigen and ND7 genes was obtained by silencing these genes during autogamy, as previously described (34). Cells were grown in a wheat grass powder (WGP; Pines International, Lawrence, KS, USA) infusion medium bacterized the day before use with Klebsiella pneumoniae, unless otherwise stated, and supplemented with 0.8 mg/l of β-sitosterol (Merck, Darmstadt, Germany). Cultivation and autogamy were carried out at 27°C as previously described (43).
Alignments and phylogenetic analyses
Protein sequences were aligned using the MUSCLE software. Phylogenetic analyses were carried out using PhyML (bootstrapping procedure, 100 bootstraps) with default parameters and trees were created by TreeDyn (http://www.phylogeny.fr) (54).
Microarray expression data
Expression data were obtained from single-channel NimbleGen microarrays covering all 39 642 annotated genes, with six different 50-mer probes per gene. Raw signals were processed using the standard RMA method (55). This includes a first step of background subtraction for each array, followed by between-array normalization which was carried out using the normalizeBetweenArrays function from the limma package (56). The latter step adjusts signals so that expression values have similar distributions across all arrays considered in the analysis (for the autogamy time course, a total of 21 microarrays including 3 or 4 biological replicates for each of the six time points). The expression level of each gene was taken as the median signal from the six probes, averaged over the biological replicates of each time point. The vegetative time point (Veg) is the average from four mass cultures containing only log-phase cells showing no sign of meiosis. Autogamy was induced by letting cultures starve; because cells enter autogamy from a fixed point of the cell cycle, which is not synchronized in vegetatively growing cultures, there is a minimal asynchrony of ∼6 h (one cell cycle) in the progression through the different cytological stages. The meiosis time point (Mei) is the average of four samples containing 20–39% of cells undergoing meiosis, and little or no fragmentation of the old MAC (see histograms in Figure 1B). The ‘Mei+F’ samples contained a similar proportion of meiotic cells (20–29%), but also 37–43% of cells with a fragmented old MAC. ‘Dev1′ samples contained 35–56% of cells with fragmented old MACs, and 35–51% of cells that already contained clearly visible new MACs (anlagen). ‘Dev2′ samples contained 73–98% of cells with visible anlagen, and the ‘Dev3′ samples were extracted ∼9 h after ‘Dev2′ samples. Microarray platform and data analyses have been described in more detail elsewhere (57) and are publicly available at the Gene Expression Omnibus database (58) under accession numbers GSE17996, GSE17997, GSE17998 and GSE18002.
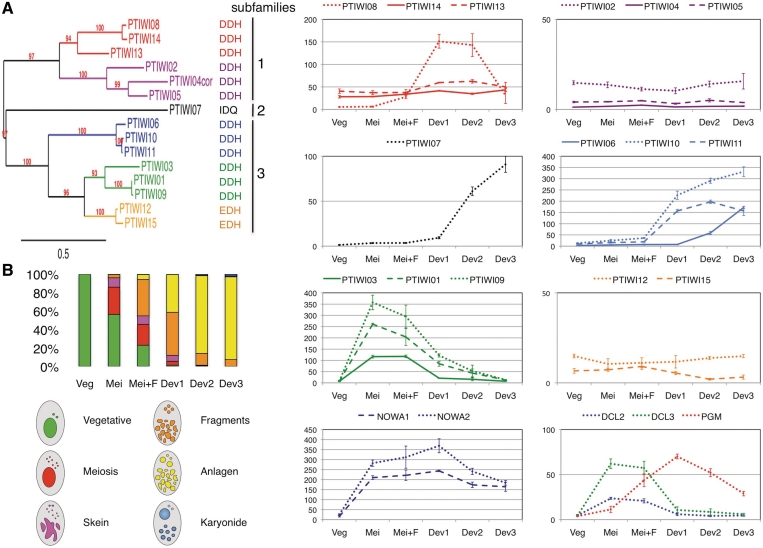
Figure 1.
Phylogenetic analysis and expression profiles of PTIWI genes. (A) Phylogenetic tree based on an alignment of deduced protein sequences. For the PTIWI04 pseudogene, a virtual protein sequence (PTIWI04cor) was created by correcting nonsense and frameshift mutations, using the PTIWI05 paralogue. Bootstrap values are indicated. The scale bar indicates the branch length corresponding to 0.5 substitution per site (inferred using the WAG model). The residues found at the positions of the slicer catalytic triad in each protein are shown on the right. (B) Expression profiles during the life cycle, as determined from NimbleGen microarray data (see ‘Materials and Methods’ section). The histograms show averages, over 3 or 4 biological replicates, of the fractions of cells in the different cytological stages (depicted and colour-coded below) during vegetative growth and at five different time points during mass autogamy. The graphs on the right show the variations in expression levels (arbitrary units, divided by 100) for all PTIWI genes and for the NOWA1-2, DCL2-3, and PGM (the putative endonuclease) genes. Error bars represent the standard errors computed from the 3 or 4 biological replicates. Graph scales were adapted to the expression levels of gene groups.
Constructs and probes
Plasmids used for T7Pol-driven dsRNA production in silencing experiments were obtained by cloning PCR products from each gene (see Supplementary Table S1A) using plasmid L4440 and Escherichia coli strain HT115 DE3, as previously described (33). The ND7, ND169 and ICL7a dsRNA fragments covered positions 873–1269, 1450–1860 and 1–580 of the PTETG500020001, GSPATG00008337001 and GSPATG00021610001 gene models, respectively. Probes used for the northern blots in Figure 2 were the same fragments as used for dsRNA production in silencing experiments. Probes used in Figure 3 and Supplementary Figure S3 are indicated in Supplementary Table S1A; the GAPDH probe covered positions 1–640 of GSPATG00013616001. The PTIWI09-GFP fusion was constructed by inserting a homemade EGFP-coding sequence, optimized for Paramecium codon usage, after codon 116 of the PTIWI09 coding sequence. The fusion was under the control of natural PTIWI09 regulatory sequences (512 bp upstream of translation start and 241 bp downstream of translation stop).
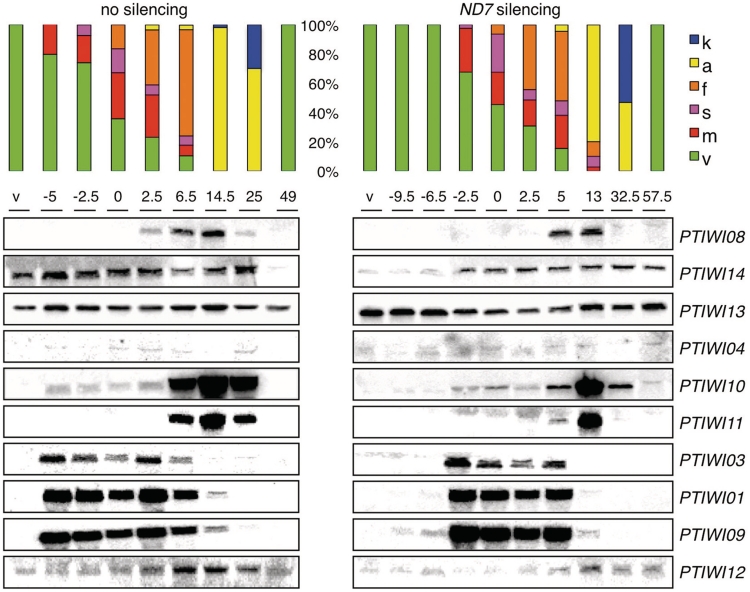
Figure 2.
Northern blot analysis of expression profiles during the life cycle. Expression profiles in vegetative cells and at different time points (h) of autogamy time courses are compared between a wild-type culture grown on Klebsiella (no silencing) and a culture submitted to dsRNA-induced silencing of the ND7 gene. The t = 0 time points are arbitrarily defined as the first samples in which at least 50% of the cells had started meiosis. The last samples in both series were obtained after refeeding post-autogamous cells enough for 2–3 divisions. Histograms show the fraction of cells in each cytological stage (same colour code as in Figure 1). The probes used for the different genes are the same segments as used for dsRNA production in silencing experiments (see Supplementary Table S1A).
