Disturbed flow-mediated PKCζ–PIASy association is critical for p53 SUMOylation and induces p53 nuclear export and endothelial cell apoptosis.
Abstract
Atherosclerosis is readily observed in regions of blood vessels where disturbed blood flow (d-flow) is known to occur. A positive correlation between protein kinase C ζ (PKCζ) activation and d-flow has been reported, but the exact role of d-flow–mediated PKCζ activation in atherosclerosis remains unclear. We tested the hypothesis that PKCζ activation by d-flow induces endothelial cell (EC) apoptosis by regulating p53. We found that d-flow–mediated peroxynitrite (ONOO−) increased PKCζ activation, which subsequently induced p53 SUMOylation, p53–Bcl-2 binding, and EC apoptosis. Both d-flow and ONOO− increased the association of PKCζ with protein inhibitor of activated STATy (PIASy) via the Siz/PIAS-RING domain (amino acids 301–410) of PIASy, and overexpression of this domain of PIASy disrupted the PKCζ–PIASy interaction and PKCζ-mediated p53 SUMOylation. En face confocal microscopy revealed increases in nonnuclear p53 expression, nitrotyrosine staining, and apoptosis in aortic EC located in d-flow areas in wild-type mice, but these effects were significantly decreased in p53−/− mice. We propose a novel mechanism for p53 SUMOylation mediated by the PKCζ–PIASy interaction during d-flow–mediated EC apoptosis, which has potential relevance to early events of atherosclerosis.
Introduction
A hallmark of atherogenesis is focalized endothelial dysfunction, which includes altered vasoregulation, activation of inflammatory processes, and compromised barrier function caused by endothelial cell (EC) apoptosis (Hu et al., 1999; Song et al., 2008). Manifestations of dysfunctional ECs are readily observed in certain areas of the arterial tree, where disturbed flow (d-flow), hence reduced time-averaged shear stress, develops (Traub and Berk, 1998; Won et al., 2007). Steady laminar flow (s-flow) promotes release of factors from ECs that inhibit coagulation, leukocyte diapedesis, and smooth muscle cell proliferation while simultaneously promoting EC survival (Garin et al., 2007; Reinhart-King et al., 2008). Conversely, d-flow alters the profile of secreted factors and EC surface molecule expression that favors the opposite effects, thereby contributing to the development of atherosclerosis (Traub and Berk, 1998). We have previously reported the critical role of PKCζ activation in EC apoptosis (Garin et al., 2007). Importantly, s-flow interferes with PKCζ signaling, thereby down-regulating the proapoptotic effect of the kinase. In contrast, unique atheroprone signals elicited by d-flow remain unclear.
Acting as a sensor for DNA damage, the transcription factor p53 is a key regulator of the life or death of a cell, depending on whether or not the cell can cope with the damage and repair it. Although the most-studied function of p53 is its role as a transcription factor that increases the expression of proapoptotic genes (Murray-Zmijewski et al., 2008), recent studies have revealed its nontranscriptional proapoptotic activities. Cytosolic p53 directly interacts with the B cell lymphoma/leukemia-2 (Bcl-2) family member proteins Bcl-xL and Bcl-2 and antagonizes their antiapoptotic function by stabilizing the outer mitochondrial membrane (Mihara et al., 2003). Interestingly, antiapoptotic effects of p53 have also been reported (Mercer et al., 2005; Garner and Raj, 2008). It is particularly interesting that p53 inhibits apoptosis of vascular smooth muscle cells and protects against atherosclerosis formation (van Vlijmen et al., 2001; Mercer et al., 2005). However, it remains unclear how these p53 functions, especially its apoptotic effect in ECs, are regulated by flow.
SUMOylation is a posttranslational modification consisting of covalent conjugation of ubiquitin-like proteins called SUMO (small ubiquitin-like modifier) to target proteins (Hilgarth et al., 2004). It is a reversible modification that affects target protein functions, such as subcellular localization, protein partnering, DNA binding, and regulation of transcription factors (Hilgarth et al., 2004). Carter et al. (2007) proposed an interesting model for p53 nuclear export and stabilization. Masking of the C-terminal nuclear export signal (NES) results in nuclear localization of unmodified p53. A low level of ubiquitination by MDM2 exposes the NES, promoting p53 to interact with protein inhibitor of activated STATy (PIASy) and further modification by SUMOylation causes p53 nuclear export (Carter et al., 2007). These data suggest an important role of SUMOylation in p53 nuclear export. In this study, we investigate the role of PKCζ and PIASy on d-flow–mediated endothelial p53 nuclear export and apoptosis, which may contribute to EC dysfunction and subsequent atherosclerotic plaque formation. Furthermore, we show that the cytoplasmic expression of p53 and EC apoptosis are increased in ECs in the area of d-flow in vivo.
Results
D-flow and ONOO−, but not s-flow, activate PKCζ
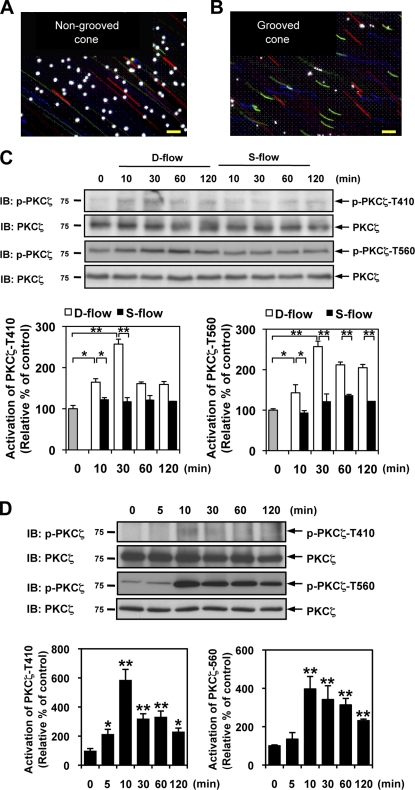
We first verified the potential role of shear stress in PKCζ activation in cultured ECs using a cone and plate type of flow apparatuses as described previously (Reinhart-King et al., 2008). To generate d-flow, we used cones with radial grooves that were 1-mm deep. Fig. 1 (A and B) shows tracks of fluorescent beads suspended in culture media when grooved and nongrooved cones were rotated at the same speed. Although the nongrooved cone created straight unidirectional tracks indicating s-flow, tracks made by the grooved cone were short and not oriented in the same direction, indicating nonlaminar movement of the media in the dish. Using these cones, we determined the effect of s- and d-flow on PKCζ activation in human umbilical vein ECs (HUVECs). D-flow increased PKCζ phosphorylation at both Thr410 and Thr560 after 10 min of stimulation (Fig. 1 C), whereas s-flow failed to activate PKCζ, although ERK5 was activated by s-flow as we reported previously (not depicted; Woo et al., 2008). PKCζ phosphorylation at Thr560 was sustained longer than phosphorylation at Thr410. These data obtained from our in vitro system are consistent with in vivo data, which showed increased PKCζ activity in d-flow area compared with s-flow area in porcine arteries (Magid and Davies, 2005), supporting the physiological relevance of our in vitro system.
Figure 1.
