Abstract
The human immunodeficiency virus type 1 (HIV-1) external envelope glycoprotein gp120 presents conserved binding sites for binding to the primary virus receptor CD4 as well as the major HIV chemokine coreceptors, CCR5 and CXCR4.
Concerted efforts are underway to understand the specific interactions between gp120 and coreceptors as well as their contribution to the subsequent membrane fusion process.
The present review summarizes the current knowledge on this biological aspect, which represents one of the key and essential points of the HIV-host cell interplay and HIV life cycle. The relevance of conformational HIV-1 Envelope proteins presented on Virus-like Particles for appropriate assessment of this molecular interaction, is also discussed.
Introduction
The molecular interaction between HIV-1 gp120, in its trimeric conformation, and the CD4 receptor on the host cell surface represents the first step of the HIV infection cycle. Upon this interaction, the co-receptor-binding site on the gp120 is exposed, enabling the binding to HIV chemokine coreceptors (mainly CCR5 or CXCR4) expressed on the surface of a subset of CD4+ lymphocytes. The binding to the coreceptors is followed by fusion of the viral and host cell membranes mediated by the HIV gp41 transmembrane glycoprotein [1-6].
Dissecting the structural changes which HIV external envelope glycoprotein gp120 molecule undergo upon molecular interactions with its cognate cellular receptor and coreceptors, provide essential information to the development of HIV-1-specific drugs, targeting the viral entry step [7-16], as well as of vaccines [17-20].
Gp120 binding to chemokine coreceptors
The HIV-1 Envelope is synthesized as the polyprotein precursor gp160, which undergoes oligomerization, disulfide bond formation and extensive glycosylation in the endoplasmic reticulum [21]. The full post-translational processing and maturation lead to proteolytical cleavage of precursor gp160 into the surface gp120 and transmembrane gp41 subunits by furin-like endo-proteases in the Golgi network [22-24]. The two subunits will assemble into a trimer consisting of three gp120 molecules associated non-covalently with three gp41 subunits.
The molecular interaction of HIV gp120 with the CD4 receptor and, subsequently, with the CCR5 or CXCR4 coreceptor leads to the insertion of the hydrophobic gp41 N-terminal region (fusion peptide) into the host cell membrane. In particular, the gp41 ectodomain trimer acquires the six-helix bundle configuration which drives in close contact the viral and cell membranes, ultimately resulting in their fusion [1,2,4,25,26]. Therefore, the binding of HIV envelope to cellular coreceptors dramatically influence the strength of viral-cell interaction and promote the conformational changes in the gp41 required to overcome the energy barrier and induce pore formation and membrane fusion.
Within the CCR5 and CXCR4 amino acid residues interacting with the gp120, most of the cysteine residues are involved in disulfide bonds formation and play a key functional role. In particular, the N-terminal and second extracellular domain (ECII) of both coreceptors seem to be critical for gp120-CD4 complex binding [27-35].
The role of coreceptors in the conformational changes of the HIV transmembrane gp41 to facilitate virus-cell membrane fusion has not yet been fully clarified, mainly due to the lack of the CCR5 and CXCR4 crystal structure and, therefore, their absence in high resolution X-ray structures solved for CD4-bound gp120 [17]. The currently accepted theory proposes that, upon the coreceptor binding to the gp120-CD4 complex, the gp41 acquires the thermostable, six-helix bundle structure that brings the two membranes together and results in fusion pore formation [36,37].
The first step is the exposure of the hydrophobic fusion peptide at the N terminus of gp41 which interacts with the target cell membrane, generating an intermediate, pre-hairpin state bridging the virus and cell membranes. The pre-hairpin then refolds into the stable, six-helix bundle core structure [38,39], releasing sufficient energy to overcome the kinetic barrier [40,41] and catalyzing the fusion of the two membranes [42]. Whether the fusion can occur with the free energy liberated during refolding of one or several trimers, is still debated [40,43] (Fig.1).
Figure 1.
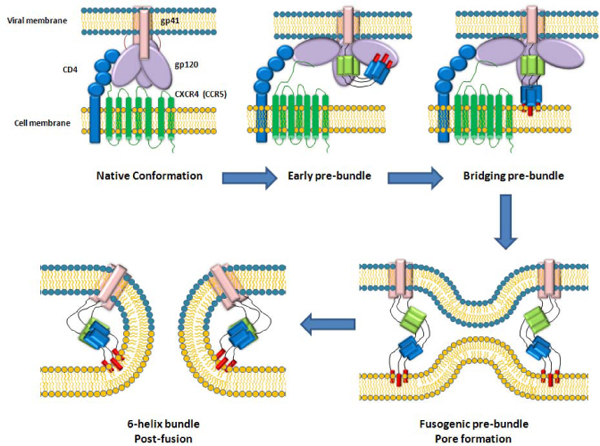
Dissection of sequential steps occurring after engagement of receptor and coreceptor by trimeric HIV envelope proteins.
In the described stepwise process, the pre-hairpin state shows a relatively long half-life [44], representing a favorable target for inhibitory peptides [45,46] as well as neutralizing antibodies specific for the gp41 HR1 and MPER regions [47-50].
Several data about the envelope/receptor interactions have been generated also for the simian counterpart of HIV (Simian Immunodeficiency Virus, SIV). Indeed, SIVmac is the natural etiological agent of the AIDS-like syndrome in Rhesus Macaques, which is the only available animal model for obtaining relevant information on AIDS pathogenesis [51-54] as well as for testing efficacy of antiviral therapeutics and vaccine candidates [55,56].
Similarly to HIV-1, SIV infection starts with the high-affinity interaction of the gp120-gp41 envelope glycoprotein (Env) complex with CD4 on the target cell surface [57,58]. However, in contrast to HIV-1, different strains of SIV preferentially use CCR5 and not CXCR4 as coreceptor for entry [59-61], although they may show promiscuity in coreceptor usage, engaging alternative coreceptors GPR15 and CXCR6/STRL33 with high efficiency [62,63].
Moreover, SIV strains have been shown to infect target cells via a CD4-independent pathway, directly interacting with CCR5 or CXCR4 coreceptor [64,65]. It has been proposed that Env protein of such strains contains multiple amino acid substitutions leading to the constitutive exposure of coreceptor binding site [66,67], similarly to what described for CD4-independent HIV-1 envelope proteins [68,69]. As consequence, CD4-independent viruses acquire a broader cell tropism, being able to infect also CD4 negative or low-expressing cells, such as macaque macrophages [70,71].
