Abstract
One of the most exciting developments in signal transduction research has been the proliferation of studies in which a biological discovery was initiated by computational modeling. Here we review the major efforts that enable such studies. First, we describe the experimental technologies that are generally used to identify the molecular components and interactions in, and dynamic behavior exhibited by, a network of interest. Next, we review the mathematical approaches that are used to model signaling network behavior. Finally, we focus on three specific instances of “model-driven discovery”: cases in which computational modeling of a signaling network has led to new insights which have been verified experimentally.
Signal transduction networks are the bridge between the extraordinarily complex extracellular environment and a carefully orchestrated cellular response. These networks are largely composed of proteins which can interact, move to specific cellular locations, or be modified or degraded. The integration of these events often leads to the activation or inactivation of transcription factors, which then induce or repress the expression of thousands of genes.
Because of this critical role in translating environmental cues to cellular behaviors, malfunctioning signaling networks can lead to a variety of pathologies. One example is cancer, in which many of the key genes found to be involved in cancer onset and development are components of signaling pathways [1, 2]. A detailed understanding of the cellular signaling networks underlying such diseases would likely be extremely useful in developing new treatments.
However, the complexity of signaling networks is such that their integrated functions cannot be determined without computational simulation. In recent years, mathematical modeling of signal transduction has led to some exciting new findings and biological discoveries. Here, we review the work that has enabled computational modeling of mammalian signaling networks, as well as the demonstrated value of such modeling. We begin by reviewing the experimental techniques commonly associated with model-building efforts, in terms of mapping network interactions as well as determining the dynamic network response to perturbation. We then discuss modeling strategies, and finally focus on three cases that dramatically illustrate the power of models to discover new biology.
Keywords: Signaling pathways, Mathematical models, Intra-cellular signaling, Regression models, Transcription factor networks
Mapping Network Interactions
Experimental interrogation and realistic mathematical modeling of a biological signaling circuit require at least partial knowledge of the elements of the system, and how these elements interact. Although the term “interactome” is generally invoked in the context of protein-protein interactions, the complete interactome describes the set of physical interactions between all biological molecules in vivo. These interactions are the basis for all biological phenomena.
Focused experimental studies have traditionally expanded the known interactome through sequential discovery and characterization of genes and proteins. Most model builders begin by obtaining this data from the published literature, in order to compile a coherent set of signaling components and interactions. Increasing the efficiency of this process would relieve a significant bottleneck in biomedical research. This can be done by either developing high throughput technologies that can identify network components and interactions en masse, or by facilitating the data mining process so that network reconstruction requires minimal human curation.
The interactome can be divided into three partially overlapping subsets: binary interactions, co-complex interactions, and functional interactions. The most widely used high throughput method for mapping binary protein interactions is yeast two hybrid (Y2H) screening [3-5]. In Y2H, interaction between bait and prey proteins reconstitutes an active transcription factor that drives expression of a reporter. Other approaches for detecting binary molecular interactions range from those that can be performed in mammalian cells [6-8] to a versatile in vitro method that uses a microfluidic chip [9-11]. Protein-DNA interactions can also be characterized with ChIP-chip [12] and ChIP-seq [13], which can detect binary interactions that may exist within a multi-molecular complex.
Affinity purification-based mass spectrometry (AP-MS) has become the standard technique for probing the space of protein co-complex interactions. Although more technically demanding than Y2H, AP-MS has the ability to track the dynamic composition of macromolecular complexes in a near physiological setting [14]. Complexes containing tagged bait proteins are isolated in the AP step, then the proteins in the complexes are identified and quantified by MS [14]. Most large scale efforts have been directed toward S. cerevisiae [15, 16], but a limited number of studies have applied AP-MS to small parts of the human interactome [17, 18].
A functional interaction is the effect of one molecule on the activity of another molecule, regardless of the path of physical interactions from one molecule to the other. Functional interactions can be inferred by gene co-expression as measured by cDNA microarrays [19]. Alternatively, by perturbing cells with cDNA or RNAi libraries, functional screens can identify genetic perturbations that affect the activity of a given signaling pathway [20, 21].
Informatics-based approaches focus both on making manually curated networks easily accessible and reconstructing networks automatically. An example of focused manual curation is a recently compiled comprehensive map of the epidermal growth factor receptor signaling system [22]. Such maps, made available in a readily useable standard such as SBML [23] or CellML [24], could be extremely valuable to modelers. Large scale efforts to manually assimilate biological knowledge include KEGG (Kyoto Encyclopedia of Genes and Genomes) [25] and Ingenuity Pathway Analysis (Ingenuity Systems). Automated reconstruction of signaling networks is generally based on probabilistic methods that can integrate data from various sources [add Sachs] [26-28].
Up to this point, both high throughput experimental technology and automated data mining are underutilized in mammalian signal transduction modeling efforts. This may be due to the general modeling preference for smaller, well documented networks over larger networks with less characterization. Until recently, the data quality of Y2H and AP-MS was broadly questioned [5, 14, 29]. This may be due to the general modeling preference for smaller, well documented networks over larger networks with less characterization. However, a thorough comparative analysis by Yu et al. suggests that both Y2H and AP-MS provide high quality interactome maps [30]. A paradigm for future studies is the work by Bouwmeester et al., who combined RNAi perturbations with AP-MS to map the physical and functional interactions of the TNFα/NF-κB signaling pathway [17]. Finally, recent work indicates that literature-curated interaction databases may be subject to inspection biases and therefore less reliable that commonly thought [30, 31]. Such findings may lead to a greater use of high throughput interactome data and automated network reconstruction in generating signaling network models.
Quantitatively Measuring Signaling Dynamics
Static, topological information is only a first step towards a complete understanding of cellular signaling. To accurately describe signal transduction, computational models must account for the intricate spatiotemporal dynamics that shape cell decisions [32]. For example, most protein signals are transmitted by changes in abundance, localization, activity, interactions, or post-translational state [33]. Technologies for experimentally measuring signaling dynamics can be broadly classified as population- or single cell-based (Table 1). Ideally, intracellular activity should be measured in single cells, because population-based assays can mask heterogeneity in single cell behaviors (e.g., all-or-none or asynchronous responses). However, the variety of measurable signals and the throughput of population level assays are generally superior to single cell approaches [34].
Table 1.