PKCζ activation by d-flow and ONOO−. (A and B) Generation of s-flow and d-flow using a cone and plate flow chamber. Tracks of fluorescent beads in a cone and plate flow chamber. S-flow was generated using a nongrooved cone (A), and d-flow was generated using a grooved cone (B). Note straight tracks with s-flow and short tracks with different orientations with d-flow, which are caused by beads going out of focus. Both cones were rotated at the same speed. Color tracks indicate time x (red), time x + 10 s (green), and time x + 20 s (blue). Bright dots are beads adhered to the dish. Exposure time is 0.4 s. Bars, 100 µm. (C and D) HUVECs were stimulated by s- or d-flow (C) or ONOO− (D) for the indicated times, and PKCζ phosphorylation was determined by Western blotting. The level of PKCζ phosphorylation was determined by taking the ratio of optical densities between antiphospho-PKCζ and anti-PKCζ bands (bar graphs) as described in Materials and methods. The experiments were performed in triplicate using three different batches of s- or d-flow or ONOO−-stimulated HUVECs (means ± SD; n = 3; *, P < 0.05; and **, P < 0.01 compared with control). Molecular masses are given in kilodaltons. IB, immunoblot.
Reactive oxygen species has been shown to react with NO and generate ONOO−, reducing NO availability and accelerating endothelial dysfunction and development of atherosclerosis (Ponnuswamy et al., 2009). Furthermore, it has been reported that ONOO− was increased by d-flow both in vitro and in vivo (Hsiai et al., 2007). To test whether some effects of d-flow could be mediated by ONOO−, we treated ECs with ONOO− and examined PKCζ activation. Indeed, ONOO− also increased PKCζ phosphorylation at both Thr410 and Thr560 within 5 min of stimulation, reaching its maximum by 10 min (Fig. 1 D), suggesting that ONOO−-mediated PKCζ kinase activation is similar to d-flow stimulation.
PKCζ activation by d-flow and ONOO− induces apoptosis
We examined whether PKCζ activation was also required for d-flow and ONOO−-mediated EC apoptosis using two different approaches. PKCζ activity was down-regulated by inhibiting its expression by siRNA and also by expressing an adenoviral dominant-negative (DN) kinase-dead form of PKCζ (Ad-DN-PKCζ). Although d-flow and ONOO− treatment increased apoptosis in control cells, a significant reduction in the number of TUNEL-positive cells was noted in PKCζ-depleted HUVECs (Fig. 2, A and B, left). When HUVECs were infected with Ad-DN-PKCζ (mutating Lys281to Met), the ONOO−-induced increase in TUNEL-positive cells and the expression of cleaved caspase 3 fragments (17/19 kD) were reduced compared with control cells (Fig. 2, B [center] and C). These data suggest the critical role of PKCζ kinase activity in d-flow and ONOO−-mediated EC apoptosis.
Figure 2.
PKCζ depletion by siRNA and DN-PKCζ inhibits d-flow and ONOO−-induced apoptosis. (A) HUVECs were transfected with control or PKCζ siRNA for 48 h and then stimulated with d-flow for 36 h followed by TUNEL staining. Images were recorded as described in Materials and methods after counterstaining with DAPI to visualize nuclei (bottom). Apoptotic nuclei appear green (top). Bars, 25 µm. (right) Quantification of apoptosis is shown as the percentage of TUNEL-positive cells. (B) HUVECs were transfected with control or PKCζ siRNA for 48 h (left) or transduced with Ad-DN-PKCζ or Ad-LacZ as a control for 24 h (center). Cells were then treated with 100 µM ONOO− or vehicle for 8 h and assayed by TUNEL staining. (right) DN-PKCζ and reduced PKCζ expression were confirmed by Western blotting with anti-PKCζ. Data are from three separate experiments using two or more different EC preparations (**, P < 0.01). (C) After transduction of Ad-DN-PKCζ or Ad-LacZ for 24 h, HUVECs were treated with 100 µM ONOO− for 9 and 18 h, and Western blotting with anti–cleaved caspase 3 was performed. DN-PKCζ expression and protein loading were assessed by Western blotting with anti-PKCζ (middle) and antitubulin (bottom). (right) Quantification of cleaved caspase 3 is expressed as the relative ratio compared with tubulin expression. Results are expressed as the relative percentage of untreated cells in the LacZ control (100%). n = 3. **, P < 0.01 compared with each control. Molecular masses are given in kilodaltons. Error bars are means ± SD.
ONOO− induces p53 nuclear export and binding to Bcl-2 in a PKCζ-dependent manner
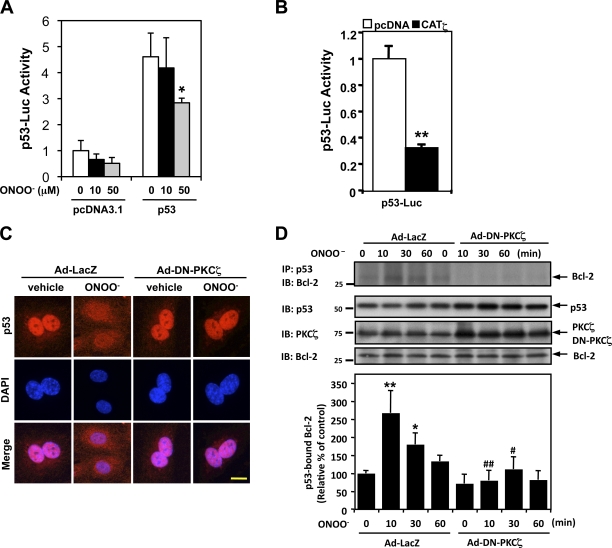
p53 promotes transcription of several proapoptotic genes (Mihara et al., 2003; Mercer et al., 2005; Bischof et al., 2006; Garner et al., 2007). To test whether it plays a role in the ONOO−-induced EC apoptosis, we first examined the effect of ONOO− on p53 transcriptional activity in HUVECs that were overexpressing p53 with or without coexpression of a constitutively active PKCζ (catalytic domain of PKCζ [CATζ]; Garin et al., 2007). To our surprise, ONOO− inhibited p53 transcriptional activity in control cells and also in those overexpressing p53 (Fig. 3 A). Consistent with these results, cells transfected with CATζ also showed a decreased p53 transcriptional activity (Fig. 3 B). These results strongly suggest that ONOO−-mediated EC apoptosis is not caused by increased p53 transcriptional activation of proapoptotic genes.
Figure 3.
PKCζ mediates ONOO−-induced p53 nuclear export and p53–Bcl-2 binding instead of the regulation of p53 transcriptional activity. (A and B) HUVECs were transfected with the p53-Luc reporter and Renilla luciferase–encoding plasmid (pRL- thymidine kinase) used as an internal control reporter together with p53–wild type or vector alone (pcDNA3.1; A). Some cells were further transfected with or without pcDNA3.1-CATζ (B). Transcriptional activity was determined by a reporter plasmid encoding 13 copies of the p53-binding sequence (p53-Luc reporter; Kern et al., 1992). After 24 h of transfection, p53 transcriptional activity was assayed using the dual-luciferase kit (B), or the cells were further treated with 10 or 50 µM ONOO− for 8 h as indicated, and luciferase activity was assayed (A). Data are representative of triplicates using two or more different preparations of ECs. *, P < 0.05; **, P < 0.01. (C) HUVECs were transduced with Ad-DN-PKCζ or Ad-LacZ as a control for 24 h, treated with vehicle or 100 µM ONOO− for 4 h, and immunostained with anti-p53 followed by DAPI counter staining for nuclei. Bar, 5 µm. (D, top) HUVECs were transduced with Ad-DN-PKCζ or Ad-LacZ for 24 h and stimulated with 100 µM ONOO– for the indicated times. p53–Bcl-2 binding was determined by coimmunoprecipitation with anti-p53 followed by immunoblotting with anti–Bcl-2. p53, PKCζ, and Bcl-2 in total cell lysates were detected by Western blotting with each specific antibody. (bottom) Quantification of p53–Bcl-2 binding expressed as the relative band intensity ratio between coimmunoprecipitated versus total Bcl-2. Results were normalized as described in Fig. 1. n = 3. *, P < 0.05 and **, P < 0.01 compared with the vehicle control, and #, P < 0.05 and ##, P < 0.01 compared with the LacZ control at each time point. Molecular masses are given in kilodaltons. Error bars indicate means ± SD. IB, immunoblot. IP, immunoprecipitation.