The molecular interaction of SIV envelope protein with the CD4 receptor and coreceptors leads to conformational changes in the gp41 ectodomain trimers and exposure of the fusion peptide which closely resemble what described for the HIV-1 counterpart [72,73].
Role of coreceptors post-translational modifications in HIV-1-mediated cell fusion
The effectiveness of CCR5 and CXCR4 as HIV coreceptors depends on the several possible conformations which may significantly influence their ability to support viral entry in different cells [74,75].
Furthermore, post-translational modifications of HIV coreceptors and, in particular, of the extracellular domains (including the N-terminal, EC I – III) and intracellular loops, may modulate the receptor turnover as well as the binding efficacy to the HIV gp120. In general, extracellular domains may undergo N-linked or O-linked glycosylation and tyrosine sulfation, while modifications of intracellular loops include palmitoylation, phosphorylation, and ubiquitination [76-79].
CCR5. The CCR5 N-terminal is relevant for the role as HIV-1 coreceptor [27,29,30] and contains several tyrosine residues which may be modified by sulfation, contributing to binding to natural ligands (MIP1-α, MIP1-β) as well as HIV-1 gp120-CD4 complexes [80-82]. In particular, sulfation of Tyrosines at position 10 and 14 seems to be a requisite for the CCR5 binding efficacy [83,84]. Moreover, CCR5 is also modified by O-linked glycosylation, preferentially on Ser-6 [85,86], although it does not seem to affect the role of CCR5 in the HIV entry [86].
As most chemokine receptors, the CCR5 carboxyl-terminus contains one or more cysteine residues compatible with receptor palmitoylation, typically located 12 to 25 amino acids away from the plasma membrane boundary [87]. Cysteine residues at amino acid positions 321, 323, and 324 undergo to palmitoylation, facilitating the CCR5 transport to the plasma membrane as well as ligand-stimulated endocytosis and affecting its ability to initiate intra-cellular signalling pathways [88-90]. However, the biological role of improved localization of CCR5 to lipid rafts for HIV entry into host cells is still disputed [88-92].
CXCR4. Similar to CCR5, also CXCR4 undergo post-translational modifications contributing to its function. CXCR4 is sulfated at three tyrosine residues in the N-terminus, with Tyr21 accounting for the majority of sulfate incorporation, although this modification doesn’t appear to modulate the CXCR4 coreceptor function for HIV-1 [83].
In contrast to CCR5, the extracellular domain of CXCR4 is post-translationally modified by N-linked glycosylation in two potential sites, Asn11 and Asn176 [93,94], although only Asn11 appears to be glycosylated in mammalian cells [94]. Mutation of Asn11 does not impair the CXCR4-mediated HIV-1 infection [95,96]; however, a Asn11-to-Glu11 mutation leads to enhanced binding of both CXCR4-specific and dual-tropic (CCR5 and CXCR4) HIV-1 isolates [78,96].
HIV-1 Envelope-coreceptor signaling
The binding of the gp120-CD4 complex to chemokine coreceptors not only mediates HIV entry but also activates intracellular signaling cascades, mimicking chemokine signaling induced by binding to cognate receptors [97,98] (reviewed in [77,99,100].
However, in addition to signaling pathways mediating cell migration, transcriptional activation, cell growth and differentiation [101-106], binding of gp120-CD4 complex to CCR5 or CXCR4 coreceptor has also been shown to trigger the activation of proline-rich tyrosine kinase (Pyk2), phosphoinositide 3-kinases (PI3K) [107,108] and CD4/CXCR4-dependent NFAT (nuclear factor of activated T cells) nuclear translocation [109]. Furthermore, gp120 was demonstrated to mediate chemotaxis, actin cytoskeleton rearrangement [108], and the activation of an actin depolymerization factor, cofilin, to increase the cortical actin dynamics in resting CD4 T cells [110]. (For a more comprehensive description of gp120-triggered chemokines coreceptor signaling, refer to Cicala and Arthos in this same supplement).
Stable trimeric forms of human immunodeficiency virus recombinant gp140
As described above, the envelope proteins on the virus surface are assembled into trimers, consisting of three gp120 molecules associated non-covalently with three gp41 subunits, which interact sequentially with the CD4 receptor and the chemokine coreceptors, ultimately leading to viral and cell membrane fusion. In the last years, mainly aiming at inducing more potent and broader anti-HIV neutralizing antibodies, several groups have been developing soluble trimers of the gp120 - gp41 Env ectodomain (i.e., lacking the transmembrane, cytoplasmic domains and named gp140) which are considered to preserve or mimic the structure of functional Env complexes [111-114]. Considering the close structural similarity of these molecules to native trimeric HIV Envelope proteins, this can represent a relevant tool also for receptor and chemokines coreceptor binding studies.
Mutations in the furin cleavage site at the gp120–gp41 junction inhibit the dissociation between the two envelope subunits [114-117], but cleavage-defective gp140 Env proteins seem to be antigenically different from fully processed Env [118-120]. In order to express processed and stable trimers of gp140, an intermolecular disulfide bond has been introduced between the gp120 and gp41 subunits to form a complex called “SOS” gp140 which, although still predominantly monomeric, strongly reacts with the broadly neutralizing b12mAb [118]. An isoleucine-to-proline substitution introduced at position 559 in the N-terminal heptad region of gp41 has been shown to increase the stability of SOS gp140 (SOSIP gp140), leading to a fully cleaved and trimeric structure with optimal antigenic properties [121,122].
Moreover, cleavage-defective gp140 Env proteins have been further modified at the C-terminus to improve trimer formation and stabilization, fusing different heterologous trimerization motifs. In particular, a 32-amino-acid form of the GCN4 transcription factor (GCN), a 27-amino-acid trimerization domain from the C-terminus of bacteriophage T4 fibritin (T4F), or a soluble trimerization domain of chicken cartilage matrix (CART) protein have been employed, showing enhanced binding to broadly neutralizing b12 and 2G12 mAbs [115,116,123]. More recently, the effectiveness of the catalytic chain of aspartate transcarbamoylase (ATCase) as trimerization domain for the HIV gp140 has been described [124,125].
Exploiting the VLP model
In addition to soluble forms, HIV gp140 trimeric complex can be presented on membrane structures including liposomes, inactivated viruses and virus-like particles (VLPs) or pseudovirions, to mimic as close as possible the native conformation [126-131].