Overview of experimental techniques for measuring dynamics of mammalian signal transduction networks.
| Method | Strengths | Limitations | Antibody-dependent | Single Cell | Ref. |
|---|---|---|---|---|---|
| Gel-based | Simple, established | Usually semi-quantitative | ✓ | [35, 36] | |
| Mass spectrometry | Multiplexing potential for peptide identification | Difficult analysis, limited dynamic data | [14, 37, 38] | ||
| Protein arrays | Targeted multiplexing of protein measurement | Antibody cross-reactivity | ✓ | [39, 40] | |
| Live cell imaging | Subcellular localization of proteins through time in the same cell | Protein functionality, low throughput | ✓ | [41, 42] | |
| Immuno-fluorescence microscopy | Subcellular localization of endogenous proteins | Fixed cells | ✓ | ✓ | [43] |
| Flow cytometry | Single cell multivariate data | Fixed cells | ✓ | ✓ | [44, 45] |
A range of dynamic cellular signals can be measured in low throughput using population level techniques based on separation by gel electrophoresis. An immunoblot (or western blot) measures protein abundance [35] and can be combined with subcellular fractionation to determine a protein's location, or with phospho-specific antibodies to detect protein phosphorylation. The electrophoretic mobility shift assay (EMSA, also called the gel shift assay) is a sensitive method to determine the DNA-binding ability of a protein in nuclear extract [36]. Gel-based in vitro kinase assays measure incorporation of radioactive 32P into a peptide fragment substrate to determine the activity of a particular kinase.
Using gel-based methods in conjunction with quantitative modeling has some limitations. Because the linear dynamic range of many gel quantification instruments is relatively small, it would be appropriate to generate standard curves to relate intensity to protein concentration. Unfortunately, this rarely occurs. Furthermore, the Bradford assay that is often used to normalize cell lysates is linear over a very narrow range of protein concentrations, but this can be improved by calculating the ratio of absorbances at two wavelengths rather than only measuring one [46]. Additionally, the time required to process cellular samples means that very short time points (e.g., 5 minutes or less) are likely to have more experimental error than longer time points.
Proteomic measurements can be scaled up using mass spectrometry or multiplexed immunoaffinity methods [47]. In addition to mapping the existence of phosphorylation sites, mass spectrometry allows one to quantify hundreds or thousands of peptides and their post-translational modifications [38, 48]. Although mass spectrometry can resolve many closely related peptides, analysis is time-consuming and dynamic experiments have been limited to a few time points [34].
Multiplexed protein microarrays use affinity reagents spotted onto solid supports similar to cDNA microarrays. These techniques are related to traditional ELISA, in which antigens or antibodies are fixed to a surface and then secondary antibodies are linked to enzymatic detection. Similar to this format, protein microarrays typically employ the sandwich format, in which separate antibodies are used for capture to a solid surface and for fluorescent detection. The specificity resulting from sandwich type assays are generally preferred for cell signaling studies [33]. A commercial suspension microbead assay also using the sandwich format shows great promise [49]. In this technique, fluorescently bar-coded beads coated with capture antibodies are quantified using flow cytometry. Comprehensive sets of antibodies for capture and detection are limiting and crosstalk can be an issue for all immunoaffinity methods. In addition, regulated protein complexing may interfere with affinity binding sites or lead to artificially higher detection signal [33]. Although the number of proteins which can be detected is not as high as mass spectrometry, protein microarray and microbeads can be customized to specifically probe relevant proteins in a signaling pathway.
Single cell measurements can be performed on living or fixed specimens. Live cell microscopy allows one to follow a cell and its progeny over timescales of seconds to days. When combined with genetically encoded fluorescent proteins, live cell imaging allows direct observation of signaling events [42]. Fluorescent proteins can be tied to the transcriptional activity of a promoter or fused to a protein of interest to measure abundance and subcellular localization [41, 50]. FRET-based sensors can be constructed from fluorescent proteins to track enzymatic activity or post-translational changes, but such sensors currently only exist for a small number of signaling pathways [51]. A technique developed by Bertrand et al. allows tracking of mRNA in living cells by expression of a fluorescent protein fused to the RNA-binding protein MS2 and addition of MS2-binding sites to the mRNA of interest [52, 53]. Bioluminescence produced by luciferase can also be used to infer gene expression in live single cells [54] and has the advantage of very low background [55]. Given the optical properties of today's fluorescent proteins, a basic fluorescence microscopy setup can distinguish up to four colors [56]. If one wishes to mimic and model the in vivo signaling dynamics as closely as possible, it is important that fusion proteins retain not only endogenous function but regulation of expression. This can be particularly difficult in mammalian systems, where regulatory elements in chromatin and mRNA are generally poorly defined [57]. Although typically considered low throughput, large scale efforts have been made to systematically tag and track the abundance and subcellular location of proteins in mammalian cells [58, 59]. Data extraction from the raw images often determines throughput, but image analysis tools are being developed to quantify images more rapidly [60].
Measuring intracellular signaling dynamics with immunofluorescence microscopy [43] or multi-color flow cytometry [45] requires cell fixation. This means that one can monitor the population dynamics on a single cell level, but cannot follow the same cell over time. Both techniques can avoid expression of fusion proteins, but rely heavily on antibodies specific for phosphorylated signaling proteins. While multi-color flow cytometry allows quantification of up to seventeen colors and two light scattering parameters, it is unable to detect localization [44]. Flow cytometry is also well-established for studying heterogeneous cell populations on the basis of cell surface marker expression. Irish et al. leveraged these strengths of flow cytometry to identify potentiated signaling pathways in subsets of cells from patients with acute myeloid leukemia [61].
In the modeling studies we surveyed for this review, immunoblots were still the dominant experimental method for determining network dynamics. However, live-cell imaging has appeared more often in recent years.
Mathematical Approaches to Network Modeling
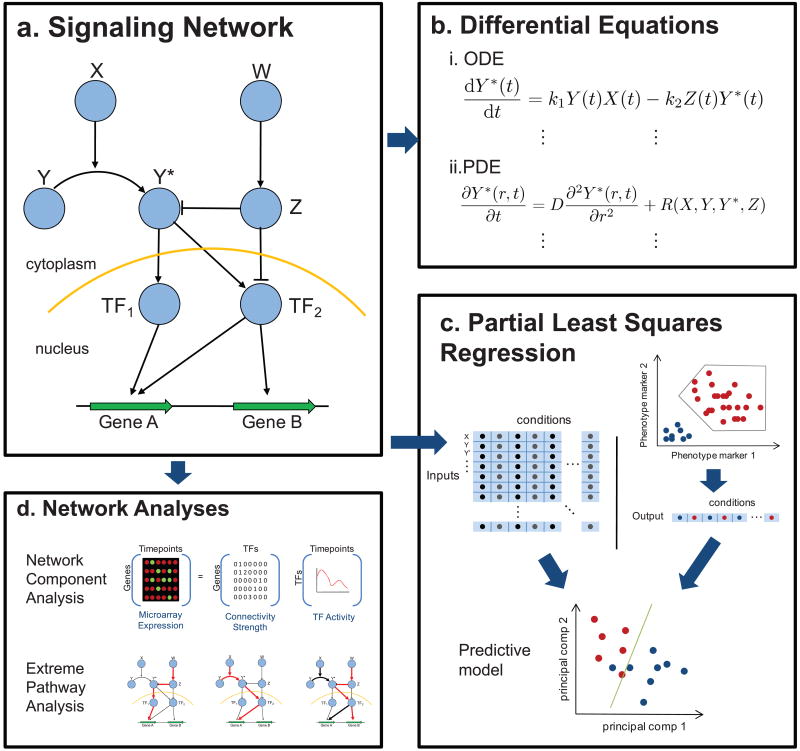
Given a signaling network which has been reconstructed and for which some dynamic information is available, a variety of mathematical approaches may be used to infer critical components or predict behaviors. Integrated modeling-experimental studies are enhanced when, some model parameters are fit using experimental data, and some model predictions are directly tested experimentally. For this reason, the scale and detail of the experimental method(s) and modeling approach should be compatible. Examples of several techniques for modeling mammalian signaling networks is given in Figure 1, and Table 2 highlights published models of mammalian signaling circuits, including the type of model and experimental methods.