It has recently been reported that cytosolic p53 directly interacts with Bcl-2 and antagonizes its antiapoptotic stabilization of the outer mitochondrial membrane (Mihara et al., 2003; Bischof et al., 2006). Because this interaction takes place in the cytoplasm, p53 may be exported from the nucleus when PKCζ is activated by ONOO−. To verify this possibility, HUVECs were stimulated with ONOO− for 4 h, and p53 localization was analyzed by immunostaining (Fig. 3 C). Interestingly, the anti-p53 staining contrast between the nucleus and the cytoplasm in ONOO−-treated cells decreased considerably, and some punctuate staining appeared in the cytoplasm (Fig. 3 C). No fluorescent signal was detected when only the secondary antibodies were used (unpublished data). ONOO−-induced anti-p53 staining in the cytoplasm was reduced in Ad-DN-PKCζ–transduced cells compared with adenovirus of LacZ (Ad-LacZ)–transduced cells, suggesting that ONOO− induced p53 export from the nucleus in a PKCζ-dependent manner.
Next, to examine whether PKCζ is involved in ONOO−-mediated p53–Bcl-2 association, we expressed Ad-DN-PKCζ in HUVECs, treated them with ONOO−, and performed a coimmunoprecipitation assay using anti-p53 followed by immunoblotting with anti–Bcl-2. Bcl-2 coimmunoprecipitated with p53 when control cells (transduced with Ad-LacZ) were treated with ONOO− for 10 min or longer, but this coimmunoprecipitation was strongly inhibited by Ad-DN-PKCζ expression (Fig. 3 D and Fig. S1, A and C). Nonimmune control IgG failed to bring down p53 and Bcl-2 (Fig. S1 A). Collectively, these results suggest a role of PKCζ in ONOO−-mediated p53 nuclear export and subsequent p53–Bcl-2 interaction.
PKCζ mediates d-flow– and ONOO−-induced p53 SUMOylation
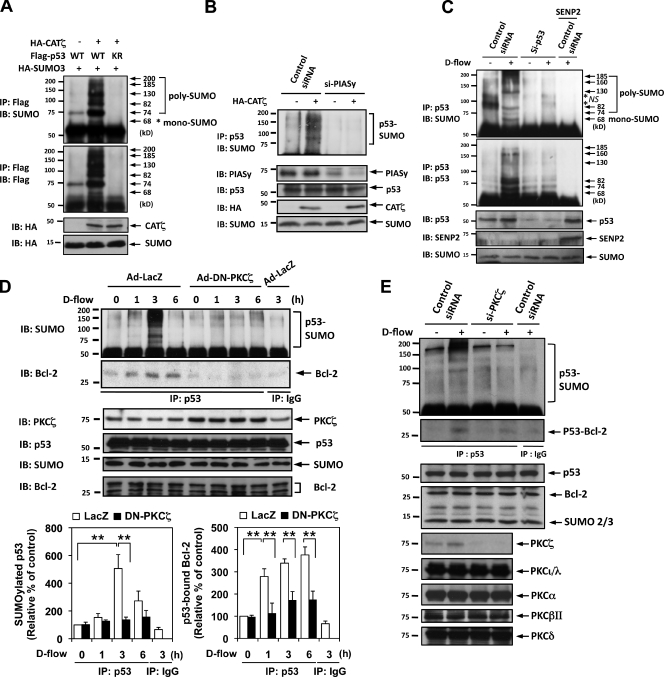
Because p53 nuclear export is positively regulated by SUMOylation (Bischof et al., 2006) and also because PKCζ regulates p53 nuclear export (present study), p53 SUMOylation may be controlled by PKCζ. To test this, we cotransfected HeLa cells with Flag-tagged p53 (Flag-p53), HA-tagged SUMO3 (HA-SUMO), and constitutively active PKCζ (CATζ) and determined p53 SUMOylation by Western blotting using anti-SUMO2/3. Several bands presumably representing mono- and poly-SUMOylated p53 with apparent molecular masses of 68, 74, 82, 130, 185, and 200 kD were noted (Fig. 4 A). These bands were also labeled by anti-Flag, indicating that they are SUMOylated Flag-p53 (Fig. 4 A, middle). p53 SUMOylation was increased in CATζ-transfected cells, suggesting the role of PKCζ activity on p53 SUMOylation. In addition, these sumoylated bands were diminished in the cells transfected with the p53-K386R SUMOylation mutant (Kwek et al., 2001), further supporting that these high mass bands were SUMOylated p53.
Figure 4.
PKCζ mediates d-flow–induced p53 SUMOylation and p53–Bcl-2 binding. (A) HeLa cells were transfected for 24 h as indicated with Flag-tagged p53, HA-tagged SUMO3, and HA-tagged CATζ. p53 SUMOylation was detected by immunoprecipitation with anti-Flag followed by Western blotting with anti-SUMO2/3 (top). Both protein expression and immunoprecipitated p53 were confirmed by anti-Flag antibody, and CATζ and SUMO expression were detected with anti-HA. Mono-SUMOylation band (∼74 kD) and poly-SUMOylation bands (>78 kD) were detected. The asterisk indicates mono-SUMOylation of p52. (B) HUVECs were transfected for 24 h with either PIASy or control siRNA as indicated, and then the cells were transfected with HA-CATζ or vector alone for another 24 h. (top) p53 SUMOylation was detected by immunoprecipitation with anti-p53 followed by Western blotting with anti-SUMO2/3. PIASy expression was confirmed by immunoblotting with anti-PIASy, and p53, HA-CATζ, and SUMO expression was confirmed with anti-p53, -HA, and -SUMO2/3, respectively. (C) HUVECs were transfected with either p53 or control siRNA as indicated for 24 h, and then the cells were transduced with an Ad-SENP2 or LacZ with a control for another 24 h. p53 SUMOylation, expression of p53, SENP2, and SUMO were determined as described in Materials and methods. The asterisks indicate nonspecific bands. (D) HUVECs were transduced with Ad-DN-PKCζ or Ad-LacZ as a control for 24 h and then stimulated with d-flow for the indicated times. p53 SUMOylation and p53–Bcl-2 binding were determined as described in Materials and methods. (left graph) Intensities of SUMOylated p53 bands at 74, 82, 130, and 185 kD were quantified by densitometry after subtracting background gel density. After normalization of each control as described in Fig. 1, results were expressed relative to the SUMOylation level in static condition (0 min; 100%). Shown are means ± SD (n = 3). **, P < 0.01 compared with the vehicle control or the LacZ control at each time point. (E) HUVECs were transfected with either PKCζ or control siRNA as indicated for 24 h and then stimulated with d-flow for 3 h. p53 SUMOylation, p53–Bcl-2 binding, expression of p53, Bcl-2, SUMO, and various PKC isoforms as indicated were determined as described in Materials and methods. Immunoblots are representative of three separate experiments. Molecular masses are given in kilodaltons. IB, immunoblot. IP, immunoprecipitation. KR, K386R. WT, wild type.