In particular, Virus-like particles (VLPs) represent a complex structure based on viral capsid proteins which self-assemble into particulate structures closely resembling immature virus particles [132-135]. VLPs are replication as well as infection incompetent, lacking regulatory proteins as well as infectious genetic material, and can be considered nanoparticles and employed to deliver antigenic structures as well as DNA molecules to antigen presenting cells, for enhanced induction of immune responses against co-administered plasmid DNA-based immunogens [136-140].
VLPs have been produced from a broad spectrum of non-enveloped and enveloped viruses [140]. In particular, the particle structure of non-enveloped VLPs can be based on single or multiple capsid proteins without a surrounding cell membrane. Examples of such VLPs are those formed by the expression of the major capsid protein of papillomaviruses, parvoviruses and polyomaviruses [135,141-143] or by multiple interacting capsid proteins of Reoviridae family [144]. Alternative strategy for generating non-enveloped virus-like-particles (VLPs) is based on the assembly of the capsid protein derived from RNA bacteriophages [145-147].
Alternatively, enveloped VLPs are based on assembled capsid proteins surrounded by cell membrane and have been developed for enveloped viruses such as hepatitis B and C virus (HBV & HCV), influenza A and retroviruses, including HIV-1 [129,132,133,148-152].
The different forms of VLPs have distinct properties for displaying antigens, given that only enveloped VLPs may display full-length monomeric or multimeric conformational proteins on their surface through trans-membrane domains [129-131,153]. Non-enveloped VLPs, on the contrary, may be employed to present mainly short peptides or protein sequences. This has been achieved either generating chimeric capsid proteins expressing foreign epitope in frame (eg. Gag:V3) [154-157], or chemically linking the foreign epitope to the assembled capsid protein [158], although HIV-1 Gag proteins fused to full-length HIV Reverse Transcriptase (RT) protein, without loosing the capability of assembling into VLPs, have been recently reported [159]. The conformational structure of full-length proteins possibly expressed on non-enveloped VLPs, however, remains to be proven.
As consequence, at present, only enveloped VLPs displaying full-length conformational proteins on their surface may be a suitable experimental model for studies on binding and interaction between HIV envelope and cellular receptor/coreceptors. They, indeed, represent the closest particle structure to native virus.
Conformational HIV Envelopes presented on particulate structures
Development of HIV-1 Pr55Gag-based VLPs expressing gp120/gp140 trimeric Envelope proteins is a goal pursued by several Groups, using different expression systems (mammalian vs baculovirus) as well as different trans-membrane domains. In particular, our group has previously developed Pr55gag-VLPs presenting a gp120 anchored through the trans-membrane (TM) portion of the Epstein-Barr virus (EBV) gp220/350, showing the formation of oligomeric structures [129] and inducing Env-specific humoral and cellular immune response [160-162]. Additional strategies have been more recently described to express trimeric forms of HIV-1 gp140 molecules on the surface of Pr55gag-VLPs [130,131]. Comparative studies showed that specific trans-membrane domains induced optimal incorporation of gp140 Env trimers onto VLP surface, retaining conserved epitopes and undergoing conformational changes upon CD4 binding [131] (Buonaguro et al., submitted).
Considering the biological and structural properties of HIV gp140 trimers presented on the VLP surface, they can be even more strategic for binding studies, providing an invaluable tool for evaluating and dissecting the whole virus-host cell interaction leading to and ending with membrane fusion [163,164] (Fig. 2).
Figure 2.
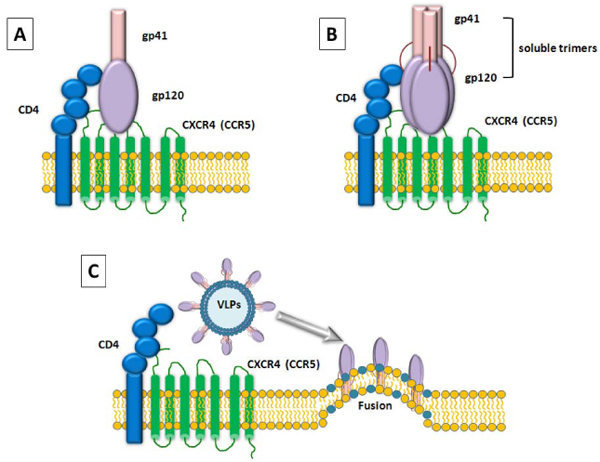
Schematic representation of HIV-1 gp120 binding to cellular receptor and coreceptors. The binding of HIV Envelope protein to CD4 receptor and chemokines coreceptors on the host cell surface is represented showing the gp120 in its monomeric form (A) and trimeric form as soluble (B) or bound to virus like particles (VLPs) (C).
Conclusions
The interaction between HIV particles and target host cells is a defined temporally sequential stepwise process, characterized by the binding of surface gp120 Envelope protein to CD4 receptor and subsequent binding of the gp120-CD4 complex to chemokines coreceptors. This will ultimately lead to membrane fusion and host cell infection.
Studies aimed at dissecting the sequential steps of this process have been and will be instrumental not only to fully understand the strategies adopted by HIV to hijack host cells for its own replication but also to develop HIV-1-specific drugs and vaccines.
The development of novel and improved molecular tools, mimicking as close as possible native Envelope trimeric structures expressed on non-infectious particulate structures (i.e. VLPs), will expand the armamentarium for HIV-host cell interaction studies. This will help in shedding further light on such a key moment of the HIV infection as well as pathogenesis.
Competing interests
The authors declare no competing financial or other interests.
Contributor Information
Maria Tagliamonte, Email: mariellatagliamonte@libero.it.
Maria Lina Tornesello, Email: mltornesello@virgilio.it.
Franco M Buonaguro, Email: irccsvir@unina.it.
Luigi Buonaguro, Email: lbuonaguro@tin.it.
Acknowledgements
The study was supported by the European Community's Seventh Framework Programme NGIN (FP7/2007-2013) under grant agreement n 201433. MT was supported by the EU NGIN project.
This article has been published as part of Journal of Translational Medicine Volume 9 Supplement 1, 2011: Differential use of CCR5 vs. CSCR4 by HIV-1. Pathogenic, Translational and Clinical Open Questions. The full contents of the supplement are available online at http://www.translational-medicine.com/supplements/9/S1.