Figure 1.
Mathematical approaches to model a signaling network.
a. A hypothetical signaling network, which transfers information from cytosolic enzymes to transcription factors to regulate gene expression.
b. The ordinary differential equation represents the phosphorylation and dephosphorylation of the Y protein. The partial differential equation models the effects of molecular diffusion and biochemical reactions with spatial dependence.
c. Measurements of the signaling network and phenotypic output (possibly measured through flow cytometry) are analyzed together to form a reduced space partial least squares regression model.
d. Network-based approaches. Network component analysis determines the linear weight of transcription factors on gene regulation. Extreme pathway analysis gives the minimal set of pathways that characterize the functional signaling in a network.
Table 2.
Models of various mammalian signaling pathways (not necessarily studied in mammalian cells), along with the major experimental technologies used when developing the model. (c) denotes a two-compartment model (cytoplasm and nucleus), (n) denotes a differential equation-based model incorporated noise.
| Signaling pathway | Model type | Experimental Verification | Ref. |
|---|---|---|---|
| β-Adrenergic receptor | ODE | - | [62] |
|
| |||
| β-Adrenergic receptor / MAPK | ODE, PDE | Live cell imaging, immunofluorescence microscopy, immunoblot | [63] |
|
| |||
| Cell Cycle | ODE | Immunoblot | [64] |
| ODE | - | [65] | |
| ODE | - | [66] | |
| Boolean | - | [67] | |
|
| |||
| Chemotaxis | ODE (n) | - | [68] |
| ODE, PDE | - | [69] | |
| PDE | - | [70] | |
|
| |||
| Circadian Clock | ODE (c, n) | Live cell imaging | [71, 72] |
| ODE (c), Gillespie | - | [73, 74] | |
|
| |||
| Cytokines / Apoptosis | PLSR | High throughput kinase assay, protein microarray, flow cytometry | [75-77] |
|
| |||
| Delta / Notch | ODE (c) | - | [78] |
| Piecewise affine hybrid | - | [79] | |
|
| |||
| EGFR / Ras | ODE, Gillespie | Immunoblot, ELISA | [80, 81] |
|
| |||
| EGFR / MAPK | ODE | - | [82] |
|
| |||
| Hedgehog | ODE, Gillespie, PDE | - | [83, 84] |
|
| |||
| JAK / STAT | ODE (c) | Immunoblot | [85] |
| ODE (c) | - | [86] | |
| Extreme pathways | - | [87] | |
| ODE (c) | Immunofluorescence microscopy, flow cytometry | [88] | |
|
| |||
| MAPK | ODE | Immunoblot, kinase assay | [89] |
|
| |||
| NF-κB | ODE (c) | EMSA, kinase assay [90, 91]; live cell imaging, immunofluorescence microscopy [50]; immunoblot | [50, 90, 91] |
| ODE (c) | EMSA, immunoblot | [92] | |
| ODE (c) | - | [93] | |
| ODE (c) | EMSA, immunoblot, kinase assay, mass spectrometry | [94] | |
|
| |||
| p53 | ODE | Immunoblot | [95] |
| ODE (c) | - | [96] | |
| ODE (n) | - | [97] | |
| ODE | Immunoblot | [98] | |
|
| |||
| Ras | ODE, Gillespie | Flow cytometry, immunoblot | [99] |
|
| |||
| TGF-β / Smad | ODE (c) | Live cell imaging, immunoblot | [100] |
|
| |||
| Wnt | ODE | Immunoblot | [101, 102] |
| PDE | Histology | [103] | |
| PDE | Histology | [104] | |
Modeling with Differential Equations
The most common modeling approach is to represent the signaling system as a set of ordinary differential equations (ODEs) using mass action kinetics, which can be integrated to determine the concentration of species over time [105].
Small sets of ODEs can either be solved exactly or by an approximate analytical solution. Using techniques such as phase space portraits, one can easily identify steady states and visualize how the dynamic behavior varies with the state of the system. Analytical methods are useful for studying the recurring modules and network motifs that constitute larger biochemical networks [106-109]. Investigation of a signaling network's positive and negative feedback loops can give insight into the network's behavior and identify important system properties such as multistability, excitability, and limit cycle oscillations [109-111]. Bifurcation analysis is helpful in understanding the transitions between dynamic behaviors that result from changes in model parameters.
As the size of the dynamical system increases, analytical and graphical approaches become increasingly difficult. Consequently, numerical integration is necessary to find the solutions of concentrations with time. In such cases sensitivity analysis is often used to determine which parameters have the greatest effect on the output of the system, and therefore require the most accurate experimental measurement [105]. Sensitivity analysis is often performed by varying one parameter at a time, while the others are held at their estimated values, which can be misleading in cases where parameters are not independent. Determining the output while simultaneously varying multiple parameters can give a wider, more integrated view of the network, but is computationally expensive for large pathways.
Many signaling networks have a significant spatial component to their behavior. For example, the activity of a transcription factor can be controlled by regulating its access to the nucleus. Spatial information can be incorporated into ODE-based models with compartmentalization (e.g., of the cytosol and nucleus). This assumes the contents within a compartment are well-mixed and requires specifying transport rates between compartments.
Although compartmental ODE modeling is often used as a simplification for PDEs [112], in some cases detailed spatial localization is important for cell signaling [32, 113]. Often spatial dependencies such as gradients and microdomains arise due to the geometry of the cell membrane and can be caused by opposing biochemical reactions that are spatially separated [63]. PDEs represent biochemical processes in space and time as reaction-diffusion equations that account for diffusion and biochemical reactions of signaling molecules. Diffusional constants in cellular environments are often unknown and therefore usually approximated [113]. The computational tools needed to solve PDEs are not widely used in biology but these tools are being developed and finding use in some modeling efforts [114]. PDE models will become increasingly valuable as live cell imaging and fluorescent reporters provide the spatial and functional measurements needed to build and validate these models.