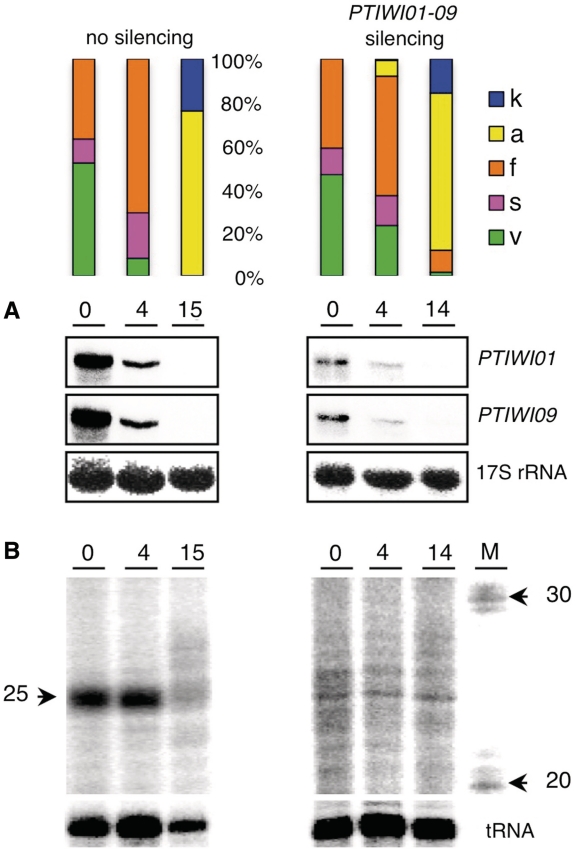
Figure 3.
Accumulation of scnRNAs during autogamy. Total RNA samples from a wild-type culture grown on Klebsiella (no silencing) and a culture submitted to dsRNA-induced silencing of PTIWI01 and PTIWI09 were extracted at different time points (hrs) of autogamy. The histograms show the fraction of cells in each cytological stage (same colour code as in Figure 1; meiosis was not documented by DAPI staining). The t = 0 time point was arbitrarily defined as the first sample in which ∼50% of cells had begun old MAC fragmentation. (A) Northern blot detection of PTIWI01 and PTIWI09 mRNAs. Signals were quantified and normalized with the 17S rRNA signal. (B) To examine scnRNA accumulation, RNA samples were 5′-end-labelled with T4 kinase and run on a 15% polyacrylamide–urea gel. The lower panel shows hybridization of the same membranes with a tRNA probe as a loading control. The size marker (M) shows the position of 20- and 30-nt RNAs.
Silencing by dsRNA feeding
DsRNA feeding media were prepared by diluting LB pre-cultures of the appropriate E. coli strains in WGP Paramecium medium containing 0.1 mg/ml ampicillin, followed by overnight growth at 37°C with shaking. On the next day, the culture was diluted 4-fold in the same medium. After 1 h of incubation at 37°C, IPTG was added at a final concentration of 0.5 mM to induce T7Pol transcription and dsRNA synthesis, and shaken for 4 h at 37°C. The medium was cooled to 27°C and supplemented with 0.8 mg/l of β-sitosterol just before use.
Microinjections
Paramecium cells from a single caryonidal clone in each experiment were microinjected in Volvic mineral water (Volvic, France) containing 0.2% bovine serum albumin (BSA), under an oil film (Nujol), while they were visualized with a phase contrast inverted microscope (Axiovert 35M, Zeiss). Column-purified (Qiagen) plasmids were linearized within the vector sequence, filtered on a 0.22 μm Ultrafree-MC filter (Millipore), precipitated with ethanol, and dissolved in filtered water. Approximately 5 pl of a 5 mg/ml solution were delivered into the MAC.
DNA and RNA extraction
About 200–400-ml cultures of exponentially growing cells at 1000 cells/ml or of autogamous cells at 2000 cells/ml were centrifuged. For DNA extraction, cell pellets were washed in 10 mM Tris–HCl (pH 7.0), re-suspended in one volume of the same buffer and quickly added to four volumes of lysis solution [0.44 M EDTA (pH 9.0), 1% SDS, 0.5% N-laurylsarcosine (Sigma) and 1 mg/ml proteinase K (Merck)]. The lysates were incubated at 55°C for at least 12 h, gently extracted with phenol and dialysed twice against TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) containing 20% ethanol and once against TE. RNA was extracted from unwashed cell pellets with the TRIzol (Invitrogen) procedure, modified by the addition of glass beads. PolyA+ RNAs were purified using the Poly(A)Purist mRNA purification kit (Ambion).
Southern blots, dot-blots and northern blot
DNA and RNA electrophoreses were carried out according to standard procedures (sambrook 2001). DNA (1–2 μg per sample) was transferred from agarose gels on Hybond N+ membranes (Amersham) in 0.4 N NaOH after depurination in 0.25 N HCl. For dot-blots, total DNA samples were denatured in 0.4 N NaOH at 65°C for 30 min and directly spotted onto the membrane. Northern blots of Figure 2 were carried out using the NorthernMax-Gly kit (Ambion). For the northern blots of Figure 3, total RNA samples (10 µg) were denatured in 1× MOPS containing 50% formamide and 3.7% formaldehyde for 5 min at 70°C before loading on 1% agarose gels. Gels were transferred to Hybond N+ membrane in 20× SSC buffer, and UV cross-linked. Hybridization was carried out in 7% SDS, 0.5 M sodium phosphate, 1% BSA and 1 mM EDTA (pH 7.2) at 60°C. Double-stranded probes were labelled by random priming with [α-32P] dATP (3000 Ci/mmol, Amersham). The tRNA and 17S rRNA oligonucleotide probes were labelled with [γ-32P] ATP (5000 Ci/mmol, Amersham) and T4 polynucleotide kinase. Membranes were then washed for at least 30 min in 0.2× SSC, 0.1% SDS at 60°C prior to image-plate exposure.
ScnRNA 5′ end labelling and electrophoresis
For scnRNA detection, total RNA samples (1.5 μg) were 5′-end-labelled by the exchange reaction of T4 polynucleotide kinase (Fermentas) with [γ-32P] ATP (5000 Ci/mmol, Amersham), denatured and run on 15% polyacrylamide–urea gel. After migration, the gel was directly exposed for 30 min on PhosphoImager screen.
RESULTS
Phylogenetic analysis and expression patterns of the 15 P. tetraurelia PTIWI genes
Annotation of the P. tetraurelia MAC genome (59) and homology searches identified 15 genes putatively encoding Argonaute proteins, which were named PTIWI01-15. After correction of assembly indels and manual reannotation, one of them (PTIWI04) was found to be a pseudogene, containing frameshift and nonsense mutations that can easily be detected by comparison with the closely related PTIWI05 paralogue. Gene models and reannotations can be found in ParameciumDB (60) using the accession numbers given in Supplementary Table S1A. The positions of the PAZ, MID and PIWI domains were deduced from an alignment with Argonaute proteins from other eukaryotes (Supplementary Figure S1 and Supplementary Table S1A). Key residues implicated in the binding of small RNA ends and in slicer activity are conserved in most of the Paramecium proteins. However, the DDH catalytic triad is mutated to IDQ in the most divergent protein (Ptiwi07), suggesting it does not have slicer activity; this may also be the case of the Ptiwi12 and Ptiwi15 proteins, which contain the variant triad EDH. A phylogenetic analysis based on this alignment indicates that all Paramecium proteins, like the Twi proteins from T. thermophila, clearly group with metazoan Piwi proteins (Supplementary Figure S2); these ciliates do not contain any member of the Ago subclade (15,61). Paramecium proteins can be divided into three subfamilies (Figure 1A). Subfamilies 1 and 3 each contain several groups of two or three closely related paralogues resulting from the last two Whole-Genome Duplications (WGDs), while subfamily 2 is limited to the very divergent Ptiwi07.