References
- Doms RW, Moore JP. HIV-1 membrane fusion: targets of opportunity. J Cell Biol. 2000;151:F9–14. doi: 10.1083/jcb.151.2.F9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry. 1999;38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Jacobson JM, Saag MS, Thompson MA, Fischl MA, Liporace R, Reichman RC, Redfield RR, Fichtenbaum CJ, Zingman BS, Patel MC, Murga JD, Pemrick SM, D'Ambrosio P, Michael M, Kroger H, Ly H, Rotshteyn Y, Buice R, Morris SA, Stavola JJ, Maddon PJ, Kremer AB, Olson WC. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. J Infect Dis. 2008;198:345–1352. doi: 10.1086/592169. [DOI] [PubMed] [Google Scholar]
- Pett SL, McCarthy MC, Cooper DA, MacRae K, Tendolkar A, Norris R, Strizki JM, Williams KM, Emery S. A phase I study to explore the activity and safety of SCH532706, a small molecule chemokine receptor-5 antagonist in HIV type-1-infected patients. Antivir Ther. 2009;14:111–115. [PubMed] [Google Scholar]
- Lalezari J, Yadavalli GK, Para M, Richmond G, Dejesus E, Brown SJ, Cai W, Chen C, Zhong J, Novello LA, Lederman MM, Subramanian GM. Safety, pharmacokinetics, and antiviral activity of HGS004, a novel fully human IgG4 monoclonal antibody against CCR5, in HIV-1-infected patients. J Infect Dis. 2008;197:721–727. doi: 10.1086/527327. [DOI] [PubMed] [Google Scholar]
- Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Armour D. The discovery of the CCR5 receptor antagonist, UK-427,857, a new agent for the treatment of HIV infection and AIDS. Prog Med Chem. 2005;43:239–271. doi: 10.1016/S0079-6468(05)43007-6. full_text. [DOI] [PubMed] [Google Scholar]
- Hendrix CW, Collier AC, Lederman MM, Schols D, Pollard RB, Brown S, Jackson JB, Coombs RW, Glesby MJ, Flexner CW, Bridger GJ, Badel K, MacFarland RT, Henson GW, Calandra G. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J Acquir Immune Defic Syndr. 2004;37:1253–1262. doi: 10.1097/01.qai.0000137371.80695.ef. [DOI] [PubMed] [Google Scholar]
- Stone ND, Dunaway SB, Flexner C, Tierney C, Calandra GB, Becker S, Cao YJ, Wiggins IP, Conley J, MacFarland RT, Park JG, Lalama C, Snyder S, Kallungal B, Klingman KL, Hendrix CW. Multiple-dose escalation study of the safety, pharmacokinetics, and biologic activity of oral AMD070, a selective CXCR4 receptor inhibitor, in human subjects. Antimicrob Agents Chemother. 2007;51:2351–2358. doi: 10.1128/AAC.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princen K, Hatse S, Vermeire K, Aquaro S, De CE, Gerlach LO, Rosenkilde M, Schwartz TW, Skerlj R, Bridger G, Schols D. Inhibition of human immunodeficiency virus replication by a dual CCR5/CXCR4 antagonist. J Virol. 2004;78:12996–13006. doi: 10.1128/JVI.78.23.12996-13006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagat JR, McCombie SW, Nazareno D, Labroli MA, Xiao Y, Steensma RW, Strizki JM, Baroudy BM, Cox K, Lachowicz J, Varty G, Watkins R. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. IV. Discovery of 1-[(4,6-dimethyl-5-pyrimidinyl)carbonyl]- 4-[4-[2-methoxy-1(R)-4-(trifluoromethyl)phenyl]ethyl-3(S)-methyl-1-piperaz inyl]- 4-methylpiperidine (Sch-417690/Sch-D), a potent, highly selective, and orally bioavailable CCR5 antagonist. J Med Chem. 2004;47:2405–2408. doi: 10.1021/jm0304515. [DOI] [PubMed] [Google Scholar]
- Baba M, Takashima K, Miyake H, Kanzaki N, Teshima K, Wang X, Shiraishi M, Iizawa Y. TAK-652 inhibits CCR5-mediated human immunodeficiency virus type 1 infection in vitro and has favorable pharmacokinetics in humans. Antimicrob Agents Chemother. 2005;49:4584–4591. doi: 10.1128/AAC.49.11.4584-4591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer PR, Ulmer JB, Rappuoli R. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol. 2008;26:659–667. doi: 10.1016/j.tibtech.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do KY, Scheid J, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010. [DOI] [PMC free article] [PubMed]
- Earl PL, Moss B, Doms RW. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Myers DJ, Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63:4670–4675. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Doms RW. The entry of entry inhibitors: a fusion of science and medicine. Proc Natl Acad Sci U S A. 2003;100:10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot DJ, Broder CC. Substitutions in a homologous region of extracellular loop 2 of CXCR4 and CCR5 alter coreceptor activities for HIV-1 membrane fusion and virus entry. J Biol Chem. 2000;275:23774–23782. doi: 10.1074/jbc.M003438200. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Locati M, Kennedy PE, Murphy PM, Berger EA. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- Doranz BJ, Lu ZH, Rucker J, Zhang TY, Sharron M, Cen YH, Wang ZX, Guo HH, Du JG, Accavitti MA, Doms RW, Peiper SC. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, Olson WC, Wu L, Mackay CR, Allaway GP, Sakmar TP, Moore JP, Maddon PJ. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/JVI.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot DJ, Zhang PF, Quinnan GV, Broder CC. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J Virol. 1999;73:6598–6609. doi: 10.1128/jvi.73.8.6598-6609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelot A, Heveker N, Adema K, Hosie MJ, Willett B, Alizon M. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J Virol. 1999;73:2576–2586. doi: 10.1128/jvi.73.4.2576-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajumo F, Thompson DA, Guo Y, Dragic T. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology. 2000;271:240–247. doi: 10.1006/viro.2000.0308. [DOI] [PubMed] [Google Scholar]
- Zhou N, Luo Z, Luo J, Liu D, Hall JW, Pomerantz RJ, Huang Z. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J Biol Chem. 2001;276:42826–42833. doi: 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]