Stochasticity can strongly influence behavior in systems with small numbers of molecules or in instances of multistability or symmetry breaking, such as when a cell must decide between two fates [115, 116]. Gene expression in mammalian cells, at least for some genes, is highly variable due to random bursts of transcription [117]. The growing realization of the importance of noise in signaling and the proliferation of single cell measurements suggests stochastic models will become more common, since they can potentially use and reproduce the variability of individual cell responses. The Gillespie algorithm and its derivatives simulate the random walk behavior of discrete molecules [99, 118]. Often a signaling pathway is modeled first with deterministic ODEs, then the same biochemical reactions are simulated with the Gillespie algorithm [74, 99]. The method is straightforward to implement but becomes computationally expensive as the number of molecules increases. Another way to incorporate stochasticity is to add a Gaussian noise term to a differential equation, resulting in a stochastic differential equation [72]. For spatial PDE models, the modeling platform Virtual Cell allows both deterministic and stochastic simulation [119].
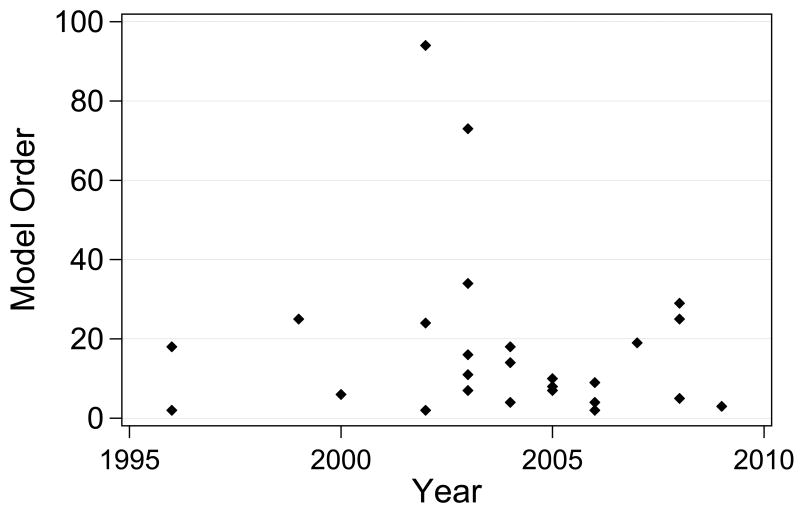
An advantage of ODE models is that they represent signal transduction circuits mechanistically, but they become impractical for extremely large networks. Figure 2 shows a timeline of the order of the differential equation-based models compiled in Table 2. None of these models have more than one hundred ODEs, and the median is ten. This is largely because mechanistic models require many parameters in the form of rate constants and initial conditions. Although some of these parameters have been directly measured experimentally, most are unknown or poorly constrained, especially in a mammalian setting. Consequently, many parameters must be estimated by making a first principles guess, by extrapolating based on homologous proteins, or by fitting the output of the model to experimental observations [120]. With too many degrees of freedom, even a model that is mechanistically unfaithful to reality can appear to fit experimental data. Additionally, when parameters of a model have been fit, it is imperative that later predictions of the model be independent of the data that was used in the fitting process.
Figure 2.
The order (number of independent variables) of differential equation-based models of mammalian signal transduction, plotted by publication year. In cases where one model's publication led to the creation of several derivative models, only the first model is included.
Large Scale Modeling Methods
To circumvent the challenges of large systems of differential equations, several methods with potential advantages in scale-up have been applied to signal transduction. These modeling approaches generally lack the detail of ODE-or PDE-based models. The trade-off between model scale and detail has been noted previously [121].
Constraint-based network analysis allows reconstruction of large systems of biochemical reactions [122]. The method has proven useful in analyzing genome-scale metabolic networks [123, 124]. This approach does not need kinetic parameters, but does require explicit enumeration of all reactions and chemical species in order to generate a stoichiometric matrix. The space of available steady state solutions is calculated subject to the constraints of the system, such as the availability of a carbon source or the maximum speed of an enzymatic reaction. One version, extreme pathway analysis, has been used to quantify the crosstalk and pathway redundancy of the JAK-STAT signaling in B cells [87]. Since calculation of the extreme pathways becomes unfeasible as the size of the network increases, Li et al. used flux balance analysis to model the Toll-like receptor network and to pinpoint eight potential drug targets [125].
Network component analysis is a method for inferring the activity of transcription factors – and by extension, some signaling pathways – from gene expression data [126]. This analysis utilizes a prior estimate of the regulatory network structure, which distinguishes it from more naïve microarray analysis methods. Network component analysis has been used to study glycerol kinase deficiency in mouse skeletal muscle [127].
Partial least squares regression (PLSR) analysis has been applied to understand complicated signaling networks that involve multiple inputs and outputs [75-77]. If the data sets (e.g., phosphorylation states, protein abundances, and kinase activities) are extensive enough, PLSR can be used even without a detailed mechanistic understanding of the underlying signaling network [2, 40, 128]. PLSR creates a linear model in which state measurements of the signaling network can be related to outputs such as cytokine secretion [75, 129]. PLSR reduces the high dimensionality of these data sets by generating principal components, which are linear combinations of the original variables that can be ranked by their ability to capture co-variation in the data. Often most of the variation in a high-dimensional data set can be represented in just a few principal components. With the PLSR formalism, it is possible to predict the outcome of previously untested experiments by measuring the global state of the signaling network [75].
Model-driven Discovery
The most exciting aspect of computational modeling of signal transduction networks is the prospect of using models to facilitate biological discovery. Model-driven discovery is appealing because computation is far less expensive and time-consuming than wet-lab experimentation. We want to further emphasize three signaling systems in which mathematical modeling has led to a deeper understanding of the biology, as verified by experiments.
p53
p53 is an intensely studied transcription factor due to its role in tumor suppression and DNA damage repair [130]. The core components of the p53 feedback loop involve p53 itself and its inhibitor protein Mdm2 [131]. Mdm2 enhances the degradation of p53, while phosphorylated p53 induces transcription of Mdm2, completing a negative feedback loop. The wider p53 signaling network also contains additional positive and negative feedback loops [131]. Given a sufficient time delay for p53-induced Mdm2 transcription, the core p53 circuit can give rise to oscillations. To model this behavior, Lev Bar-Or et al. created a set of ODEs to describe the system and added a hypothetical intermediate that directs Mdm2 transcription with kinetics governed by a Hill function [95]. They verified with immunoblots that the levels of Mdm2 and p53 oscillate in response to DNA damage-inducing irradiation. Although the model parameters were estimated rather than measured or fit to experimental data, Lev Bar-Or et al. were able to infer qualitative behaviors such as the broadening of the p53 response to weaker DNA damage signals. In addition, they saw a difference in p53-Mdm2 dynamics between 3T3 and MCF-7 cell lines and proposed a likely change in the model that could account for the differences. However, such a prediction is difficult to verify without quantitative measurements of the parameters and without the identification of the intermediate.