An important role of PIASy as a SUMO E3 ligase in p53 SUMOylation has been reported (Bischof et al., 2006). To confirm that the 4–5 bands are SUMOylated p53, HUVECs with or without CATζ expression were transfected with PIASy siRNA. This should inhibit p53 SUMOylation, and indeed, PIASy depletion eliminated the appearance of these bands (Fig. 4 B). Coimmunoprecipitation using nonimmune IgG yielded no SUMOylated bands (Fig. S1 A). To see whether endogenous p53 is SUMOylated, we specifically inhibited p53 expression using siRNA and found significantly reduced SUMOylation levels in the 68-, 74-, 82-, 130-, 160-, and 185-kD bands (Fig. 4 C). Endogenous p53 SUMOylation bands were less discrete, likely caused by the combined effects of poly-SUMOylation and ubiquitination as previously described (Carter et al., 2007). SENP2 (sentrin-specific protease 2) is a de-SUMOylation enzyme that is important for both processing new SUMO proteins for conjugation as well as deconjugating SUMO from SUMOylated proteins (Cheng et al., 2004; Yeh, 2009; Witty et al., 2010). To verify the identity of the endogenous p53 SUMOylation band, we also transduced ECs with Ad-LacZ control or adenoviral SENP2 (Ad-SENP2), stimulated them with d-flow, and evaluated changes in these multiple bands (Fig. 4 C). Transduction of Ad-SENP2 completely reduced d-flow–induced p53 SUMOylation. These results together support that the 68-, 74-, 82-, 130-, 160-, 185-, and 200-kD bands are SUMOylated p53.
When HUVECs were treated with various concentrations of ONOO− for 10 min, p53 SUMOylation increased in a dose-dependent manner (Fig. S1, A and B). We also found that d-flow significantly increased p53 SUMOylation after 2 h of stimulation (Fig. 4 D and Fig. S1 D). Next, we investigated whether PKCζ played a role in p53 SUMOylation by ONOO− and d-flow. This SUMOylation was inhibited in cells expressing Ad-DN-PKCζ (Fig. 4 D and Fig. S1 C), suggesting a role of PKCζ in p53 SUMOylation in ONOO− and d-flow–stimulated cells. In addition, Bcl-2 coimmunoprecipitated with p53 when control cells (transduced with Ad-LacZ) were treated with d-flow, but this coimmunoprecipitation was strongly inhibited by Ad-DN-PKCζ transduction (Fig. 4 D), which was also observed in ONOO−-treated cells (Fig. 3 D).
To test whether PKCζ plays a unique role on p53 SUMOylation and binding between p53 and Bcl-2, we transfected ECs with PKCζ siRNA and studied its effect on d-flow–mediated p53 SUMOylation and p53–Bcl-2 binding. As shown in Fig. 4 E, we found that PKCζ siRNA specifically inhibited PKCζ expression but not other PKC isoforms, including PKCι/λ, PKCα, PKCβII, and PKCδ (Fig. 4 E). Under this condition, d-flow effects on p53 SUMOylation and p53–Bcl-2 binding were significantly reduced, indicating the unique and critical role of PKCζ on d-flow–mediated p53 SUMOylation and its subsequent events.
ONOO− production by d-flow mediates PKCζ activation and p53 SUMOylation
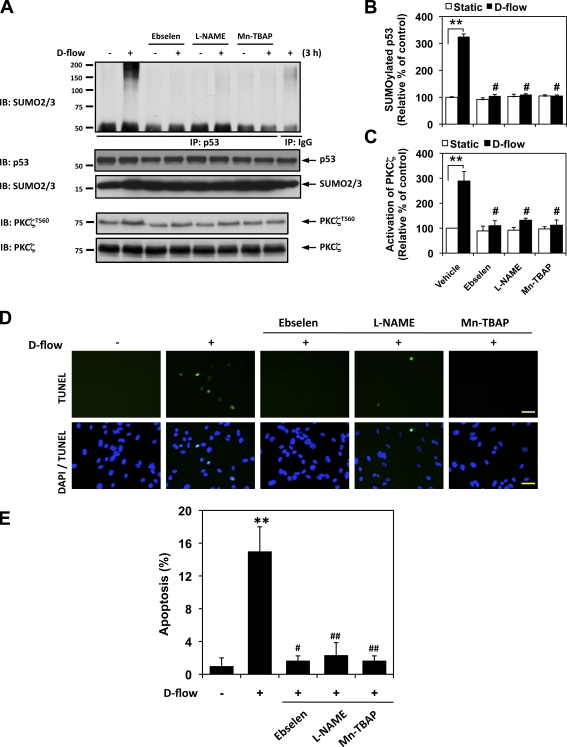
We investigated the contribution of ONOO− production on the d-flow–mediated PKCζ activation and p53 SUMOylation and found that an ONOO− scavenger, ebselen, a nonselective inhibitor of NO synthesis, N-nitro-l-arginine methyl ester (L-NAME), superoxide dismutase mimetic, and an ONOO− scavenger, Mn (III)tetrakis(4-benzoic acid)porphyrin chloride (Mn-TBAP), significantly inhibited d-flow–mediated PKCζ activation as well as p53 SUMOylation (Fig. 5, A–C). Moreover, these compounds strongly inhibited d-flow–induced EC apoptosis (Fig. 5, D and E), strongly suggesting a critical role of ONOO− production in the d-flow–mediated signaling and subsequent apoptosis.
Figure 5.
ONOO− mediates d-flow–induced PKCζ activation, p53 SUMOylation, and EC apoptosis. (A) ONOO− mediates d-flow–induced PKCζ activation and p53 SUMOylation. HUVECs were pretreated by 5 µM ebselen, 20 µM L-NAME, and 10 µM Mn-TBAP for 30 min and exposed to d-flow for 3 h. PKCζ phosphorylation at Thr560 and p53 SUMOylation were determined as described in Materials and methods. (B and C) Densitometry analyses of p53 SUMOylation (B) and PKCζ phosphorylation (C) were performed as described in Fig. 1. **, P < 0.01 compared with the vehicle control in static condition, and #, P < 0.01 compared with the vehicle control in d-flow stimulation for 3 h. (D and E) HUVECs were pretreated by each inhibitor for 30 min and exposed to d-flow for 36 h followed by TUNEL staining as described in Materials and methods (D), and quantification of apoptosis is shown as the percentage of TUNEL-positive cells (E). Bars, 30 µm. Data are from three separate experiments using two or more different EC preparations (**, P < 0.01 compared with the vehicle control in static condition, and #, P<0.05 and ##, P<0.01 compared with the vehicle control in d-flow stimulation for 36 h). Error bars show means ± SD. Molecular masses are given in kilodaltons. IB, immunoblot. IP, immunoprecipitation.