- Golding H, Zaitseva M, de RE King LR, Manischewitz J, Sidorov I, Gorny MK, Zolla-Pazner S, Dimitrov DS, Weiss CD. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J Virol. 2002;76:6780–6790. doi: 10.1128/JVI.76.13.6780-6790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:77–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83:1523–1531. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- Luftig MA, Mattu M, Di GP, Geleziunas R, Hrin R, Barbato G, Bianchi E, Miller MD, Pessi A, Carfi A. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat Struct Mol Biol. 2006;13:740–747. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry. 2007;46:1398–1401. doi: 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Daniel MD, Sehgal PK, Desrosiers RC, Hunt RD, Waldron LM, MacKey JJ, Schmidt DK, Chalifoux LV, King NW. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- Estes JD, Gordon SN, Zeng M, Chahroudi AM, Dunham RM, Staprans SI, Reilly CS, Silvestri G, Haase AT. Early resolution of acute immune activation and induction of PD-1 in SIV-infected sooty mangabeys distinguishes nonpathogenic from pathogenic infection in rhesus macaques. J Immunol. 2008;180:6798–6807. doi: 10.4049/jimmunol.180.10.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, Roques P, Butor C, Silvestri G, Giavedoni LD, Lebon P, Barre-Sinoussi F, Benecke A, Muller-Trutwin MC. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G. Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS? J Med Primatol. 2005;34:243–252. doi: 10.1111/j.1600-0684.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- Shedlock DJ, Silvestri G, Weiner DB. Monkeying around with HIV vaccines: using rhesus macaques to define 'gatekeepers' for clinical trials. Nat Rev Immunol. 2009;9:717–728. doi: 10.1038/nri2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rompay KK. Evaluation of antiretrovirals in animal models of HIV infection. Antiviral Res. 2010;85:159–175. doi: 10.1016/j.antiviral.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Kestler HW III, Naidu YN, Kodama T, King NW, Daniel MD, Li Y, Desrosiers RC. Use of infectious molecular clones of simian immunodeficiency virus for pathogenesis studies. J Med Primatol. 1989;18:305–309. [PubMed] [Google Scholar]
- Naidu YM, Kestler HW III, Li Y, Butler CV, Silva DP, Schmidt DK, Troup CD, Sehgal PK, Sonigo P, Daniel MD. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhou P, Ho DD, Landau NR, Marx PA. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Amedee A, Miller K, Doranz BJ, Endres M, Sharron M, Samson M, Lu ZH, Clements JE, Murphey-Corb M, Peiper SC, Parmentier M, Broder CC, Doms RW. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci U S A. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F, Pohlmann S, Hamacher M, Means RE, Kraus T, Uberla K, Di MP. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMx174 cells for efficient entry. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HK, Unutmaz D, Kewalramani VN, Littman DR. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- Edinger AL, Hoffman TL, Sharron M, Lee B, O'Dowd B, Doms RW. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- Dehghani H, Puffer BA, Doms RW, Hirsch VM. Unique pattern of convergent envelope evolution in simian immunodeficiency virus-infected rapid progressor macaques: association with CD4-independent usage of CCR5. J Virol. 2003;77:6405–6418. doi: 10.1128/JVI.77.11.6405-6418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Hibbitts S, Simmons G, McKnight A, zevedo-Pereira JM, Moniz-Pereira J, Clapham PR. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer BA, Altamura LA, Pierson TC, Doms RW. Determinants within gp120 and gp41 contribute to CD4 independence of SIV Envs. Virology. 2004;327:16–25. doi: 10.1016/j.virol.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Pohlmann S, Davis C, Meister S, Leslie GJ, Otto C, Reeves JD, Puffer BA, Papkalla A, Krumbiegel M, Marzi A, Lorenz S, Munch J, Doms RW, Kirchhoff F. Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors. J Virol. 2004;78:3223–3232. doi: 10.1128/JVI.78.7.3223-3232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci U S A. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche CC, Hoffman TL, Romano J, Haggarty BS, Edwards TG, Matthews TJ, Doms RW, Hoxie JA. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/JVI.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Rosenzweig M, Desrosiers RC. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/JVI.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustchina E, Hummer G, Bewley CA, Clore GM. Differential inhibition of HIV-1 and SIV envelope-mediated cell fusion by C34 peptides derived from the C-terminal heptad repeat of gp41 from diverse strains of HIV-1, HIV-2, and SIV. J Med Chem. 2005;48:3036–3044. doi: 10.1021/jm049026h. [DOI] [PubMed] [Google Scholar]
- Malashkevich VN, Chan DC, Chutkowski CT, Kim PS. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci U S A. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie JA, Clapham PR. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock GJ, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J Biol Chem. 2003;278:3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- Zaitseva M, Peden K, Golding H. HIV coreceptors: role of structure, posttranslational modifications, and internalization in viral-cell fusion and as targets for entry inhibitors. Biochim Biophys Acta. 2003;1614:51–61. doi: 10.1016/S0005-2736(03)00162-7. [DOI] [PubMed] [Google Scholar]
- Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal. 2004;16:1201–1210. doi: 10.1016/j.cellsig.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Wang J, Babcock GJ, Choe H, Farzan M, Sodroski J, Gabuzda D. N-linked glycosylation in the CXCR4 N-terminus inhibits binding to HIV-1 envelope glycoproteins. Virology. 2004;324:140–150. doi: 10.1016/j.virol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Sloane AJ, Raso V, Dimitrov DS, Xiao X, Deo S, Muljadi N, Restuccia D, Turville S, Kearney C, Broder CC, Zoellner H, Cunningham AL, Bendall L, Lynch GW. Marked structural and functional heterogeneity in CXCR4: separation of HIV-1 and SDF-1alpha responses. Immunol Cell Biol. 2005;83:129–143. doi: 10.1111/j.1440-1711.2004.01304.x. [DOI] [PubMed] [Google Scholar]