The expression patterns of PTIWI genes during the life cycle were examined by hybridization of oligonucleotide microarrays covering all annotated genes with cDNAs from exponentially growing vegetative cells or from five different time points after the onset of autogamy in mass cultures (Figure 1B). The results were confirmed for 10 genes by hybridization of Northern blots with specific probes (Figure 2). Groups of closely related genes generally showed similar expression patterns. Most genes in subfamily 1, which includes groups PTIWI08-14-13 and PTIWI02-04-05, are constitutively expressed throughout the life cycle, albeit at relatively modest levels; transcript levels for the PTIWI04 pseudogene are hardly detectable. The only exception is PTIWI08, which, unlike its paralogue from the last WGD (PTIWI14, 96% similar at the protein level), shows little or no expression during vegetative growth but is specifically expressed after meiosis, during development of new macronuclei. PTIWI08 thus appears to have recently acquired a development-specific expression pattern that distinguishes it from all other members of this subfamily.
In contrast, most genes in subfamily 3 are specifically expressed during the sexual phase of the life cycle, although different groups show markedly different patterns. Genes of the first group (PTIWI06-10-11) are silent during vegetative growth and meiosis, but become massively expressed after the onset of macronuclear development, with only minor differences among them. A similar pattern is seen for the only subfamily 2 member, PTIWI07. Genes of the second group (PTIWI03-01-09) are also silent during vegetative growth, but are turned on to high levels immediately upon meiosis. Microarray and northern blot data indicate that expression of these genes is very transient, stopping before the onset of MAC development and expression of PTIWI06-10-11. This meiosis-specific pattern is similar to that seen for the Dicer-like genes DCL2 and DCL3 (Figure 1B), which have been implicated in the biogenesis of scnRNAs (36). The third and odd group in subfamily 3 (PTIWI12-15), which is more closely related to PTIWI03-01-09, is constitutively expressed at low levels.
To determine whether the dsRNA feeding technique commonly used for RNAi has any effect on the expression of PTIWI genes, RNA samples were also extracted from a culture fed with an E. coli strain producing dsRNA homologous to the ND7 gene, during vegetative growth and at different stages of autogamy. The efficiency of ND7 post-transcriptional silencing was verified by the trichocyst non-discharge (tric-) phenotype (62). A northern blot analysis of these samples, compared to the Klebsiella-fed control culture (Figure 2), revealed a modest upregulation of PTIWI13 during vegetative growth (2- to 3-fold as determined by normalization with the 17S rRNA signal, not shown). This was confirmed by a microarray analysis of vegetative samples from the two cultures (Supplementary Table S2). The latter also showed a significant upregulation of PTIWI05 in the dsRNA-fed culture, though this was not tested on northern blots. Expression of the nine other genes tested did not appear to be affected at any stage.
Testing the functions of PTIWI genes using dsRNA-induced silencing
To study the potential roles of PTIWI genes in homology-dependent gene silencing during vegetative growth and in genome rearrangements during MAC development, we used the dsRNA feeding method to silence each of them, either alone or in all possible combinations of the 2 or 3 more closely related paralogues. dsRNA sequences (403–726 bp) were chosen to maximize silencing specificity (Supplementary Table S1B). Using RNAi to knockdown genes involved in the RNAi pathway is in principle a self-defeating process, but in practice this was shown to produce significant effects in some systems; theoretical modelling suggests the outcome might be primarily determined by the efficiency of RNAi in each system (63). In P. tetraurelia, recursive RNAi has been used successfully to identify the Dicer and RNA-dependent RNA polymerase genes involved in siRNA-mediated silencing (36,38). It should be borne in mind, however, that such tests are conclusive only if an effect is observed. In contrast, using siRNA-mediated silencing to knockdown genes required for scnRNA function is not expected to raise any problem if the PTIWI genes involved in the two pathways are entirely distinct, as seems to be the case for Dicer and Dicer-like genes (36).
PTIWI genes involved in dsRNA-induced and transgene-induced silencing
To identify PTIWI genes involved in transgene-induced silencing, the ND7 gene was silenced by transformation of the MAC with high copy numbers of a non-expressible transgene containing only the ND7 coding sequence, between translation start and stop codons. A cell line showing the tric- phenotype was then fed E. coli clones producing dsRNA for each of the PTIWI genes. Each culture was tested for reversion of the tric- phenotype, which may occur if a PTIWI gene is required for ND7 silencing. Full reversion was observed after 2 days in cultures that were fed PTIWI08, PTIWI14 or PTIWI13 dsRNAs, either alone or in all possible combinations, while the tric- phenotype was maintained in all other cases (Table 1). Because PTIWI08 is not detectably expressed during vegetative growth (Figures 1 and 2), the effect of PTIWI08 dsRNA may be due to cross-silencing of the PTIWI14 gene, to which it is 77.4% identical, with segments of perfect identity up to 19 bp (Supplementary Table S1B). This was indeed verified in another experiment (see below).
Table 1.
Summary of the phenotypic effects of dsRNA-induced silencing of PTIWI genes
| PTIWI genes silenced | Reversion of ND7 silencing |
Viability of sexual progeny | |
|---|---|---|---|
| Transgene-induced | dsRNA-induced | ||
| PTIWI01 | − | − | Viable |
| PTIWI03 | − | − | Viable |
| PTIWI09 | − | − | Viable |
| PTIWI01-03 | − | − | Viable |
| PTIWI01-09 | − | − | Non-viable |
| PTIWI03-09 | − | − | Viable |
| PTIWI01-03-09 | − | − | Non-viable |
| PTIWI10 | − | − | Viable |
| PTIWI11 | − | − | Viable |
| PTIWI06 | − | − | Viable |
| PTIWI10-11 | − | − | Viable |
| PTIWI10-06 | − | − | Viable |
| PTIWI11-06 | − | − | Viable |
| PTIWI10-11-06 | − | − | Viable |
| PTIWI12 | − | − | Viable |
| PTIWI15 | − | − | Viable |
| PTIWI12-15 | − | ± | Viable |
| PTIWI04 | − | − | Viable |
| PTIWI05 | − | − | Viable |
| PTIWI02 | − | − | Viable |
| PTIWI04-05 | − | − | Viable |
| PTIWI04-02 | − | − | Viable |
| PTIWI05-02 | − | − | Viable |
| PTIWI04-05-02 | − | − | Viable |
| PTIWI08 | + | − | Viable |
| PTIWI14 | + | − | Viable |
| PTIWI13 | + | + | Viable |
| PTIWI08-14 | + | − | Viable |
| PTIWI08-13 | + | + | Viable |
| PTIWI14-13 | + | + | Viable |
| PTIWI08-14-13 | + | + | Viable |
| PTIWI07 | − | − | Viable |
| No silencing | − | − | Viable |
The 2nd and 3rd columns indicate cases of full (+) or partial (±) reversion of the trichocyst non-discharge phenotype resulting from transgene-induced or dsRNA-induced silencing of the ND7 reporter gene during vegetative growth. The 4th column indicates the viability of the post-autogamous progeny of cells silenced for the different PTIWI genes.
In a different test, silencing was first established by ND7 dsRNA feeding, and tric- cells were then transferred to media containing a mix of E. coli clones producing ND7 or PTIWI dsRNA in equal amounts. A complete reversion of the tric- phenotype was observed after 2 days of simultaneous feeding of PTIWI13 dsRNA, but not of PTIWI08 or PTIWI14 dsRNAs; a weaker reversion (partial trichocyst discharge) was observed after 3 days when PTIWI12 and PTIWI15 were co-silenced (Table 1). A larger scale experiment was then designed to confirm these results with a different reporter gene and to monitor the steady-state mRNA levels for the targeted PTIWI genes. A mass culture of tric- cells silenced for the ND169 gene (64) by dsRNA feeding was split into seven parts which were fed bacterial mixes containing 50% of ND169 dsRNA and 50% of PTIWI08, 14, 13, 12, 15, 12 and 15, or, as a control, ICL7a [an irrelevant centrin gene (65)] dsRNAs. Exactly the same phenotypic effects were observed as with the ND7 reporter.