PIASy mediates p53 SUMOylation by ONOO− and d-flow, and p53 SUMOylation plays a critical role in d-flow–mediated apoptosis
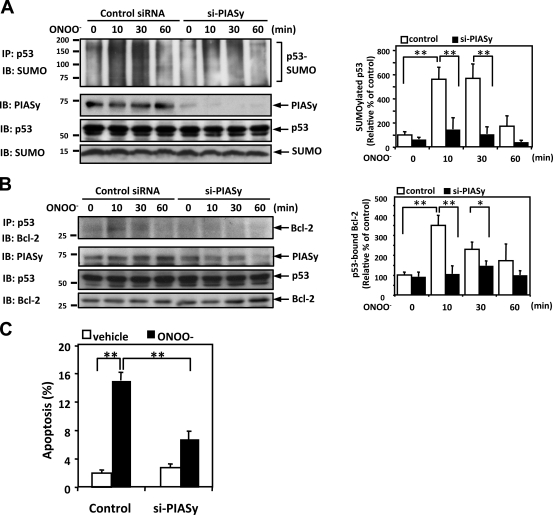
Because PIASy (SUMO E3 ligase) is involved in p53 SUMOylation in fibroblasts (Bischof et al., 2006), we wondered whether it was involved in the ONOO−- and d-flow–elicited p53 SUMOylation in HUVECs. When HUVECs were transfected with PIASy siRNA, the ONOO− and d-flow–dependent p53 SUMOylation was inhibited (Fig. 6 A and Fig. 7 A). Using the PIASy-depleted cells, we studied the role of PIASy in the ONOO−-elicited p53–Bcl-2 association and found that the PIASy depletion inhibited this interaction (Fig. 6 B), suggesting that PIASy, which SUMOylates p53, is involved in ONOO−-induced p53 nuclear export and subsequent p53–Bcl-2 binding in ECs. To see whether PIASy has a role in the ONOO− and d-flow–induced EC apoptosis, ECs were transfected with PIASy siRNA, challenged by ONOO− or d-flow, and assayed for apoptosis. The number of TUNEL-positive cells and expression of cleaved caspase 3 induced by ONOO− or d-flow were down-regulated by PIASy siRNA (Fig. 6 C and Fig. 7, B and C).
Figure 6.
ONOO− induces p53 SUMOylation and p53–Bcl-2 binding via PIASy activation. (A and B) HUVECs were transfected with PIASy siRNA (si-PIASy) or control siRNA for 48 h and then stimulated with 100 µM ONOO– for the indicated times. p53 SUMOylation (A) and p53–Bcl-2 binding (B) were determined as described in Materials and methods. (left) PIASy and p53 expressions were detected by Western blotting with appropriate specific antibodies. Densitometric analyses of p53 SUMOylation (A) and p53–Bcl-2 binding (B) were performed as described in Fig. 1. (C) HUVECs were transfected with PIASy or control siRNA for 48 h. After treatment with 100 µM ONOO− for 8 h, apoptotic nuclei were detected by TUNEL staining. Data are expressed as mean percentages ± SD from three independent experiments. *, P < 0.05; **, P < 0.01. Molecular masses are given in kilodaltons. IB, immunoblot. IP, immunoprecipitation.
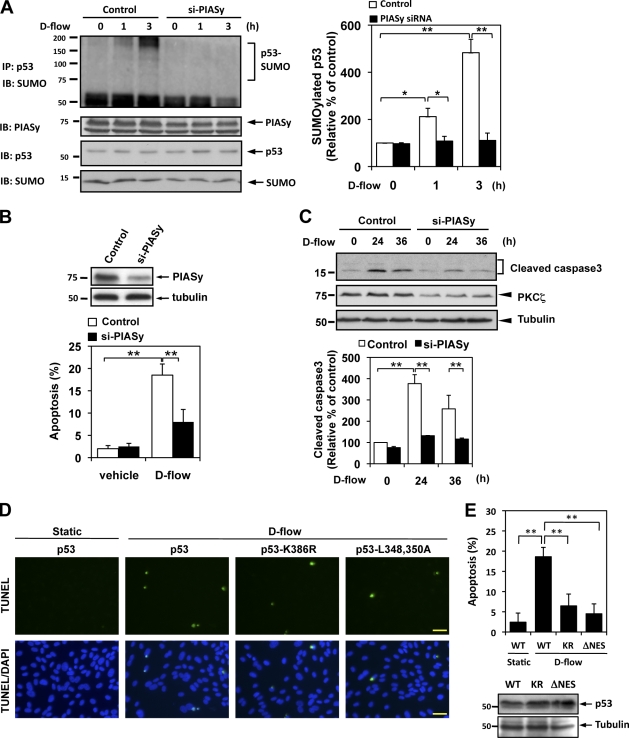
Figure 7.
D-flow induces p53 SUMOylation and apoptosis via PIASy activation. (A) HUVECs were transfected with PIASy siRNA (si-PIASy) or control siRNA for 48 h and then stimulated with d-flow for the indicated times. p53 SUMOylation, expression of PIASy, p53, and SUMO2/3 were detected as described in Material and methods. Densitometric analyses of p53 SUMOylation were performed as described in Fig. 1. (B and C) HUVECs were transfected with PIASy or control siRNA for 48 h. After treatment with d-flow for 36 h, apoptotic nuclei were detected by TUNEL staining (B, bottom), and Western blotting with anti–cleaved caspase 3 (C, top) was performed. Immunoblots of PIASy conformed depletion of PIASy by the specific siRNA (B, top). Densitometry analysis of cleaved caspase 3 expression was performed as described in Fig. 2 C (bottom). The experiments were performed in triplicate using three different batches of d-flow–stimulated HUVECs. (D) HUVECs were transduced with an adenovirus vector containing p53, p53-K386R (KR; sumoylation defect mutant), or p53-ΔNES (L348,350A; NES mutant) for 24 h and then stimulated with d-flow for 36 h followed by TUNEL staining as described in Materials and methods. (E, top) Quantification of apoptosis shown as the percentage of TUNEL-positive cells. Bars, 30 µm. (bottom) Equal expressions of p53, p53-K386R, and p53-ΔNES were analyzed by Western blotting in ECs. Data are from three separate experiments using two or more different EC preparations. Error bars show means ± SD; *, P < 0.05; **, P < 0.01. Molecular masses are given in kilodaltons. IB, immunoblot. IP, immunoprecipitation. WT, wild type.