Extending this work, Lahav et al. observed p53-Mdm2 dynamics in single cells using fluorescent fusion proteins and time-lapse microscopy [132]. Importantly, the Mdm2 fusion protein was driven by the human Mdm2 promoter. Contrary to the damped oscillations observed in population level studies, oscillations in single cells were undamped. The strength of irradiation correlated with the fraction of responding cells, while the amplitude of oscillations in single cells was independent of DNA damage signal input. The response is considered digital, as increased DNA damage increases the number of pulses rather than their amplitude. To explain the digital response and sustained oscillations, Ciliberto et al. proposed a model incorporating positive feedback in the p53-Mdm2 loop [96], while Ma et al. added positive feedback to an upstream step in the DNA repair pathway [97].
With longer time-lapse movies, Geva-Zatorsky et al. observed p53-Mdm2 oscillations lasting for over three days [133]. The amplitude of these oscillations showed greater variability than the frequency. In order to model this behavior, the authors examined six possible mathematical models (including models similar to those described above) and eliminated those that could not produce sustained oscillations or that were very sensitive to parameter values. With an additional stochastic noise term only one of the remaining models could reproduce the observed variability in oscillations. This noise was limited to protein production and not other processes such as degradation.
To further explore the nature of the oscillatory dynamics, Batchelor and coworkers studied two kinases involved in p53 activation, ATM and Chk2 [98]. It was known that ATM phosphorylates Chk2 and then both ATM and Chk2 phosphorylate p53, lessening p53 inhibition by Mdm2. Using immunoblots and immunofluorescence, they found that Chk2 shows undamped oscillations similar to p53 and that these activation pulses depended on p53. Targeted RNAi and small molecule perturbations confirmed the dependence of p53 oscillations on ATM and Chk2. Upon inhibition of ATM, oscillations of p53 were not observed even if the first p53 pulse had been initiated. These results are contrary to a model in which oscillations are driven solely by the p53-Mdm2 loop. This led Batchelor et al. to propose a model that included an unknown inhibitor of ATM and Chk2 that is activated by p53. Negative feedback in this form could explain the pulses in ATM and Chk2. The model predicted that the unknown inhibitor would oscillate in a similar fashion to p53 and be expressed at low levels in resting cells. A known interactor with ATM/Chk2/p53 called Wip1 fit these criteria. RNAi against Wip1 abolished oscillations in ATM and Chk2 and an immunoblot showed oscillations in Wip1 lagging behind p53. Thus, ATM/Chk2/p53/Wip1 is involved in a second feedback loop that governs p53 oscillations.
NF-κB
The nuclear factor-κB (NF-κB) family of transcription factors regulates the expression of genes involved in various biological processes from immune system development and inflammation to cell proliferation [134]. Dysregulation of NF-κB is associated with chronic inflammatory disease and cancer progression, in addition to many other pathologies [135, 136]. The best studied NF-κB molecule is the heterodimer p65:p50, which is held inactive in the cytosol by three IκB proteins, IκBα, IκBβ, and IκBε. In the canonical pathway a stimulus leads to the activation of IκB kinase (IKK), which phosphorylates the IκBs, precipitating their rapid ubiquitination and degradation. Free NF-κB then translocates to the nucleus, where it binds DNA and induces expression of hundreds of target genes. For example, IκBα expression is strongly induced by NF-κB, creating a negative feedback loop. Considerable complexity is laced throughout these reactions, but this represents the relatively agreed upon core of the system [134].
Hoffmann et al. observed that the DNA-binding activity of NF-κB in response to tumor necrosis factor-α (TNFα) resembles damped oscillations, as measured by EMSA with murine embryonic fibroblasts (MEFs) [90]. To try to explain the observed dynamics and untangle the roles of the three IκBs, Hoffmann et al. modeled the core circuit response to TNFα using a system of coupled differential equations. More than half of the model parameters were constrained by data in the published literature. Experimental data from mouse cells expressing only one of the three IκB proteins further constrained the model. These studies suggested that IκBα is responsible for fast NF-κB activation, while IκBβ and IκBε help dampen oscillations of NF-κB. Interestingly, values for IκBβ and IκBε mRNA synthesis parameters from the double knockouts did not match those derived from the wild type behavior, pointing to genetic compensation that is not encapsulated in the model.
Model simulations predicted that NF-κB activation for long stimulations lasts for the duration of the stimulus, while short pulses induce a level of NF-κB activation that is relatively independent of pulse duration. This suggested that NF-κB could induce expression of some genes after very short exposure to TNFα (<15 minutes), while other genes could require longer stimulation times. Indeed, IP-10 mRNA can be detected after only 30 minutes of TNFα exposure, while induction of the gene RANTES requires more than two hours of exposure to TNFα in wild type MEFs. However, Hoffmann et al. found that in IκBα-/- MEFs, in which transient stimulation results in prolonged nuclear NF-κB, RANTES can be induced by TNFα stimulations lasting only 15 minutes [90]. Thus, the computational model gave insight into how the IκB proteins regulate the output of the signaling network by controlling the dynamics of NF-κB.
Nelson et al. studied the dynamics of the NF-κB network in live HeLa cells and SK-N-AS cells (human S-type neuroblastoma cells with constitutive NF-κB activity [137]) using ectopic expression of NF-κB and IκBα fluorescent fusion proteins [50]. In response to TNFα, single cells exhibited asynchronous oscillations in NF-κB nuclear localization. Cells co-expressing control EGFP showed more regular, higher frequency oscillations than cells co-expressing IκBα-EGFP under the constitutive CMV immediate early promoter. Transfection of cells with IκBα-EGFP driven by a NF-κB-responsive promoter confirmed that increasing the strength of negative feedback reduces the oscillation frequency of nuclear NF-κB, consistent with simulations of the model developed by Hoffmann et al. [90]. Although the original computational model considers all free nuclear NF-κB as active, Nelson et al. observed that constitutive nuclear localization effected by leptomycin B results in only transient luciferase reporter expression along with rapid NF-κB dephosphorylation [50]. Thus, persistent target gene expression seems to require oscillatory behavior of NF-κB localization and phosphorylation.
The NF-κB computational model continues to be expanded and refined, leading to new insights into the behavior of the system. Modeling the network upstream of IKK helped to reveal the role of A20 in mediating crosstalk between TNFα and other inflammatory stimuli [138]. Integrated experimental and computational studies of the interactions between the canonical and non-canonical NF-κB pathways suggested that altered IκB homeostasis could result in an inflammatory response to developmental stimuli [94].
Another expansion of the NF-κB model led to the elucidation of an extracellular component to the network. While NF-κB localization dynamics are oscillatory in response to TNFα, lipopolysaccharide (LPS) causes stable activation of NF-κB as measured by EMSA. LPS signaling through TLR4 goes through a MyD88-dependent pathway and a Trif-dependent pathway, each of which acting individually in Trif-/- or MyD88-/- MEFs leads to oscillatory dynamics [92]. When given the time courses of IKK kinase assays, the Hoffmann model correctly reproduces the observed NF-κB translocation dynamics in wild type MEFs stimulated with LPS or TNFα [91]. Covert et al. fit the dynamics of the two pathways in the knockout MEFs to predict the kinetics of IKK activity, which suggested that a 30 minute time delay caused the slower activation of the Trif-dependent pathway. Subsequent experiments confirmed that the Trif-dependent pathway requires expression and secretion of TNFα, apparently mediated by IRF3 [92]. Thus, while TNFα activates NF-κB once, the qualitatively different LPS response is achieved by an autocrine or paracrine loop that activates IKK twice.