Northern blot analyses of total RNA samples from these cultures, or from cultures of wild-type cells fed the same PTIWI dsRNAs, revealed a ∼4- to 5-fold decrease of mRNA levels for the most abundantly expressed PTIWI13 gene, specifically upon PTIWI13 dsRNA feeding (Supplementary Figure S3A). Quantification of the PTIWI14 mRNA, although less reliable because of its lower abundance, also indicated that it was reduced by PTIWI14 dsRNA. The detection of shorter molecules, presumably resulting from mRNA cleavage, further provided qualitative evidence for the degradation of PTIWI14 and PTIWI12 mRNAs by cognate dsRNAs. The analysis also confirmed that PTIWI08 dsRNA could indeed target PTIWI14 mRNA. Quantification of the PTIWI15 and ND169 mRNAs was more difficult, and even a Northern blot of purified poly-A+ RNAs failed to provide conclusive evidence for PTIWI15 knockdown by the cognate dsRNA (Supplementary Figure S3B). Nevertheless, ND169 mRNA levels were reduced ∼2- to 3-fold upon ND169 dsRNA feeding, and, consistent with the phenotypes observed, showed a modest increase upon co-silencing of PTIWI13 or PTIWI12-15 (∼0.5-fold, relative to other double-silencing samples).
We conclude that Ptiwi13 is involved in both transgene- and dsRNA-induced silencing, while Ptiwi14 appears to be specifically involved in the former, and the Ptiwi12–15 pair in the latter. The fact that joint targeting of PTIWI12 and 15 is required for partial suppression of the ND7 or ND169 silencing phenotypes suggests that these genes encode redundant functions, and that cross-silencing between the two highly similar genes, if it occurs at all, is not sufficient to cause a detectable effect when only one is targeted.
PTIWI genes involved in developmental genome rearrangements
Because the meiosis-specific scnRNA pathway is essential for correct genome rearrangements in the developing MAC (36), knocking down PTIWI genes involved in scnRNA function during sexual events is expected to result in non-viable progeny. As a preliminary screen, wild-type cells were fed dsRNA for each of the PTIWI genes, or combinations of the 2 or 3 more closely related paralogues, for about four vegetative divisions before starvation triggered autogamy. Cytologically normal new MACs developed in all cases, as monitored by DAPI staining. Post-autogamous cell populations were then refed with Klebsiella to test viability. Non-viable progeny was observed only in combinations including both PTIWI01 and PTIWI09 dsRNAs (Table 1), prompting us to carry out a more detailed analysis of these two genes, and of the closely related PTIWI03. Consistent with their meiosis-specific expression, continuous silencing of these genes during ≥20 vegetative divisions, alone or in combinations of 2 or 3, did not result in any obvious phenotype. In contrast, applying dsRNA feeding for only 4 divisions, immediately followed by autogamy, confirmed the preliminary test: after isolation of 12 post-autogamous cells from each silencing test into Klebsiella medium, all progeny from the PTIWI01-09 and PTIWI01-09-03 combinations showed abnormal phenotypes and most of them (8/12 and 9/12, respectively) died after 3–5 divisions, while progeny from other combinations or single-gene silencing were comparable to unsilenced controls (Supplementary Table S3). This suggests that PTIWI01 and PTIWI09 are involved in the development of functional new MACs, which progressively become essential during the first vegetative divisions, as the non-replicating fragments of the functional old MAC become too few in each cell.
To test the role of these genes in scnRNA accumulation, total RNA was extracted at different time points during autogamy of large-scale cultures, with or without silencing of PTIWI01 and PTIWI09 by dsRNA feeding. After the onset of meiosis, progression through the different cytological stages was similar in the two cultures (Figure 3). Northern blot quantification showed that the double silencing reduced PTIWI01 and PTIWI09 mRNA amounts ∼4-fold and ∼6-fold, respectively, compared with the control (t = 0 time point, Figure 3A). To examine scnRNAs, total RNA samples were radiolabelled at the 5′-end and run on a 15% polyacrylamide–urea gel. A prominent ∼25-nt band was observed during early autogamy in the control, but little or no scnRNA accumulation was detected at similar stages in the PTIWI01-09 knockdown (Figure 3). We conclude that the proteins encoded by these genes are required for production or stabilization of scnRNAs.
Different assays were used to examine genome rearrangements in the developing new MACs after silencing of these genes. IES excision was first tested by PCR on small amounts of DNA extracted from post-autogamous cell populations, using pairs of primers located in the flanking sequences of two different IESs. PCR products from MIC DNA cannot be detected in such assays because of the high MAC:MIC ploidy ratio (200:1); in addition to the IES-excised products that are always amplified from the old MAC, IES-containing products are observed only if unexcised IES copies accumulate in the developing new MACs. This was the case only in post-autogamous cells from the PTIWI01-09 and PTIWI01-09-03 silencing combinations (Figure 4A), suggesting lethality was indeed due to defects in genome rearrangements.
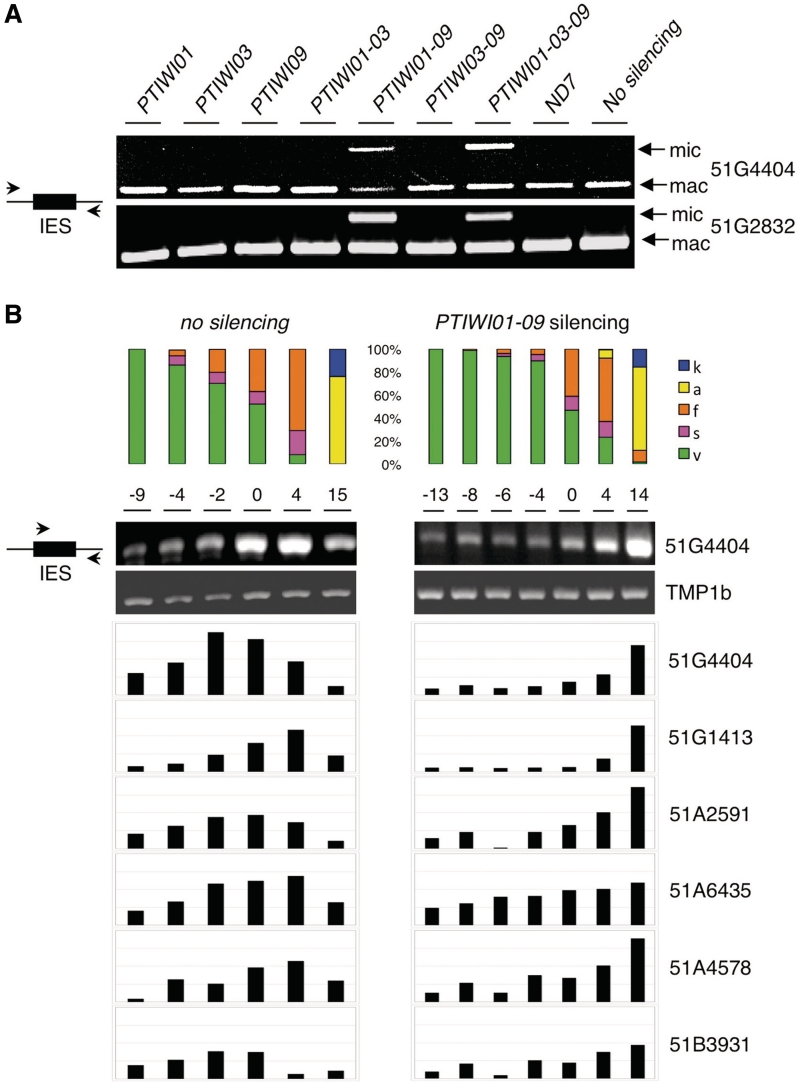
Figure 4.