To establish the role of p53 SUMOylation on d-flow–mediated EC apoptosis, we transduced ECs with an adenovirus containing wild-type p53 (Ad-WT-p53) or the p53 SUMOylation site mutant (p53-K386R) and examined d-flow–mediated apoptosis compared with wild-type p53 in ECs. First, we confirmed that the adenoviral p53-K386R (Ad-p53-K386R) mutant significantly decreased p53 SUMOylation (Fig. 4 A). Next, we found that d-flow increased apoptosis in ECs transduced by Ad-WT-p53, but transduction of the Ad-p53-K386R mutant significantly inhibited it (Fig. 7, D and E). Because nuclear export of p53 is important for p53–Bcl-2 binding, we also used an NES mutation (L348,350A; ΔNES; O’Keefe et al., 2003) and studied d-flow–mediated EC apoptosis. As shown in Fig. S2 (A and B), in contrast to wild-type p53, we found that both p53-K386R and the ΔNES mutant stayed in the nucleus after d-flow stimulation, and the ΔNES mutant significantly inhibited d-flow–mediated apoptosis compared with wild type (Fig. 7, D and E), suggesting the critical role of p53 SUMOylation and nuclear export of p53 on d-flow–mediated EC apoptosis. Collectively, these results suggest the critical role of PIASy-mediated p53 SUMOylation and subsequent nuclear export of p53 in the ONOO− and d-flow–induced EC apoptosis (Fig. S2, A and B).
p53 SUMOylation is induced through PKCζ binding to PIASy instead of PIASy phosphorylation by PKCζ
As PIASy played a critical role in CATζ-mediated p53 SUMOylation (Fig. 4 B), PKCζ might directly phosphorylate PIASy and activate its E3 SUMO ligase activity. To test this possibility, we incubated recombinant PKCζ with GST-tagged PIASy fragments and found that none of the fragments was phosphorylated by PKCζ, whereas autophosphorylation of PKCζ was detected (Fig. S3). Although it does not phosphorylate PIASy, PKCζ may still interact with PIASy. We cotransfected HeLa cells with HA-tagged PKCζ and myc-tagged PIASy and performed a coimmunoprecipitation assay in which PKCζ and PIASy were coimmunoprecipitated (unpublished data). To confirm this interaction between endogenous PKCζ and PIASy, we stimulated HUVECs with ONOO− for the indicated times and performed coimmunoprecipitation using anti-PKCζ. PIASy was indeed coimmunoprecipitated by anti-PKCζ, and ONOO− stimulation increased the PKCζ–PIASy interaction (Fig. 8 A). Next, to determine the PIASy-binding regions of PKCζ, we generated four PKCζ-truncated mutants and evaluated their association with PIASy using a mammalian two-hybrid assay (Fig. 8, B and C). Plasmids encoding the GAL4–DNA-binding domain and PKCζ (full length or one of the truncated forms) were constructed using the pBIND vector. A plasmid containing VP16-PIASy was constructed using the pACT vector. As expected, wild-type PKCζ bound to PIASy, and ONOO− increased this association (Fig. 8 B). More importantly, however, we found that the C-terminal kinase domain (aa 401–587) was required for the PKCζ–PIASy association (Fig. 8 C). Next, to determine the PKCζ binding site of PIASy, we coexpressed GST-fused PKCζ and Flag-tagged PIASy fragments in HeLa cells and performed coimmunoprecipitation using anti-Flag. We found that PIASy fragment 3 (Fr 3; aa 301–410), which contains the Siz/PIAS RING domain, interacted with PKCζ (Fig. 8 D). In a separate experiment, we examined this fragment could interfere with the PKCζ–PIASy association. HeLa cells were cotransfected with wild-type PKCζ and PIASy together with PIASy Fr 3. Consistent with the coimmunoprecipitation results, PIASy Fr 3 significantly inhibited the PKCζ–PIASy interaction (Fig. 8 E). To demonstrate the importance of the PKCζ–PIASy interaction for p53 SUMOylation, ECs expressing PIASy fragments were stimulated by d-flow. As shown in Fig. 8 F, only PIASy Fr 3 inhibited d-flow–mediated p53 SUMOylation, suggesting that p53 SUMOylation depends on PKCζ–PIASy binding.
Figure 8.
PKCζ–PIASy association is critical for p53 SUMOylation and p53–Bcl-2 binding. (A) HUVECs were stimulated with 100 µM ONOO– for the indicated times and subjected to immunoprecipitation with anti-PIASy followed by Western blotting with anti-PKCζ (top). (B and C) Association between PKCζ and PIASy was tested by a mammalian two-hybrid assay. HeLa cells were transfected with plasmids containing Gal4-PKCζ wild type and VP16-PIASy (B) or truncated mutants of VP16-PIASy (C) as well as the Gal4-responsive luciferase reporter pG5-luc. After 24 h of transfection, cells were stimulated with 100 µM ONOO− or vehicle for 16 h, and luciferase activity was quantified. Luciferase activity was normalized with the Renilla luciferase (Luc.) activity (Woo et al., 2008). Data are representative of three experiments using two or more different preparations of ECs (means ± SD; **, P < 0.01). (D) PIASy binding to PKCζ occurs via a domain consisting of aa 301–410 of PIASy. HeLa cells were transfected with each of the Flag-tagged PIASy fragments, and then pull-down assays were preformed using anti-Flag and IgG Sepharose beads in the presence of GST-fused recombinant PKCζ. Association of PIASy fragments with GST-PKCζ was assayed by Western blotting with anti-PKCζ. (bottom) PIASy fragment expression was detected by Western blotting with anti-Flag. (E) PIASy Fr3, but not Fr4, inhibited PKCζ–PIASy association. HUVECs were cotransfected with HA-tagged PKCζ wild type, Myc-tagged PIASy wild type, and Flag-tagged PIASy Fr3 or Fr4 for 24 h. Myc-PIASy wild type was immunoprecipitated with anti-Myc followed by immunoblotting with anti-HA (top). The expression of PKCζ, PIASy, and PIASy fragments was detected by Western blotting with specific antibodies. Data are representative of three independent experiments. (F) HUVECs were transfected with Flag-tagged PIASy Fr3 or Fr4 or control vectors for 24 h and then stimulated by d-flow for 3 h. p53 was immunoprecipitated using anti-p53, and d-flow–induced p53 SUMOylation was analyzed by immunoblotting with anti-SUMO2/3 (top). The expression of p53, SUMO, and PIASy fragments was detected by Western blotting with specific antibodies. Data are representative of three independent experiments. Molecular masses are given in kilodaltons. IB, immunoblot. IP, immunoprecipitation. WT, wild type.
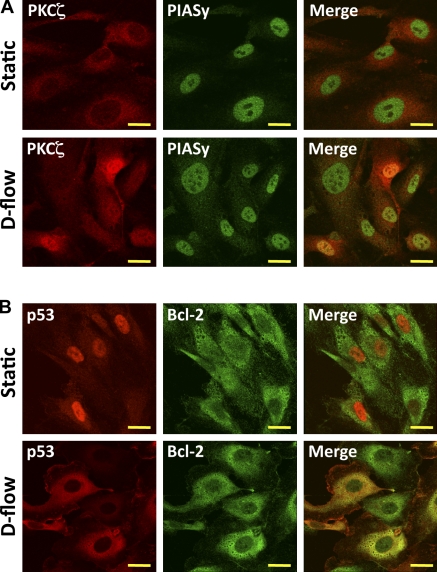
Morphological evidence for the PKCζ–PIASy and p53–Bcl-2 association was provided by confocal microscopy of coimmunostained ECs with or without flow stimulation (Fig. 9 A). Cells cultured without flow expressed PKCζ mainly in the cytosol and PIASy in the nucleus as reported previously (Sachdev et al., 2001; Li et al., 2004). However, after d-flow stimulation, PKCζ and PIASy were colocalized in the nucleus. As for the p53 and Bcl-2 colocalization, p53 was localized in the nucleus of unstimulated cells, whereas Bcl-2 was mainly outside the nucleus as previously reported (Zhong et al., 1993; Ghosh et al., 2004). D-flow stimulation caused significant nuclear export of p53 and colocalization with Bcl-2. These data support d-flow–induced association of PKCζ–PIASy and p53–Bcl-2 in ECs.