Apoptosis signaling
Apoptosis, or programmed cell death, is regulated by a variety of extracellular signals and their downstream pathways [139]. Some of these signals can have opposite effects depending on cell type and signaling context [140]. For example, the TNFα pathway can lead to cell death through caspase activation, or induce pro-survival signals through NF-κB. There is also considerable crosstalk between extracellular inputs such as cytokines.
In such cases, PLSR can take into account the global state of signaling to determine the cellular response to combinatorial inputs. Janes et al. probed the signaling network involving EGF, TNFα, and insulin to build a predictive model of the apoptotic response in HT-29 cells [75]. Twelve apoptosis markers were measured under these stimulation conditions [141]. With this data, a PLSR model was trained to predict the apoptosis markers given a set of 19 signaling measurements over 13 time points [75]. To test the predictive ability of the model, the authors perturbed the network by applying either a blocking antibody to the TGFα receptor or an IL-1 receptor antagonist. They then repeated the signaling network measurements and used the PLSR classifier to predict the result. The model accurately predicted the apoptotic responses even though these perturbations interfered with autocrine signals not explicitly encoded in the model. Two of the principal component vectors from the PLSR model correlated well with pro-survival and pro-apoptosis signals. Thus, Janes et al. could represent the effects of multiple stimuli or network perturbations in terms of the projection of signaling data along pro-survival and pro-apoptosis axes. In a related study, Janes et al. (2006) used informative principal components to visualize the contribution of EGF, TNFα, and insulin to the activity of various signaling kinases and receptors [76]. The signaling map showed that some pathway specific components were activated to a similar extent by other ligands. This shared influence was shown to be a result of an ordered series of autocrine cytokine secretion.
Many signaling models concentrate on the response of a specific cell type and it is often unknown whether the model is valid in other cell types. To investigate this, Miller-Jensen et al. measured the dynamics of kinase activity for various signaling pathways and the apoptotic response in HT-29 cells treated with TNFα and adenovirus [77]. A PLSR model was trained to learn the relation between this input and output data. HT-29 and HeLa cells exhibit distinct signaling network and apoptotic responses after combined adenoviral and TNFα treatment. However, the computational model correctly predicted the apoptotic response in HeLa cells and MCF-10A cells given the kinase activity measurements for those respective cell types. The model also correctly predicted that specific inhibition of Akt by the PI(3)K inhibitor LY294002 would affect apoptosis in HT-29 cells but not in HeLa cells. However, the model underestimated the level of TNFα-induced apoptosis in HeLa cells when IKK activity was blocked by a neutralizing antibody. Upon closer examination, Miller-Jensen et al. noticed that early IKK activity in HeLa cells was significantly higher than in the HT-29 training data, which prevented the PLSR model from learning the extent of the relationship between early IKK activity and apoptosis. Further experiments confirmed that early IKK activity is anti-apoptotic in HeLa cells. Thus, even though the cells respond differently to the same extracellular stimuli, these epithelial cell types use common effectors, e.g., kinase substrates and transcription factors, to integrate multiple signals to produce a phenotypic output.
Conclusion
Our main goal in writing this review was to highlight the impact that integrated computational-experimental studies are having on our understanding of mammalian signal transduction. Accordingly, we described the current experimental and computational approaches to understanding mammalian signal transduction, and focused on how these approaches have been successfully used together to facilitate biological discovery.
In compiling the data for this paper, we made two interesting observations that relate to the future of this field. First, we noticed that models fell into two groups: refining and expansive. The refining models are related to pre-existing models, with new parameter values and relatively minor changes. The expansive modeling efforts move into new biological territory, a previously un-modeled signaling pathway. While proven models are key to an iterative discovery program [142], in the future it will be imperative that the number of expansive models increase. Deriving maximal benefit from expansive models will require rigorous experimental testing of model predictions. Fortunately, this is likely to occur as more young scientists are learning both biology and mathematics, and interaction between biologists and computational scientists is on the rise.
Our second observation was that most models are relatively small. To be sure, small models of signal transduction can make nontrivial predictions and provide meaningful insight. However, the incoming experimental data indicates that signaling pathways that are traditionally studied separately are highly interconnected. This can be seen in Table 2 as well, with the numerous models that focus on different parts of the network involving EGFR, β-adrenergic receptor, Ras, and MAPK. As a result, it is important that in the coming years, models incorporate more signals and downstream responses – that they become more integrative with respect to biological function. Recent work suggests this trend has already begun [143, 144]. This shift will require the development of new methods to assimilate and organize large data sets, and to model network behaviors at a larger scale and in more detail.
Finally, we highlighted three important examples, where the success of the experimental biology was a direct result of applying computational modeling to the system. Table 2 holds many more such examples. Taken together, these studies highlight the important role that systems biology is beginning to play in elucidating mammalian signaling networks. As modeling efforts in signal transduction become more expansive and integrative, we expect to see a dramatic rise in our understanding of the complex links between environmental cues and cellular responses, which will have a direct impact on our ability to treat pathology.
Acknowledgments
The authors would like to acknowledge a Bio-X Graduate Fellowship to J.J.H., a Stanford Graduate Fellowship to T.K.L., and a K99/R00 award to M.W.C. (CA125994-01A1).
Contributor Information
Jacob J Hughey, Department of Bioengineering, Stanford University.
Timothy K Lee, Department of Bioengineering, Stanford University.
Markus W Covert, Email: mcovert@stanford.edu, Department of Bioengineering, Stanford University.