Effects of PTIWI01 and PTIWI09 silencing on IES excision. (A) Total DNA was extracted after autogamy from cells grown on Klebsiella (no silencing) or silenced for ND7 or for genes of the PTIWI01-03-09 group, as indicated, and PCR amplified using pairs of primers located in the flanking sequences of IESs 51G4404 and 51G2832. The excised version (mac) can always be amplified from the fragments of the old MAC, while unexcised copies (mic) can be detected only when they accumulate in the new MACs. (B) Total DNA samples were extracted at different time points (h) from the same autogamy time courses as in Figure 3, and semi-quantitative PCR analyses were performed for 6 IESs with one primer in the IES and the other in the flanking sequence. The gel is shown only for IES 51G4404 (see Supplementary Figure S4 for other gel images). The histograms represent the amounts of unexcised IES copies, after normalization with the intensity of a control PCR amplifying the macronuclear TMP1b gene. 51G4404, 51G2832 and 51A2591 are maternally controlled IESs, while 51G1413, 51A6435 and 51A4578 are not (43). The status of 51B3931 is unknown.
These results were then confirmed and extended to other IESs using DNA samples extracted at different time points from the two large-scale autogamous cultures used for scnRNA analysis. Semi-quantitative PCR tests were performed for six different IESs using one primer within the IES sequence and the other in the flanking sequence, so that only unexcised copies can be amplified. In the control time course, the unexcised signal rose transiently above the background level due to amplification of MIC DNA, reflecting the few rounds of amplification of the zygotic genome that take place in the developing new MACs before IES excision starts (66). In contrast, the unexcised signal rose continuously in the PTIWI01-09 knockdown and reached a maximum in the last time point, a stage at which IES excision was mostly complete in the control (Figure 4B and Supplementary Figure S4). Importantly, the silencing of PTIWI01 and PTIWI09 was found to impair excision not only for mcIESs such as 51G4404 and 51A2591, but also for non-mcIESs such as 51G1413, 51A6435 and 51A4578.
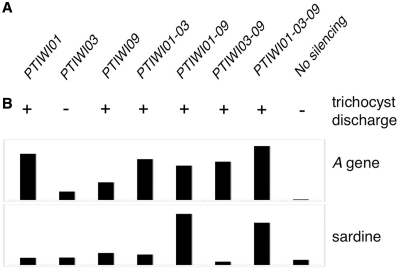
We further tested the role of the PTIWI03-01-09 group of genes in the imprecise DNA elimination mechanism that is responsible for the deletion of MIC transposable elements during MAC development (67) and for maternally inherited deletions of non-essential cellular genes, which can be induced experimentally (34). A cell line reproducibly deleting the the A51 surface antigen and ND7 genes from the MAC genome at each sexual generation was fed dsRNAs for each of these genes alone, or in all possible combinations, for about 4 divisions prior to autogamy. Reversion of the ND7 MAC deletion in post-autogamous cell populations was assessed by phenotypic testing: in all silencing combinations involving either PTIWI01 or PTIWI09, but not in the PTIWI03 single silencing nor in the unsilenced control, the occurrence of trichocyst discharge indicated that the ND7 gene was at least partially maintained in the new MAC (Figure 5A). Total DNA from the same post-autogamous cell populations was then loaded on a dot-blot, which was successively hybridized with probes specific for the Sardine transposon, for the A51 gene, and for mitochondrial DNA as a loading control (Supplementary Figure S5). Quantification of the signals revealed that, as observed with ND7, the MAC deletion of the A51 gene reverted in all silencing combinations involving either PTIWI01 or PTIWI09; the relatively modest effect of the PTIWI09 single silencing could be due to a lower efficiency of silencing in that test since the PTIWI01-03 and PTIWI03-09 double silencing tests gave very similar results (Figure 5B). In contrast, elimination of the Sardine transposon was impaired only in silencing combinations involving both PTIWI01 and PTIWI09, as observed for IES excision.
Figure 5.
Effects of PTIWI01-03-09 silencing on transposon elimination and maternal inheritance of macronuclear deletions. (A) Reversion of the trichocyst non-discharge phenotype in post-autogamous progeny of cells carrying a macronuclear deletion of the ND7 gene and submitted during autogamy to dsRNA-induced silencing of PTIWI genes, as indicated. Reversion (+) indicates amplification of the ND7 gene in the new MACs. (B) Dot-blot quantification of the Sardine transposon and of the A surface antigen gene in post-autogamous progeny of cells carrying a macronuclear deletion of the A gene and submitted during autogamy to dsRNA-induced silencing of PTIWI genes, as indicated (see Supplementary Figure S5 for dot-blot image). The signals obtained with the Sardine and A-gene probes were normalized with those of a mitochondrial DNA probe.
Subcellular localization of Ptiwi09p during autogamy
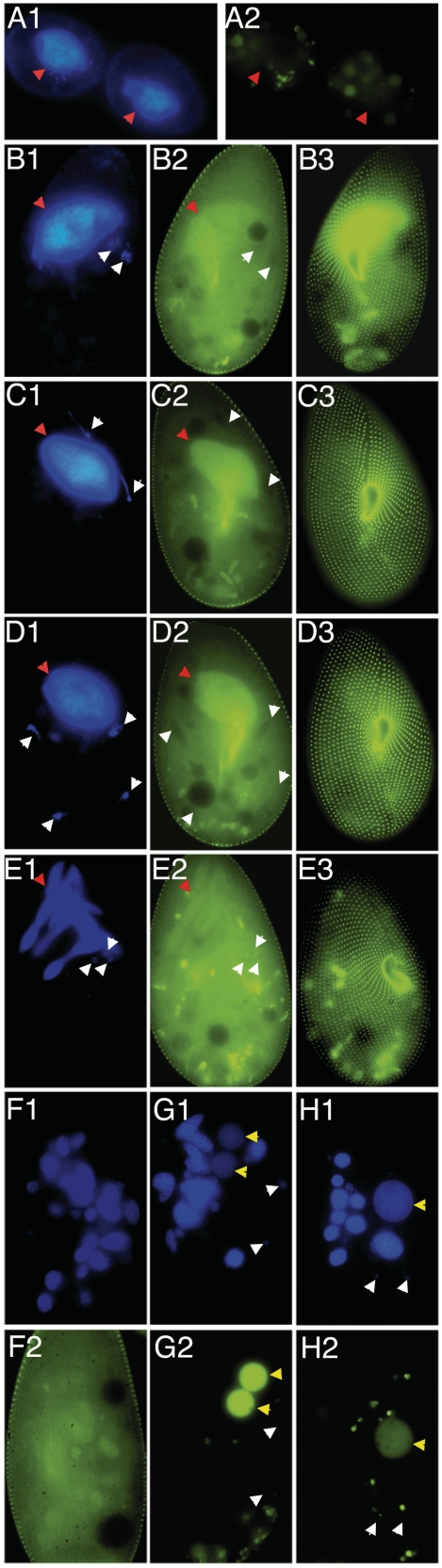
To study the subcellular localization of Ptiwi09p, a GFP fusion was constructed by inserting the GFP coding sequence into the PTIWI09 gene, upstream of the PAZ domain (after codon 116). Expression of the fusion gene was under the control of the natural PTIWI09 up- and downstream sequences. After microinjection of the construct into the MAC of vegetative cells, no fluorescence could be detected during vegetative growth of transformed clones (Figure 6A). When autogamy was induced by starvation, GFP fluorescence first appeared in the cytoplasm (Figure 6B). It then accumulated to some extent in the old MAC during the crescent stage (i.e. prophase of meiosis I, Figure 6C), but was always excluded from micronuclei and later from their meiotic products. Surprisingly, the GFP fusion also localized to basal bodies. This localization pattern remained essentially unchanged throughout meiosis I, meiosis II and skein formation (Figure 6D–E). When fragmentation of the old MAC was complete (Figure 6F), GFP fluorescence started to decrease at these locations and progressively relocalized to the new MACs as they developed, until all of the fusion protein was concentrated there (Figure 6G). Fluorescence finally faded away from the new MAC after the caryonidal division (Figure 6H).