- Simpson LS, Zhu JZ, Widlanski TS, Stone MJ. Regulation of chemokine recognition by site-specific tyrosine sulfation of receptor peptides. Chem Biol. 2009;16:153–161. doi: 10.1016/j.chembiol.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/S0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- Lam SN, Acharya P, Wyatt R, Kwong PD, Bewley CA. Tyrosine-sulfate isosteres of CCR5 N-terminus as tools for studying HIV-1 entry. Bioorg Med Chem. 2008;16:10113–10120. doi: 10.1016/j.bmc.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Vasilieva N, Schnitzler CE, Chung S, Robinson J, Gerard NP, Gerard C, Choe H, Sodroski J. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry2. J Biol Chem. 2000;275:33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- Cormier EG, Persuh M, Thompson DA, Lin SW, Sakmar TP, Olson WC, Dragic T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc Natl Acad Sci U S A. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry 3. Cell. 1999;96:667–676. doi: 10.1016/S0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- Bannert N, Craig S, Farzan M, Sogah D, Santo NV, Choe H, Sodroski J. Sialylated O-glycans and sulfated tyrosines in the NH2-terminal domain of CC chemokine receptor 5 contribute to high affinity binding of chemokines. J Exp Med. 2001;194:1661–1673. doi: 10.1084/jem.194.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan S, Petrovic A, Locati M, Kim YO, Weissman D, Murphy PM. A membrane-proximal basic domain and cysteine cluster in the C-terminal tail of CCR5 constitute a bipartite motif critical for cell surface expression. J Biol Chem. 2001;276:40133–40145. doi: 10.1074/jbc.M105722200. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Wittamer V, Vanderwinden JM, Boom A, Renneboog B, Lee B, Le PE, El AL, Govaerts C, Vassart G, Doms RW, Parmentier M. Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways. J Biol Chem. 2001;276:23795–23804. doi: 10.1074/jbc.M100583200. [DOI] [PubMed] [Google Scholar]
- Percherancier Y, Planchenault T, Valenzuela-Fernandez A, Virelizier JL, renzana-Seisdedos F, Bachelerie F. Palmitoylation-dependent control of degradation, life span, and membrane expression of the CCR5 receptor. J Biol Chem. 2001;276:31936–31944. doi: 10.1074/jbc.M104013200. [DOI] [PubMed] [Google Scholar]
- Kraft K, Olbrich H, Majoul I, Mack M, Proudfoot A, Oppermann M. Characterization of sequence determinants within the carboxyl-terminal domain of chemokine receptor CCR5 that regulate signaling and receptor internalization. J Biol Chem. 2001;276:34408–34418. doi: 10.1074/jbc.M102782200. [DOI] [PubMed] [Google Scholar]
- Steffens CM, Hope TJ. Mobility of the human immunodeficiency virus (HIV) receptor CD4 and coreceptor CCR5 in living cells: implications for HIV fusion and entry events. J Virol. 2004;78:9573–9578. doi: 10.1128/JVI.78.17.9573-9578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popik W, Alce TM, Au WC. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J Virol. 2002;76:4709–4722. doi: 10.1128/JVI.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Long D, Doranz BJ, Rucker J, Jirik FR, Doms RW. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot DJ, Chen H, Dimitrov DS, Broder CC. N-linked glycosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J Virol. 2000;74:4404–4413. doi: 10.1128/JVI.74.9.4404-4413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736–23744. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- Thordsen I, Polzer S, Schreiber M. Infection of cells expressing CXCR4 mutants lacking N-glycosylation at the N-terminal extracellular domain is enhanced for R5X4-dualtropic human immunodeficiency virus type-1. BMC Infect Dis. 2002;2:31. doi: 10.1186/1471-2334-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D, Rabin RL, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber JM, Fauci AS. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- Davis CB, Dikic I, Unutmaz D, Hill CM, Arthos J, Siani MA, Thompson DA, Schlessinger J, Littman DR. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768:952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yoder A. Chemokine coreceptor signaling in HIV-1 infection and pathogenesis. PLoS Pathog. 2009;5:e1000520. doi: 10.1371/journal.ppat.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotsios Y, Whittaker GC, Westwick J, Ward SG. The CXC chemokine stromal cell-derived factor activates a Gi-coupled phosphoinositide 3-kinase in T lymphocytes. J Immunol. 1999;163:5954–5963. [PubMed] [Google Scholar]
- Vicente-Manzanares M, Rey M, Jones DR, Sancho D, Mellado M, Rodriguez-Frade JM, del Pozo MA, Yanez Mo M, de Ana AM, Martinez A, Merida I, Sanchez-Madrid F. Involvement of phosphatidylinositol 3-kinase in stromal cell-derived factor-1 alpha-induced lymphocyte polarization and chemotaxis. J Immunol. 1999;163:4001–4012. [PubMed] [Google Scholar]
- Vicente-Manzanares M, Cabrero JR, Rey M, Perez-Martinez M, Ursa A, Itoh K, Sanchez-Madrid F. A role for the Rho-p160 Rho coiled-coil kinase axis in the chemokine stromal cell-derived factor-1alpha-induced lymphocyte actomyosin and microtubular organization and chemotaxis. J Immunol. 2002;168:400–410. doi: 10.4049/jimmunol.168.1.400. [DOI] [PubMed] [Google Scholar]
- Ganju RK, Brubaker SA, Meyer J, Dutt P, Yang Y, Qin S, Newman W, Groopman JE. The alpha-chemokine, stromal cell-derived factor-1alpha, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J Biol Chem. 1998;273:23169–23175. doi: 10.1074/jbc.273.36.23169. [DOI] [PubMed] [Google Scholar]
- Porcile C, Bajetto A, Barbieri F, Barbero S, Bonavia R, Biglieri M, Pirani P, Florio T, Schettini G. Stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12) stimulates ovarian cancer cell growth through the EGF receptor transactivation. Exp Cell Res. 2005;308:241–253. doi: 10.1016/j.yexcr.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Barbero S, Bonavia R, Bajetto A, Porcile C, Pirani P, Ravetti JL, Zona GL, Spaziante R, Florio T, Schettini G. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res. 2003;63:1969–1974. [PubMed] [Google Scholar]
- Francois F, Klotman ME. Phosphatidylinositol 3-kinase regulates human immunodeficiency virus type 1 replication following viral entry in primary CD4+ T lymphocytes and macrophages. J Virol. 2003;77:2539–2549. doi: 10.1128/JVI.77.4.2539-2549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanian K, Harriague J, Decrion C, Lagane B, Shorte S, Baleux F, Virelizier JL, renzana-Seisdedos F, Chakrabarti LA. CXCR4-tropic HIV-1 envelope glycoprotein functions as a viral chemokine in unstimulated primary CD4+ T lymphocytes. J Immunol. 2004;173:7150–7160. doi: 10.4049/jimmunol.173.12.7150. [DOI] [PubMed] [Google Scholar]