Figure 9.
D-flow–induced PKCζ–PIASy association in nuclei and p53–Bcl-2 binding in the cytosol. (A and B) HUVECs were stimulated with either static or d-flow for 3 h and immunoassayed with antibodies of mouse anti-PKCζ and rabbit anti-PIASy (A) or mouse anti-p53 and rabbit anti–Bcl-2 (B). After d-flow stimulation, yellow in the merged images represent colocalization between PKCζ and PIASy in nuclei or p53 and Bcl-2 in cytosol. Images were recorded using a confocal microscope equipped with a Plapon 60× 1.42 NA oil lens objective. Shown are representative images from cells analyzed from three independent experiments in which ≥30 cells were analyzed per experiment. Bars, 10 µm.
En face immunohistochemistry of PKCζ, p53, nitrotyrosine, and apoptosis in mouse aorta
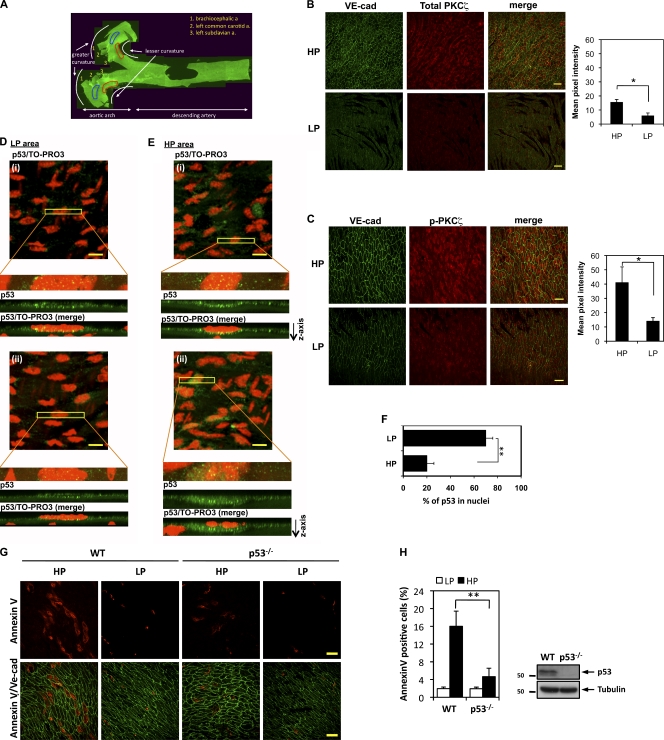
PKCζ activation in the ECs of the lesser curvature of the aortic arch in porcine aorta was recently reported (Magid and Davies, 2005). Because activation of this kinase is proatherogenic, we investigated the expression of PKCζ, phosphorylated PKCζ, a nitrotyrosine-containing protein, p53, and apoptotic ECs using en face aorta preparations and confocal microscopy. Aortas from male wild-type C57BL/6 mice (6–8 wk old) fed with normal chow were isolated after perfusion fixation and en face preparations were made. We focused on areas designated as high probability (HP) regions (lesser curvature of aortic arch) and low probability (LP) regions (greater curvature of aortic arch) for atherogenesis (Fig. 10 A) as described previously (Iiyama et al., 1999) and in the Materials and methods section. When the endothelium was double stained with anti-PKCζ or antiphospho-PKCζ together with anti–vascular endothelial (VE)-cadherin as an EC marker, we found that the expression of total PKCζ increased in the HP area (Fig. 10 B), and especially phospho-PKCζ was significantly higher in the HP area compared with the LP area (Fig. 10 C). Quantification of these data obtained from five mice supports this conclusion (Fig. 10, B and C, bar graph). These results confirm our in vitro results that show d-flow–dependent activation of PKCζ (Fig. 1).
Figure 10.
Increases in phosphorylated and total PKCζ and nonnuclear p53 expression within the d-flow regions (HP areas) and decreased apoptosis in ECs of p53−/− mice. (A) A representative epifluorescence image of the whole specimen. Fixed aortas of wild-type mice were cut longitudinally, and the arch region was further cut into two halves. Areas of d-flow (HP area; lesser curvature) are outlined in red, and neighboring areas of s-flow (LP area) are lined in blue. a, artery. (B and C) En face preparations were double stained with anti–VE-cadherin (VE-cad; used as an EC marker) and an anti–total PKCζ antibody (B) or phospho-PKCζ T560 antibody (C). X-y axis images were collected at 0.5-µm increments so that a z stack of ∼4-µm thickness from the luminal surface was obtained. From each image background, fluorescence intensity was subtracted, and the pixel number of the stained region per unit area of the endothelium in HP and LP area within the aortic arch was determined (n = 3). Areas of d-flow (HP areas; lesser curvature) show both increased total and phospho-PKCζ expression compared with the neighboring areas of s-flow (LP area). Bars, 20 µm. Bar graphs show quantification of total (B) and phospho (C)-PKCζ in HP and LP areas. Data are shown as means ± SEM; *, P < 0.05. (D and E) Increased cytoplasmic p53 localization in HP area ECs. Aortic arches were immunostained for endothelial p53 (green). Nuclei were stained using TO-PRO3 (red). From initial stacked x-y axis images (top), a narrow rectangular area crossing an EC was selected for x-y-z scanning at 0.1-µm increments. Images below the clipped images show rectangular z-axis images. Two representative sets of images (i and ii) are shown for LP (D) and HP (E) areas. Bars, 10 µm. (F) Quantification of nuclear p53. The pixel number of nuclear and nonnuclear regions per cell was determined, and the ratio of nuclear/total intensity was calculated from 60 cells from each HP and LP area (four cells/field, five fields/mouse, and a total of three mice). (G) The number of d-flow–mediated annexin V–positive cells (red) in the HP area is decreased in the mice deficient for p53 (p53 knockout) compared with the wild-type mouse aorta. Anti–VE-cadherin staining (green) was used as a marker for ECs. Bars, 100 µm. (H, left) Quantification of apoptotic cells. Percentages of annexin V–positive cells in LP and HP areas determined from 7-wk wild-type and p53-deficient mice (n = 3 each) are shown. In HP area, the number of annexin V–positive cells are significantly decreased in the p53 knockout compared with the wild type. **, P < 0.01. (right) Deletion of p53 was confirmed by Western blotting with anti-p53 using a lung protein lysate. Molecular masses are given in kilodaltons. Data are shown as means ± SEM. WT, wild type.
Next, aortas were immunostained with anti-p53 and then with TO-PRO3 for nucleus staining, and confocal microscopy was used to acquire a z series of fluorescence images. No significant fluorescent signal was observed in aorta samples treated with nonimmune rabbit IgG (not depicted) or with antigen-preabsorbed anti-p53 (Fig. S5 A). In LP areas, p53 was localized mainly in the nucleus (Fig. 10, D and F). In contrast, in HP areas, significant levels of anti-p53 staining were detected outside the nucleus, although anti-p53 staining was still associated with the nucleus. This cytoplasmic staining was localized primarily to the area underneath the nucleus (Fig. 10, E and F). These results appear to suggest that d-flow causes p53, which is present mainly in the nucleus in ECs exposed to s-flow (i.e., LP area), to translocate into the cytoplasm especially to the area directly below the nucleus.