Figure 6.
Expression and dynamic localization pattern of a Ptiwi09-GFP fusion during autogamy. Pictures numbered 1 show DAPI staining; pictures numbered 2 and 3 show GFP fluorescence within the cell body and at the cell surface, respectively. Arrowheads indicate the different types of nuclei: red, old MAC before fragmentation; white, MICs and their meiotic products; yellow, new MAC anlagen. A, vegetative cell during division; B, beginning of meiosis I; C, meiosis I; D, beginning of meiosis II; E, skein formation; F, fragmentation of the old MAC; G, new MAC development; H, karyonide after the first vegetative division.
DISCUSSION
Evolutionary diversification of P. tetraurelia PTIWI genes
The phylogenetic analysis of the Ptiwi protein family presented in Supplementary Figure S2 indicates that P. tetraurelia, like the related T. thermophila, contains only members of the Piwi subclade of Argonaute proteins. The topology of the tree further shows that each of the three subfamilies of P. tetraurelia proteins groups with specific T. thermophila homologs (91–98% bootstrap values) and that recurrent gene duplications have occurred in each of the two oligohymenophorean lineages since their divergence. Further relationships can be established by comparing the expression profiles of T. thermophila Twi proteins (49,68) with those determined here. As detailed below, these suggest that, while the different subfamilies may have broadly conserved functions, intra-lineage gene duplications have probably led to functional specialization, notably through changes in expression patterns.
Subfamily 1 is related to a subfamily of Twi proteins shown to bind ∼23–24-nt endogenous sRNAs (49). The cytoplasmic protein Twi2 is the most abundant in vegetative cells and is upregulated upon expression of hairpin RNA, suggesting it may be involved in siRNA-mediated silencing (48), as well as during conjugation. Other members are specifically expressed during MAC development (Twi9 and Twi10) or in starved cells (Twi7) (68). Some of these features are also seen in Paramecium subfamily 1: Ptiwi13 and 14 are the most abundant proteins in vegetative cells, and were shown here to be involved in siRNA-mediated silencing; Ptiwi13 is upregulated in response to dsRNA uptake, and Ptiwi08 is expressed during MAC development. Thus, although the full sets of functions ensured by these subfamilies remain unknown, they may largely overlap.
The only member of subfamily 2, the non-catalytic Ptiwi07, appears to be an ortholog of Twi8 (bootstrap 96%), which was shown to bind 3′-end modified, ∼23–24-nt sRNAs and to localize in the MAC (49). However, these genes may have diverged in function since Twi8 is expressed at highest levels during vegetative growth, while expression of Ptiwi07 is restricted to late MAC development.
Subfamily 3 forms a group with Twi1 and Twi11 (bootstrap 98%) in which most proteins are specifically expressed during sexual events at very high levels. Interestingly, in both species some genes are expressed very early during meiosis (Ptiwi03-01-09 and Twi1) while others are induced during MAC development (Ptiwi06-10-11 and Twi11), when expression of the early genes decreases. Although the phylogenetic tree does not support the orthology of early or late genes of both species, similar expression patterns may underlie similar functions. Indeed Twi1, the only Tetrahymena protein with an experimentally demonstrated function, is loaded with scnRNAs during meiosis, is required for genome rearrangements in the developing MAC (50,51), and thus appears to be a functional homolog of Ptiwi01 and 09. Ptiwi12 and 15 are the only subfamily 3 members showing constitutive expression throughout the life cycle, albeit at low levels. Their similarity to the Ptiwi03-01-09 group (bootstrap 100%) suggests they have arisen through Paramecium-specific gene duplications and have acquired functions unrelated to sexual events (see below).
Distinct mechanisms underlie transgene- and dsRNA-induced silencing
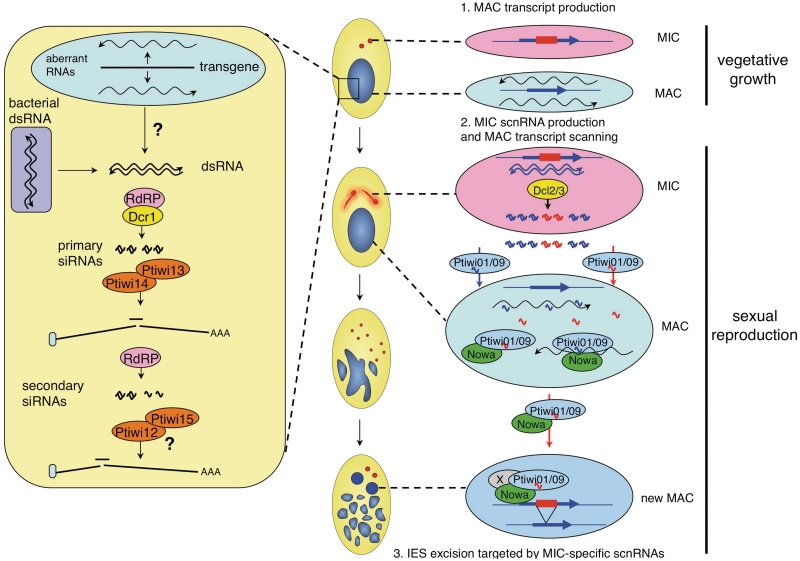
We have shown that Ptiwi13 is required for both transgene- and dsRNA-induced silencing, while Ptiwi14 appears to be specifically involved in the former, and Ptiwi12 and 15 in the latter. Although both silencing methods were previously shown to result in the accumulation of ∼23-nt siRNAs Figure 7 (34,35), which appear to depend on the Dicer gene DCR1 (36), the present results indicate that the mechanisms involved are at least partially distinct. Interestingly, the same conclusion was drawn in a study of the roles of P. tetraurelia RdRPs, which showed that the two processes require different RdRP genes and that the associated siRNAs are of different types (38). Confirming the results of a previous small-scale sequencing study (36), it was found that dsRNA-induced siRNAs comprise two distinct classes: ∼23-nt primary siRNAs that are processed from both strands of the ingested dsRNA, and a faster migrating class (∼22 nt) with a strictly antisense polarity, believed to represent secondary siRNAs produced by an RdRP activity from the targeted mRNA. In contrast, only ∼23-nt siRNAs were found to accumulate in transgene-induced silencing; however, these were shown to differ from dsRNA-induced primary siRNAs in that they carry a modification of the 3′-terminal ribose (38). It is, therefore, tempting to speculate about the Ptiwi proteins that bind these different types of siRNAs.
Figure 7.
Schematic representation of the roles of Ptiwi proteins in sRNA pathways. SiRNA pathway (left): In vegetative cells, dsRNA may be formed by the pairing of sense and antisense aberrant transcripts during transgene-induced silencing, or imported from the food vacuoles during dsRNA-induced silencing. Primary siRNAs generated through the action of Dcr1 and different RdRPs (38) are then be loaded onto Ptiwi13 and/or Ptiwi14, resulting in cleavage of homologous mRNAs at least in the case of dsRNA-induced silencing. In that case, further RdRP activity using the mRNA as a template would generate antisense secondary siRNAs which may be loaded onto Ptiwi12 and Ptiwi15 to amplify the silencing response. ScnRNA pathway (right): promiscuous bidirectional transcription of the MAC genome occurs at low levels during vegetative growth. Upon meiosis, bidirectional transcription of the MIC genome forms dsRNA which is cleaved into scnRNA duplexes by Dcl2 and Dcl3. The guide strands of scnRNAs are then loaded onto Ptiwi01 and Ptiwi09 in the cytoplasm and transported to the old MAC where they would scan nascent transcripts with the help of the Nowa1 and 2 proteins. ScnRNAs able to find a perfect match would be sequestered or degraded, so that only those homologous to MIC-specific sequences would later be available to target epigenetic modifications on these sequences in the developing new MAC, eventually leading to their elimination.