References
- 1.Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4(3):167–74. doi: 10.1016/s1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- 2.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6(2):146–55. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 3.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 4.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88(21):9578–82. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol. 2006;17(4):387–93. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9(4):789–98. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 7.Remy I, Michnick SW. Mapping biochemical networks with protein-fragment complementation assays. Methods Mol Biol. 2004;261:411–26. doi: 10.1385/1-59259-762-9:411. [DOI] [PubMed] [Google Scholar]
- 8.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307(5715):1621–5. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 9.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315(5809):233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 10.Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26(9):1019–27. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerber D, Maerkl SJ, Quake SR. An in vitro microfluidic approach to generating protein-interaction networks. Nat Methods. 2009;6(1):71–4. doi: 10.1038/nmeth.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83(3):349–60. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 14.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8(8):645–54. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 15.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 16.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 17.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 18.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278(5338):680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 20.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002;21(19):5184–94. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441(7089):106–10. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 22.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005 0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd CM, Halstead MD, Nielsen PF. CellML: its future, present and past. Prog Biophys Mol Biol. 2004;85(2-3):433–50. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303(5659):799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Probabilistic model of the human protein-protein interaction network. Nat Biotechnol. 2005;23(8):951–9. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan BS, Shah NH, Flannick JA, Abeliuk E, Novak AF, Batzoglou S. Current progress in network research: toward reference networks for key model organisms. Brief Bioinform. 2007;8(5):318–32. doi: 10.1093/bib/bbm038. [DOI] [PubMed] [Google Scholar]
- 29.Fields S. High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 2005;272(21):5391–9. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–10. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, Vandenhaute J, Galli M, Yazaki J, Hill DE, Ecker JR, Roth FP, Vidal M. Literature-curated protein interaction datasets. Nat Methods. 2009;6(1):39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7(3):165–76. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32(Suppl):526–32. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 34.Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S. Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol. 2006;7(11):803–12. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849–61. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104(14):5860–5. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1(5):252–62. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 39.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2(2):RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5(4):310–20. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weijer CJ. Visualizing signals moving in cells. Science. 2003;300(5616):96–100. doi: 10.1126/science.1082830. [DOI] [PubMed] [Google Scholar]
- 42.Megason SG, Fraser SE. Imaging in systems biology. Cell. 2007;130(5):784–95. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4(8):648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 45.Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20(2):155–62. doi: 10.1038/nbt0202-155. [DOI] [PubMed] [Google Scholar]
- 46.Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem. 1996;236(2):302–8. doi: 10.1006/abio.1996.0171. [DOI] [PubMed] [Google Scholar]
- 47.Schmelzle K, White FM. Phosphoproteomic approaches to elucidate cellular signaling networks. Curr Opin Biotechnol. 2006;17(4):406–14. doi: 10.1016/j.copbio.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics. 2008;8(21):4433–43. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nolan JP, Mandy F. Multiplexed and microparticle-based analyses: quantitative tools for the large-scale analysis of biological systems. Cytometry A. 2006;69(5):318–25. doi: 10.1002/cyto.a.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704–8. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3(12):906–18. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 52.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–45. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 53.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13(2):161–7. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14(24):2289–95. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9(1):123–8. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 56.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2(12):905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 57.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, Makeev VJ, Mironov AA, Noble WS, Pavesi G, Pesole G, Regnier M, Simonis N, Sinha S, Thijs G, van Helden J, Vandenbogaert M, Weng Z, Workman C, Ye C, Zhu Z. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23(1):137–44. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 58.Garcia Osuna E, Hua J, Bateman NW, Zhao T, Berget PB, Murphy RF. Large-scale automated analysis of location patterns in randomly tagged 3T3 cells. Ann Biomed Eng. 2007;35(6):1081–7. doi: 10.1007/s10439-007-9254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322(5907):1511–6. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 60.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 62.Saucerman JJ, Brunton LL, Michailova AP, McCulloch AD. Modeling beta-adrenergic control of cardiac myocyte contractility in silico. J Biol Chem. 2003;278(48):47997–8003. doi: 10.1074/jbc.M308362200. [DOI] [PubMed] [Google Scholar]
- 63.Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, Taubenfeld SM, Alberini CM, Schaff JC, Blitzer RD, Moraru II, Iyengar R. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133(4):666–80. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pomerening JR, Sontag ED, Ferrell JE., Jr Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5(4):346–51. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 65.Qu Z, Weiss JN, MacLellan WR. Regulation of the mammalian cell cycle: a model of the G1-to-S transition. Am J Physiol Cell Physiol. 2003;284(2):C349–64. doi: 10.1152/ajpcell.00066.2002. [DOI] [PubMed] [Google Scholar]
- 66.Novak B, Tyson JJ. A model for restriction point control of the mammalian cell cycle. J Theor Biol. 2004;230(4):563–79. doi: 10.1016/j.jtbi.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 67.Faure A, Naldi A, Chaouiya C, Thieffry D. Dynamical analysis of a generic Boolean model for the control of the mammalian cell cycle. Bioinformatics. 2006;22(14):e124–31. doi: 10.1093/bioinformatics/btl210. [DOI] [PubMed] [Google Scholar]
- 68.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112(Pt 17):2867–74. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 69.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82(1 Pt 1):50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: a balanced inactivation model. Proc Natl Acad Sci U S A. 2006;103(26):9761–6. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leloup JC, Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc Natl Acad Sci U S A. 2003;100(12):7051–6. doi: 10.1073/pnas.1132112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129(3):605–16. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forger DB, Peskin CS. A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci U S A. 2003;100(25):14806–11. doi: 10.1073/pnas.2036281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forger DB, Peskin CS. Stochastic simulation of the mammalian circadian clock. Proc Natl Acad Sci U S A. 2005;102(2):321–4. doi: 10.1073/pnas.0408465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310(5754):1646–53. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 76.Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124(6):1225–39. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 77.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448(7153):604–8. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 78.Collier JR, Monk NA, Maini PK, Lewis JH. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J Theor Biol. 1996;183(4):429–46. doi: 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- 79.Ghosh R, Tomlin C. Lecture Notes in Computer Science. Springer; Berlin / Heidelberg: 2001. Lateral Inhibition through Delta-Notch Signaling: A Piecewise Affine Hybrid Model; pp. 232–246. [Google Scholar]
- 80.Kholodenko BN, Demin OV, Moehren G, Hoek JB. Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem. 1999;274(42):30169–81. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]