- Cicala C, Arthos J, Censoplano N, Cruz C, Chung E, Martinelli E, Lempicki RA, Natarajan V, VanRyk D, Daucher M, Fauci AS. HIV-1 gp120 induces NFAT nuclear translocation in resting CD4+ T-cells. Virology. 2006;345:105–114. doi: 10.1016/j.virol.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Yoder A, Yu D, Dong L, Iyer SR, Xu X, Kelly J, Liu J, Wang W, Vorster PJ, Agulto L, Stephany DA, Cooper JN, Marsh JW, Wu Y. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, VanDorsten K, Vojtech L, Barnett SW, Stamatatos L. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J Virol. 2003;77:2310–2320. doi: 10.1128/JVI.77.4.2310-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000;74:4746–4754. doi: 10.1128/JVI.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000;74:5716–5725. doi: 10.1128/JVI.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Broder CC, Long D, Lee SA, Peterson J, Chakrabarti S, Doms RW, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure10. J Virol. 2000;74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1 3. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, Ketas T, Sanders RW, Maddon PJ, Olson WC, Moore JP. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79:8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajah S, Puffer BA, Lee FH, Zhu P, Li Y, Wyatt R, Roux KH, Doms RW, Burton DR. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res Hum Retroviruses. 2008;24:301–314. doi: 10.1089/aid.2007.0158. [DOI] [PubMed] [Google Scholar]
- Chen B, Cheng Y, Calder L, Harrison SC, Reinherz EL, Skehel JJ, Wiley DC. A chimeric protein of simian immunodeficiency virus envelope glycoprotein gp140 and Escherichia coli aspartate transcarbamoylase. J Virol. 2004;78:4508–4516. doi: 10.1128/JVI.78.9.4508-4516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SX, Idiart RJ, Mariano EB, Chen H, Jiang P, Xu L, Ostrow KM, Wrin T, Phung P, Binley JM, Petropoulos CJ, Ballantyne JA, Whalen RG. Effect of trimerization motifs on quaternary structure, antigenicity, and immunogenicity of a noncleavable HIV-1 gp140 envelope glycoprotein. Virology. 2009;395:33–44. doi: 10.1016/j.virol.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grovit-Ferbas K, Hsu JF, Ferbas J, Gudeman V, Chen IS. Enhanced binding of antibodies to neutralization epitopes following thermal and chemical inactivation of human immunodeficiency virus type 1 3. J Virol. 2000;74:5802–5809. doi: 10.1128/JVI.74.13.5802-5809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner C, Mirzabekov T, Sodroski J, Wyatt R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins 1. J Virol. 2002;76:3511–3521. doi: 10.1128/JVI.76.7.3511-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race E, Frezza P, Stephens DM, Davis D, Polyanskaya N, Cranage M, Oxford JS. An experimental chemically inactivated HIV-1 vaccine induces antibodies that neutralize homologous and heterologous viruses 3. Vaccine. 1995;13:54–60. doi: 10.1016/0264-410X(95)80011-2. [DOI] [PubMed] [Google Scholar]
- Buonaguro L, Buonaguro FM, Tornesello ML, Mantas D, Beth-Giraldo E, Wagner R, Michelson S, Prevost M-C, Wolf H, Giraldo G. High efficient production of Pr55gag Virus-like Particles expressing multiple HIV-1 epitopes, including a gp120 protein derived from an Ugandan HIV-1 isolate of subtype A. Antiviral Research. 2001;49:35–47. doi: 10.1016/S0166-3542(00)00136-4. [DOI] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, de Vries RP, Wiley C, Zharkikh I, Schulke N, Roux KH, Montefiori DC, Burton DR, Binley JM. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366:245–262. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BZ, Liu W, Kang SM, Alam M, Huang C, Ye L, Sun Y, Li Y, Kothe DL, Pushko P, Dokland T, Haynes BF, Smith G, Hahn BH, Compans RW. Incorporation of high levels of chimeric human immunodeficiency virus envelope glycoproteins into virus-like particles. J Virol. 2007;81:10869–10878. doi: 10.1128/JVI.00542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBOJ. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanohara A, Imamura T, Araki M, Sugawara K, Ohtomo N, Matsubara K. Expression of hepatitis B virus core antigen gene in Saccharomyces cerevisiae: synthesis of two polypeptides translated from different initiation codons. J Virol. 1986;59:176–180. doi: 10.1128/jvi.59.1.176-180.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus- like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S, Niikura M, Li TC, Takeda N, Kusagawa S, Takebe Y, Miyamura T, Yasutomi Y. DNA vaccine-encapsulated virus-like particles derived from an orally transmissible virus stimulate mucosal and systemic immune responses by oral administration. Gene Ther. 2004;11:628–635. doi: 10.1038/sj.gt.3302193. [DOI] [PubMed] [Google Scholar]
- Malboeuf CM, Simon DA, Lee YE, Lankes HA, Dewhurst S, Frelinger JG, Rose RC. Human papillomavirus-like particles mediate functional delivery of plasmid DNA to antigen presenting cells in vivo. Vaccine. 2007;25:3270–3276. doi: 10.1016/j.vaccine.2007.01.067. [DOI] [PubMed] [Google Scholar]
- Touze A, Coursaget P. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res. 1998;26:1317–1323. doi: 10.1093/nar/26.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YF, Zhang YQ, Xu XM, Song GX. Papillomavirus virus-like particles as vehicles for the delivery of epitopes or genes. Arch Virol. 2006;151:2133–2148. doi: 10.1007/s00705-006-0798-8. [DOI] [PubMed] [Google Scholar]
- Buonaguro L, Tornesello ML, Buonaguro FM. Virus-Like Particles as Particulate Vaccines. Curr HIV Res. 2010. [DOI] [PubMed]
- Fernandez-San MA, Ortigosa SM, Hervas-Stubbs S, Corral-Martinez P, Segui-Simarro JM, Gaetan J, Coursaget P, Veramendi J. Human papillomavirus L1 protein expressed in tobacco chloroplasts self-assembles into virus-like particles that are highly immunogenic. Plant Biotechnol J. 2008;6:427–441. doi: 10.1111/j.1467-7652.2008.00338.x. [DOI] [PubMed] [Google Scholar]