In the case of dsRNA-induced silencing, Ptiwi13 may bind the ∼23-nt primary siRNAs and its potential slicer activity may be responsible for mRNA cleavage, which was found to occur in the region targeted by siRNAs (37). The specific involvement of Ptiwi12 and 15 in that process suggests that they could bind the secondary siRNAs. Aside from the very divergent Ptiwi07, these are the only Paramecium proteins, which deviate from the canonical DDH catalytic triad, as is the case of many of the Wagos that bind secondary siRNAs in C. elegans (3). However, only biochemical tests will tell whether the EDH triad of Ptiwi12 and 15 is unable to support slicer activity. If so, these proteins could contribute to silencing by recruiting other nucleases to the targeted mRNAs, as proposed for the non-catalytic Wagos (69), or by inhibiting their translation. An alternative possibility is that these proteins also bind the primary siRNAs and are required for the production, rather than the binding, of secondary siRNAs, by recruiting RdRPs to the targeted mRNA. Whatever the case may be, our recursive RNAi tests do not allow us to conclude as to the extent of their contribution to silencing. The residual reporter silencing observed may reflect the impossibility to completely silence PTIWI12 and 15 by dsRNA feeding, since these genes are involved in the process; on the other hand, it could be due to the action of PTIWI13 and primary siRNAs, which would occur upstream of secondary siRNA synthesis and should not be affected by the silencing of PTIWI12 and 15.
While it is also tempting to assume that Ptiwi14 associates with 3′-modified siRNAs, the precise role of Ptiwi proteins, and in particular of Ptiwi13, in transgene-induced silencing is less clear, because siRNA-mediated mRNA cleavage has not been documented in that case. One possibility is that the process transiently depends on unmodified ∼23-nt siRNAs bound by Ptiwi13, in such a low steady-state amount that they escape detection. Since indirect evidence from the RdRP study (38) suggests that transgene-induced silencing might not be entirely post-transcriptional, Ptiwi14 and modified siRNAs could possibly affect transcription of the target gene in the nucleus.
The Ptiwi14 and Ptiwi08 proteins are 96% similar and are therefore likely to have similar biochemical activities. The PTIWI08 expression profile indicates that these activities are globally upregulated ∼4-fold during MAC development, suggesting that endogenous mechanisms akin to the transgene-induced siRNA pathway normally operate at that stage. This is reminiscent of the upregulation or specific expression of several Twi2-subfamily proteins in T. thermophila (49,68). However, specific roles for endogenous siRNAs during sexual events have not been documented so far in these ciliates. In this study, the silencing of PTIWI08 (or the entire PTIWI08-14-13 group) during autogamy did not result in any obvious phenotype, but the experimental design used may not have been as efficient for silencing genes during MAC development as it proved to be at the earlier stage of meiosis (see below).
PTIWI01 and PTIWI09 are involved in developmental genome rearrangements
Our preliminary screen identified the meiosis-specific PTIWI01 and 09 genes as being required for the survival of sexual progeny. The two encoded proteins are 99% identical and are likely to have the same or very similar functions. Cells that were fed both dsRNAs for a few divisions before the onset of meiosis showed cytologically normal nuclear reorganization throughout autogamy, indicating that these proteins are not required for meiosis, karyogamy, mitotic divisions of the zygotic nucleus or amplification of the genome in the developing new MACs. The death of post-autogamous progeny after a few vegetative divisions may be entirely attributed to non-functional new MACs. Indeed, the silencing of PTIWI01 and 09 was found to impair the excision of all IESs tested, as well as the elimination of the Sardine transposon. The double silencing also completely prevented the accumulation of the ∼25-nt scnRNAs that are produced by the Dicer-like proteins Dcl2 and Dcl3 during early meiosis of the MIC (36). This strongly suggests that scnRNAs are immediately loaded onto the Ptiwi01 and 09 proteins upon exit from the MIC, and that this is required for their stability.
The dynamic localization pattern of a Ptiwi09-GFP fusion suggests that Ptiwi01 and 09 mediate both types of pairing interactions implied by the genome-scanning model Figure 7 (36,39). The fusion protein was detected in the cytoplasm during meiosis, where it could be loading scnRNAs, and accumulated in the old MAC, likely reflecting the scanning of nascent transcripts by scnRNAs. GFP fluorescence later relocalized to the new MACs as they developed, where MIC-specific scnRNAs are thought to target sequences to be eliminated, again through an interaction with nascent transcripts. The same localization pattern is observed for the Twi1 protein in T. thermophila (50), as well as for the Ema1 helicase, which has been proposed to facilitate the pairing of scnRNAs with nascent transcripts in both types of nuclei (51). In P. tetraurelia the meiosis-specific protein Nowa1, which is believed to act in the scnRNA pathway because it is required for elimination of mcIESs and transposons, shows a very similar localization pattern (35). Nowa1 and the closely related Nowa2 contain ‘FRG’ repeats that may bind nascent transcripts, interspersed with ‘GGWG’ repeats that have been shown in other systems to form an ‘Ago hook’ binding Argonaute proteins (70–74). Thus, Nowa1 and 2 likely interact with Ptiwi01 and 09 to assist them in pairing interactions, both in the old MAC during meiosis and in the developing new MACs. Intriguingly, Ptiwi09-GFP also localized to basal bodies during early stages of autogamy. While this would need to be tested with the native Ptiwi09 to rule out a fusion or overexpression artefact, this localization is not a binding property of the GFP itself and was observed at the same stages for a Dcl2-GFP fusion (V. Serrano and E. Meyer, unpublished data). Together with old data indicating the presence of RNA in basal bodies (75), this raises the possibility that these structures play some role in the scnRNA pathway.
Consistent with Ptiwi01 and Ptiwi09 having the same functions, the feeding of each dsRNA alone prior to autogamy had the same subtle effects. While this did not compromise the viability of sexual progeny or the elimination of tested IESs and transposons, it was sufficient to induce at least a partial reversion of maternally inherited MAC deletions of cellular genes. It also frequently caused new MACs to be determined for mating type E when the old MAC was determined for mating type O (not shown), which likely results from defective rearrangements in a mating-type gene (76). These milder effects indicate that each dsRNA was not able to completely cross-silence the other gene, although some level of cross-silencing probably occurred because of the relatively high sequence similarity (∼85%, with identical segments up to 19 bp). Exactly the same differences were observed between the effects of single and double silencing of the DCL2 and/or DCL3 genes (36). Together these observations suggest that higher amounts of scnRNA-loaded Ptiwi01/09 proteins are required to target the deletion of cellular genes, as compared to IESs and transposons, in the developing MAC.
It is interesting to note that PTIWI01 and 09 are required for the excision of both mcIESs and non-mcIESs. In contrast, NOWA1 and 2 are required only for the excision of mcIESs (35), and the same is true of DCL2 and DCL3 (36; H. Touzi and S. Duharcourt, Personal communication). Thus, Ptiwi01 and 09 are involved in more genome rearrangements than those thought to be regulated by the scnRNA pathway. Similarly, the Twi1 protein of T. thermophila is required for the deletion of more DNA elements than is Ema1 (51). In addition to scnRNAs, the Ptiwi01 and 09 proteins may be loaded with some other type of sRNAs, not submitted to selection by the scanning of old MAC transcripts, that would enable them to target the deletion of non-mcIESs in the developing MAC irrespective of the presence of homologous sequences in the old MAC.
No phenotype was observed after the silencing of genes of the late group, PTIWI06-10-11. Their expression patterns suggest functions in MAC development, but do not strongly support a role in genome rearrangements. Indeed, their expression peaks later than that of the PiggyMac gene (PGM in Figure 1), the putative endonuclease which appears to be essential for all types of DNA elimination (77), whereas one would expect earlier expression for proteins involved in sRNA-directed chromatin modifications guiding rearrangements, as observed for PTIWI01 and 09. PTIWI06-10-11 could be involved in other epigenetic functions, possibly related to gene expression programming, which could have escaped detection in our simple tests. However, it is also possible that the experimental scheme used was not very efficient for the silencing of late genes. Indeed, the dsRNA feeding technique relies on continuous uptake of dsRNA from food bacteria, but meiosis can only be induced by starvation. Results presented here and elsewhere indicate that the stock of accumulated siRNAs is sufficient to silence genes expressed early during sexual events, but it may not be sufficient during late stages of MAC development.