- 81.Resat H, Ewald JA, Dixon DA, Wiley HS. An integrated model of epidermal growth factor receptor trafficking and signal transduction. Biophys J. 2003;85(2):730–43. doi: 10.1016/s0006-3495(03)74516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20(4):370–5. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 83.Lai K, Robertson MJ, Schaffer DV. The sonic hedgehog signaling system as a bistable genetic switch. Biophys J. 2004;86(5):2748–57. doi: 10.1016/S0006-3495(04)74328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saha K, Schaffer DV. Signal dynamics in Sonic hedgehog tissue patterning. Development. 2006;133(5):889–900. doi: 10.1242/dev.02254. [DOI] [PubMed] [Google Scholar]
- 85.Swameye I, Muller TG, Timmer J, Sandra O, Klingmuller U. Identification of nucleocytoplasmic cycling as a remote sensor in cellular signaling by databased modeling. Proc Natl Acad Sci U S A. 2003;100(3):1028–33. doi: 10.1073/pnas.0237333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamada S, Shiono S, Joo A, Yoshimura A. Control mechanism of JAK/STAT signal transduction pathway. FEBS Lett. 2003;534(1-3):190–6. doi: 10.1016/s0014-5793(02)03842-5. [DOI] [PubMed] [Google Scholar]
- 87.Papin JA, Palsson BO. The JAK-STAT signaling network in the human B-cell: an extreme signaling pathway analysis. Biophys J. 2004;87(1):37–46. doi: 10.1529/biophysj.103.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahdavi A, Davey RE, Bhola P, Yin T, Zandstra PW. Sensitivity analysis of intracellular signaling pathway kinetics predicts targets for stem cell fate control. PLoS Comput Biol. 2007;3(7):e130. doi: 10.1371/journal.pcbi.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93(19):10078–83. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298(5596):1241–5. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 91.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309(5742):1857–61. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 92.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309(5742):1854–7. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 93.Lipniacki T, Paszek P, Brasier AR, Luxon B, Kimmel M. Mathematical model of NF-kappaB regulatory module. J Theor Biol. 2004;228(2):195–215. doi: 10.1016/j.jtbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 94.Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128(2):369–81. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.LevBar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A. 2000;97(21):11250–5. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ciliberto A, Novak B, Tyson JJ. Steady states and oscillations in the p53/Mdm2 network. Cell Cycle. 2005;4(3):488–93. doi: 10.4161/cc.4.3.1548. [DOI] [PubMed] [Google Scholar]
- 97.Ma L, Wagner J, Rice JJ, Hu W, Levine AJ, Stolovitzky GA. A plausible model for the digital response of p53 to DNA damage. Proc Natl Acad Sci U S A. 2005;102(40):14266–71. doi: 10.1073/pnas.0501352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30(3):277–89. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136(2):337–51. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmierer B, Tournier AL, Bates PA, Hill CS. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci U S A. 2008;105(18):6608–13. doi: 10.1073/pnas.0710134105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1(1):E10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wawra C, Kuhl M, Kestler HA. Extended analyses of the Wnt/beta-catenin pathway: robustness and oscillatory behaviour. FEBS Lett. 2007;581(21):4043–8. doi: 10.1016/j.febslet.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 103.Amonlirdviman K, Khare NA, Tree DR, Chen WS, Axelrod JD, Tomlin CJ. Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science. 2005;307(5708):423–6. doi: 10.1126/science.1105471. [DOI] [PubMed] [Google Scholar]
- 104.Sick S, Reinker S, Timmer J, Schlake T. WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism. Science. 2006;314(5804):1447–50. doi: 10.1126/science.1130088. [DOI] [PubMed] [Google Scholar]
- 105.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8(11):1195–203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 106.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9(12):981–91. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100(21):11980–5. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma'ayan A, Cecchi GA, Wagner J, Rao AR, Iyengar R, Stolovitzky G. Ordered cyclic motifs contribute to dynamic stability in biological and engineered networks. Proc Natl Acad Sci U S A. 2008;105(49):19235–40. doi: 10.1073/pnas.0805344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14(2):140–8. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 110.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310(5747):496–8. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferrell JE, Xiong W. Bistability in cell signaling: How to make continuous processes discontinuous, and reversible processes irreversible. Chaos. 2001;11(1):227–236. doi: 10.1063/1.1349894. [DOI] [PubMed] [Google Scholar]
- 112.Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG. Systems analysis of Ran transport. Science. 2002;295(5554):488–91. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- 113.Neves SR. Models of spatially restricted biochemical reaction systems. J Biol Chem. 2008 doi: 10.1074/jbc.R800058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slepchenko BM, Schaff JC, Macara I, Loew LM. Quantitative cell biology with the Virtual Cell. Trends Cell Biol. 2003;13(11):570–6. doi: 10.1016/j.tcb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 115.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–7. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gillespie D. Exact Stochastic Simulation of Coupled Chemical Reactions. Journal of Physical Chemistry. 1977;81(25):2340–2361. [Google Scholar]
- 119.Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19(10):401–6. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- 120.Moles CG, Mendes P, Banga JR. Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res. 2003;13(11):2467–74. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ideker T, Lauffenburger D. Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends Biotechnol. 2003;21(6):255–62. doi: 10.1016/S0167-7799(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 122.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6(2):99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 123.Wiback SJ, Mahadevan R, Palsson BO. Reconstructing metabolic flux vectors from extreme pathways: defining the alpha-spectrum. J Theor Biol. 2003;224(3):313–24. doi: 10.1016/s0022-5193(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 124.Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO. Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol. 2002;184(16):4582–93. doi: 10.1128/JB.184.16.4582-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li F, Thiele I, Jamshidi N, Palsson BO. Identification of potential pathway mediation targets in Toll-like receptor signaling. PLoS Comput Biol. 2009;5(2):e1000292. doi: 10.1371/journal.pcbi.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100(26):15522–7. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rahib L, MacLennan NK, Horvath S, Liao JC, Dipple KM. Glycerol kinase deficiency alters expression of genes involved in lipid metabolism, carbohydrate metabolism, and insulin signaling. Eur J Hum Genet. 2007;15(6):646–57. doi: 10.1038/sj.ejhg.5201801. [DOI] [PubMed] [Google Scholar]
- 128.Janes KA, Albeck JG, Peng LX, Sorger PK, Lauffenburger DA, Yaffe MB. A high-throughput quantitative multiplex kinase assay for monitoring information flow in signaling networks: application to sepsis-apoptosis. Mol Cell Proteomics. 2003;2(7):463–73. doi: 10.1074/mcp.M300045-MCP200. [DOI] [PubMed] [Google Scholar]
- 129.Janes KA, Kelly JR, Gaudet S, Albeck JG, Sorger PK, Lauffenburger DA. Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol. 2004;11(4):544–61. doi: 10.1089/cmb.2004.11.544. [DOI] [PubMed] [Google Scholar]
- 130.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 131.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 132.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36(2):147–50. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 133.Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, Yarnitzky T, Liron Y, Polak P, Lahav G, Alon U. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:2006 0033. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 135.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25(51):6831–43. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 136.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 137.Bian X, Opipari AW, Jr, Ratanaproeksa AB, Boitano AE, Lucas PC, Castle VP. Constitutively active NFkappa B is required for the survival of S-type neuroblastoma. J Biol Chem. 2002;277(44):42144–50. doi: 10.1074/jbc.M203891200. [DOI] [PubMed] [Google Scholar]
- 138.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O'Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22(15):2093–101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Strasser A, O'Connor L, Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–45. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 140.Wallach D, Kovalenko AV, Varfolomeev EE, Boldin MP. Death-inducing functions of ligands of the tumor necrosis factor family: a Sanhedrin verdict. Curr Opin Immunol. 1998;10(3):279–88. doi: 10.1016/s0952-7915(98)80166-0. [DOI] [PubMed] [Google Scholar]
- 141.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4(10):1569–90. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 142.Kitano H. Systems biology: a brief overview. Science. 2002;295(5560):1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 143.Kim D, Rath O, Kolch W, Cho KH. A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways. Oncogene. 2007;26(31):4571–9. doi: 10.1038/sj.onc.1210230. [DOI] [PubMed] [Google Scholar]
- 144.Toettcher JE, Loewer A, Ostheimer GJ, Yaffe MB, Tidor B, Lahav G. Distinct mechanisms act in concert to mediate cell cycle arrest. Proc Natl Acad Sci U S A. 2009;106(3):785–90. doi: 10.1073/pnas.0806196106. [DOI] [PMC free article] [PubMed] [Google Scholar]