- Lopez de Turiso JA, Cortes E, Martinez C, Ruiz de YR, Simarro I, Vela C, Casal I. Recombinant vaccine for canine parvovirus in dogs. J Virol. 1992;66:2748–2753. doi: 10.1128/jvi.66.5.2748-2753.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam S, Snezhkov E, Bishop DH. Formation of poliovirus-like particles by recombinant baculoviruses expressing the individual VP0, VP3, and VP1 proteins by comparison to particles derived from the expressed poliovirus polyprotein. Virology. 1993;192:512–524. doi: 10.1006/viro.1993.1067. [DOI] [PubMed] [Google Scholar]
- French TJ, Roy P. Synthesis of bluetongue virus (BTV) corelike particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J Virol. 1990;64:1530–1536. doi: 10.1128/jvi.64.4.1530-1536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovska TM, Cielens I, Dreilinna D, Dislers A, Baumanis V, Ose V, Pumpens P. Recombinant RNA phage Q beta capsid particles synthesized and self-assembled in Escherichia coli. Gene. 1993;137:133–137. doi: 10.1016/0378-1119(93)90261-Z. [DOI] [PubMed] [Google Scholar]
- Peabody DS, Manifold-Wheeler B, Medford A, Jordan SK, do Carmo CJ, Chackerian B. Immunogenic display of diverse peptides on virus-like particles of RNA phage MS2. J Mol Biol. 2008;380:252–263. doi: 10.1016/j.jmb.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J, Overbeek GP, van den Worm SH, Ackermann HW, van DJ. Nucleotide sequence of a ssRNA phage from Acinetobacter: kinship to coliphages. J Gen Virol. 2002;83:1523–1533. doi: 10.1099/0022-1317-83-6-1523. [DOI] [PubMed] [Google Scholar]
- Yamshchikov GV, Ritter GD, Vey M, Compans RW. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology. 1995;214:50–8. doi: 10.1006/viro.1995.9955. [DOI] [PubMed] [Google Scholar]
- Baumert TF, Ito S, Wong DT, Liang TJ. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol. 1998;72:3827–36. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleer WJ, Buynak EB, Maigetter RZ, Wampler DE, Miller WJ, Hilleman MR. Human hepatitis B vaccine from recombinant yeast. Nature. 1984;307:178–180. doi: 10.1038/307178a0. [DOI] [PubMed] [Google Scholar]
- Kang SM, Song JM, Quan FS, Compans RW. Influenza vaccines based on virus-like particles. Virus Res. 2009;143:140–146. doi: 10.1016/j.virusres.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Deml L, Flieβbach H, Wanner G, Wolf H. Assembly and extracellular release of chimeric HIV-1 Pr55gag retrovirus-like particles. Virology. 1994;200:162–175. doi: 10.1006/viro.1994.1175. [DOI] [PubMed] [Google Scholar]
- Deml L, Kratochwil G, Osterrieder N, Knuchel R, Wolf H, Wagner R. Increased incorporation of chimeric human immunodeficiency virus type 1 gp120 proteins into Pr55gag virus-like particles by an Epstein-Barr virus gp220/350-derived transmembrane domain. Virology. 1997;235:10–25. doi: 10.1006/viro.1997.8669. [DOI] [PubMed] [Google Scholar]
- Tobin GJ, Li GH, Fong SE, Nagashima K, Gonda MA. Chimeric HIV-1 virus-like particles containing gp120 epitopes as a result of a ribosomal frameshift elicit gag- and SU-specific murine cytotoxic T-lymphocyte activities. Virology. 1997;236:307–315. doi: 10.1006/viro.1997.8745. [DOI] [PubMed] [Google Scholar]
- Griffiths JC, Harris SJ, Layton GT, Berrie EL, French TJ, Burns NR, Adams SE, Kingsman AJ. Hybrid human immunodeficiency virus Gag particles as an antigen carrier system: Induction of cytotoxic T-cell and humoral responses by a Gag:V3 fusion. J Virol. 1993;67:3191–3198. doi: 10.1128/jvi.67.6.3191-3198.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale CJ, Liu XS, De Rose R, Purcell DF, Anderson J, Xu Y, Leggatt GR, Frazer IH, Kent SJ. Chimeric human papilloma virus-simian/human immunodeficiency virus virus-like-particle vaccines: immunogenicity and protective efficacy in macaques. Virology. 2002;301:176–87. doi: 10.1006/viro.2002.1589. [DOI] [PubMed] [Google Scholar]
- Weber J, Cheinsong-Popov R, Callow D, Adams S, Patou G, Hodgkin K, Martin S, Gotch F, Kingsman A. Immunogenicity of the yeast recombinant p17/p24:Ty virus-like particles (p24-VLP) in healthy volunteers. Vaccine. 1995;13:831–834. doi: 10.1016/0264-410X(94)00061-Q. [DOI] [PubMed] [Google Scholar]
- Schmitz N, Dietmeier K, Bauer M, Maudrich M, Utzinger S, Muntwiler S, Saudan P, Bachmann MF. Displaying Fel d1 on virus-like particles prevents reactogenicity despite greatly enhanced immunogenicity: a novel therapy for cat allergy. J Exp Med. 2009;206:1941–1955. doi: 10.1084/jem.20090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey RJ, Tanzer FL, Meyers A, Pillay S, Lynch A, Shephard E, Williamson AL, Rybicki EP. Chimaeric HIV-1 subtype C Gag molecules with large in-frame C-terminal polypeptide fusions form virus-like particles. Virus Res. 2008;133:259–268. doi: 10.1016/j.virusres.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Buonaguro L, Racioppi L, Tornesello ML, Arra C, Visciano ML, Biryahwaho B, Sempala SDK, Giraldo G, Buonaguro FM. Induction of neutralizing antibodies and CTLs in Balb/c mice immunized with Virus-like Particles presenting a gp120 molecule from a HIV-1 isolate of clade A (HIV-VLPAs). Antiviral Research. 2002;54:189–201. doi: 10.1016/S0166-3542(02)00004-9. [DOI] [PubMed] [Google Scholar]
- Buonaguro L, Visciano ML, Tornesello ML, Tagliamonte M, Biryahwaho B, Buonaguro FM. Induction of systemic and mucosal cross-clade neutralizing antibodies in BALB/c mice immunized with human immunodeficiency virus type 1 clade A virus-like particles administered by different routes of inoculation. J Virol. 2005;79:7059–7067. doi: 10.1128/JVI.79.11.7059-7067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L, Devito C, Tornesello ML, Schroder U, Wahren B, Hinkula J, Buonaguro FM. DNA-VLP prime-boost intra-nasal immunization induces cellular and humoral anti-HIV-1 systemic and mucosal immunity with cross-clade neutralizing activity. Vaccine. 2007;25:5968–5977. doi: 10.1016/j.vaccine.2007.05.052. [DOI] [PubMed] [Google Scholar]
- Triyatni M, Saunier B, Maruvada P, Davis AR, Ulianich L, Heller T, Patel A, Kohn LD, Liang TJ. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J Virol. 2002;76:9335–9344. doi: 10.1128/JVI.76.18.9335-9344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L, Tornesello ML, Tagliamonte M, Gallo RC, Wang LX, Kamin-Lewis R, Abdelwahab S, Lewis GK, Buonaguro FM. Baculovirus-derived human immunodeficiency virus type 1 virus-like particles activate dendritic cells and induce ex vivo T-cell responses. J Virol. 2006;80:9134–9143. doi: 10.1128/JVI.00050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