Piwi protein functions are not limited to sexual processes
This technique nevertheless allowed us to assign functions to 6 of the 14 potentially functional genes. Two of them mediate the action of scnRNAs, which show intriguing similarities to metazoan piRNAs: both classes are longer than siRNAs, have a strong 5′U bias, and are specifically required for sexual reproduction. Both have also been implicated in the control of transposable elements, although ciliate scnRNAs target their elimination in the developing somatic MAC but are not known to contribute to repression in the germline MIC, where no mRNA is produced. Another difference is that the biosynthesis of scnRNAs depends on Dicer-like proteins, while that of piRNAs does not. PiRNAs were initially defined in metazoans by their interaction with Piwi-subclade proteins, but a more biologically relevant definition could perhaps be based on their essential roles in sexual reproduction; such meiosis-specific sRNAs may even be universally conserved in eukaryotes. The results presented here indicate that four different P. tetraurelia Piwi proteins mediate homology-dependent regulation of genes in the somatic MAC during vegetative growth and likely associate with different types of siRNAs. Thus, as previously observed with Ago proteins in plants, Piwi proteins can evolve to ensure a wide range of functions at all stages of the life cycle, through mechanisms that range from PTGS to epigenetic modification of the genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche (ANR-08-BLAN-0233); Fondation pour la Recherche Médicale (Equipe FRM grant to E.M.); Centre National de la Recherche Scientifique (ATIP and ATIP Plus grants to M.B.). A.K. was recipient of doctoral fellowships from the Ministère de l’Education Supérieure et de la Recherche and from the Fondation pour la Recherche Médicale. Funding for open access charge: Agence Nationale de la Recherche (ANR-08-BLAN-0233).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank O. Arnaiz and J. Nowak for their help, and all members of the lab for fruitful and stimulating discussions. We are grateful to J. Cohen and his team because a part of this work was done in his lab. We thank L. Sperling, J. Beisson, M. Simon, S. Marker, and S. Duharcourt for critical reading of the manuscript.
REFERENCES
- 1.Nowotny M, Yang W. Structural and functional modules in RNA interference. Curr. Opin. Struct. Biol. 2009;19:286–293. doi: 10.1016/j.sbi.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker JS, Parizotto EA, Wang M, Roe SM, Barford D. Enhancement of the seed-target recognition step in RNA silencing by a PIWI/MID domain protein. Mol. Cell. 2009;33:204–214. doi: 10.1016/j.molcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 9.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 10.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 11.Janowski BA, Hu J, Corey DR. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nat. Protoc. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- 12.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Chan SW. Inputs and outputs for chromatin-targeted RNAi. Trends Plant Sci. 2008;13:383–389. doi: 10.1016/j.tplants.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Sigova A, Rhind N, Zamore PD. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18:2359–2367. doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol. Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 18.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 21.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc. Natl Acad. Sci. USA. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotelnikov RN, Klenov MS, Rozovsky YM, Olenina LV, Kibanov MV, Gvozdev VA. Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res. 2009;37:3254–3263. doi: 10.1093/nar/gkp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 27.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol. Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Yao MC, Duharcourt S, Chalker DL. Genome-wide rearrangements of DNA in ciliates. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 730–755. [Google Scholar]
- 30.Gratias A, Betermier M. Developmentally programmed excision of internal DNA sequences in Paramecium aurelia. Biochimie. 2001;83:1009–1022. doi: 10.1016/s0300-9084(01)01349-9. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz F, Vayssie L, Klotz C, Sperling L, Madeddu L. Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell. 1998;9:931–943. doi: 10.1091/mbc.9.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galvani A, Sperling L. Transgene-mediated post-transcriptional gene silencing is inhibited by 3′ non-coding sequences in Paramecium. Nucleic Acids Res. 2001;29:4387–4394. doi: 10.1093/nar/29.21.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galvani A, Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- 34.Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowacki M, Zagorski-Ostoja W, Meyer E. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 36.Lepere G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- 38.Marker S, Le Mouel A, Meyer E, Simon M. Distinct RNA-dependent RNA polymerases are required for RNAi triggered by double-stranded RNA versus truncated transgenes in Paramecium tetraurelia. Nucleic Acids Res. 2010;38:4092–4107. doi: 10.1093/nar/gkq131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duharcourt S, Lepere G, Meyer E. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009;25:344–350. doi: 10.1016/j.tig.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Meyer E, Chalker DL. Epigenetics of ciliates. In: Allis CD, Jenuwein T, Reinberg D, Caparros MC, editors. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 127–150. [Google Scholar]
- 42.Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- 43.Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol. Cell. Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer E. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 1992;6:211–222. doi: 10.1101/gad.6.2.211. [DOI] [PubMed] [Google Scholar]
- 45.Meyer E, Butler A, Dubrana K, Duharcourt S, Caron F. Sequence-specific epigenetic effects of the maternal somatic genome on developmental rearrangements of the zygotic genome in Paramecium primaurelia. Mol. Cell. Biol. 1997;17:3589–3599. doi: 10.1128/mcb.17.7.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 48.Howard-Till RA, Yao MC. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol. Cell. Biol. 2006;26:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 51.Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl Acad. Sci. USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 56.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 57.Arnaiz O, Gout JF, Betermier M, Bouhouche K, Cohen J, Duret L, Kapusta A, Meyer E, Sperling L. Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia. BMC Genomics. 2010;11:547. doi: 10.1186/1471-2164-11-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 60.Arnaiz O, Cain S, Cohen J, Sperling L. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007;35:D439–D444. doi: 10.1093/nar/gkl777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skouri F, Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell. 1997;8:1063–1071. doi: 10.1091/mbc.8.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall WF. Modeling recursive RNA interference. PLoS Comput. Biol. 2008;4:e1000183. doi: 10.1371/journal.pcbi.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Froissard M, Keller AM, Dedieu JC, Cohen J. Novel secretory vesicle proteins essential for membrane fusion display extracellular-matrix domains. Traffic. 2004;5:493–502. doi: 10.1111/j.1600-0854.2004.00194.x. [DOI] [PubMed] [Google Scholar]
- 65.Gogendeau D, Klotz C, Arnaiz O, Malinowska A, Dadlez M, de Loubresse NG, Ruiz F, Koll F, Beisson J. Functional diversification of centrins and cell morphological complexity. J. Cell Sci. 2008;121:65–74. doi: 10.1242/jcs.019414. [DOI] [PubMed] [Google Scholar]
- 66.Betermier M, Duharcourt S, Seitz H, Meyer E. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol. Cell. Biol. 2000;20:1553–1561. doi: 10.1128/mcb.20.5.1553-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Mouel A, Butler A, Caron F, Meyer E. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot. Cell. 2003;2:1076–1090. doi: 10.1128/EC.2.5.1076-1090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 72.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bednenko J, Noto T, DeSouza LV, Siu KW, Pearlman RE, Mochizuki K, Gorovsky MA. Two GW repeat proteins interact with Tetrahymena thermophila argonaute and promote genome rearrangement. Mol. Cell. Biol. 2009;29:5020–5030. doi: 10.1128/MCB.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dippell RV. Effects of nuclease and protease digestion on the ultrastructure of Paramecium basal bodies. J. Cell Biol. 1976;69:622–637. doi: 10.1083/jcb.69.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer E, Keller AM. A Mendelian mutation affecting mating-type determination also affects developmental genomic rearrangements in Paramecium tetraurelia. Genetics. 1996;143:191–202. doi: 10.1093/genetics/143.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Betermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.