We also examined whether ECs in the HP area underwent more apoptosis than ECs in the LP area. Aortas of 7-wk-old C57BL/6 mice were immunostained first for VE-cadherin (Fig. S4 A, red) followed by the TUNEL staining (Fig. S4 A) or were coimmunostained for annexin V (Fig. S4 B, red) and VE-cadherin (Fig. S4 B, green). Although, in the LP area, TUNEL- and annexin V–positive cells were rarely detected, such cells were frequently detected in the HP area (Fig. S4, A and B). Next, we studied the role of p53 in EC apoptosis in the 7-wk-old p53-deficient C57BL/6 mice (p53−/−; Fig. 10, G and H). p53 was absent in lung ECs isolated from p53−/− mice (Fig. 10 H, right). Interestingly, annexin V–positive cells in the HP area were significantly decreased in the p53−/− mice (Fig. 10, G and H), supporting the critical role of p53 on d-flow–mediated apoptosis in vivo. Because we found the critical role of ONOO− in d-flow–mediated PKCζ activation, p53 SUMOylation, and apoptosis, we investigated the expression of nitrotyrosine-containing proteins by double staining aortas with anti–VE-cadherin and antinitrotyrosine (Fig. S5, B–D). Antinitrotyrosine staining was significantly higher in HP areas than in LP areas. Our data collectively suggest that d-flow induces ONOO− production, PKCζ activation, and nuclear export of p53, which increases EC apoptosis, and may prime ECs in the d-flow (HP) area to become susceptible to atherogenesis under the influence of various systemic risk factors.
Discussion
Several atheroprotective signals activated by s-flow have been identified, but atheroprone signaling activated by d-flow is not well understood. The main finding of this study is that d-flow–mediated ONOO− production activates PKCζ and induces p53 SUMOylation, p53 nuclear export, p53–Bcl-2 binding, and subsequent apoptosis. p53 appears to have both yin and yang effects on blood vessel physiology. Recent studies show its protective effect against cell death (Tian et al., 2000; Garner and Raj, 2008). Because p53 positively regulates the expression of p21, an antiapoptotic protein (Garner and Raj, 2008), p53 may exert its antiapoptotic effect by increasing p21 expression. Previously, Lin et al. (2000) have reported that laminar shear stress increased p53 expression and JNK-mediated p53 phosphorylation, which leads to endothelial growth arrest via increasing GADD45 and p21cip1 expression. Therefore, it is important to emphasize here that p53 can induce growth arrest by inhibiting apoptosis via regulating p21. The interplay between cell cycle arrest and apoptosis is possibly critical in maintaining endothelial function because p53 may be able to reduce cells with DNA damages from apoptosis by preventing entry into the S phase (Garner and Raj, 2008). However, it remains unclear how p53 determines which of these two activities, pro- or antiapoptotic, to implement. Of note, most of the p53 antiapoptotic effects have been explained by its nuclear localization, as nuclear p53 protects cells from apoptosis, especially under low stress conditions (Tian et al., 2000; Garner and Raj, 2008). Indeed, our current study has revealed the antiapoptotic localization of p53 in ECs in the LP area exposed to steady laminar stress, which we consider to be a low stress condition. In contrast, ECs exposed to d-flow and ONOO− had increased cytoplasmic p53 localization (i.e., nuclear export of p53), which enhanced EC apoptosis via increased p53–Bcl-2 binding.
Little information is available regarding the pathological significance of p53 subcellular localization, especially in ECs. Our study suggests that PKCζ activation by d-flow and ONOO− and subsequent p53 nuclear export promote EC apoptosis. This notion is consistent with an earlier study showing that human cytomegalovirus infection of ECs induces p53 accumulation in the cytoplasm and apoptosis via a p53-dependent pathway (Shen et al., 2004). Together, these results strongly suggest the involvement of cytoplasmic p53 in EC apoptosis (Utama et al., 2006). Our en face confocal data are consistent with this idea because increased cytoplasmic anti-p53 staining and increased TUNEL and annexin V stainings are found in ECs located in the area exposed to d-flow (i.e., HP area).
In our in vitro study, we found the importance of ONOO− on d-flow–mediated PKCζ activation, p53 SUMOylation, and EC apoptosis using ebselen, L-NAME, and Mn-TBAP (Fig. 5). In addition, we found that nitrotyrosine staining was significantly increased in d-flow (HP) areas in vivo (Fig. S5, B–D), where PKCζ activation, nuclear exports of p53, and EC apoptosis were increased. We used 10–100 µM ONOO− as the final concentration, which is in agreement with the concentration of ONOO− used in previous studies by other investigators (Alvarez et al., 2004; Levrand et al., 2005; Szabó et al., 2007). Because ONOO− is a transient intermediate in free radical chemistry and is highly reactive, it is difficult to measure actual concentrations of ONOO− in vivo. However, 10–100 µM ONOO− are thought to be physiological because it has been suggested that the rates of ONOO− production in vivo in specific compartments have been estimated to be as high as 50–100 µM/min (Alvarez et al., 2004; Levrand et al., 2005; Szabó et al., 2007). In addition, it has also been estimated that the rate of ONOO− generation may reach up to 1 mM/min in an inflamed organ (lung) in vivo (Ischiropoulos et al., 1992). ONOO− has several possible ways to activate PKCζ. One of the major effects of ONOO− is protein tyrosine nitration, which can regulate a variety of kinases (Liaudet et al., 2009). First, ONOO− can activate various receptor tyrosine kinases (Klotz et al., 2000; Zhang et al., 2000). Second, ONOO− can activate Src by displacing Tyr527 from its binding site to the SH2 domain (Roskoski, 2005). Finally, ONOO− can inhibit phosphatases via oxidation of cysteine-bound thiols (Takakura et al., 1999). Although the direct effect of ONOO− on PKCζ remains unclear, these molecules may be involved in signaling events upstream of PKCζ. Therefore, we hypothesize that activation of a variety of kinases may be involved in ONOO−-mediated PKCζ activation. Further studies are necessary to determine the major signaling events.
In addition to regulating cell physiology through kinase activities, active kinases can also regulate cell physiology by altering their interaction with other molecules (Akaike et al., 2004; Boggon and Eck, 2004). PKCζ contains the pseudosubstrate autoinhibitory sequence (aa 116–122), and the release of the kinase domain (aa 268–587) from this autoinhibitory domain leads to PKCζ activation (Newton, 2001; Smith et al., 2003). In this study, we were unable to show PKCζ-mediated phosphorylation of PIASy in our in vitro kinase assay (Fig. S3), but we found that activation of PKCζ increased the PKCζ–PIASy interaction (Fig. 8). Using a mammalian two-hybrid assay, we then determined the C-terminal kinase domain of PKCζ (aa 401–587) was a PIASy binding site (Fig. 8 C). Of note, the deletion of the N-terminal autoinhibitory domain of PKCζ (aa 1–200) increased PKCζ–PIASy association, suggesting that this domain is a negative regulator not only of its kinase activity but also its binding ability with PIASy. Therefore, releasing the PKCζ N-terminal inhibitory effect (i.e., activation of PKCζ) is critical for both kinase activation and PKCζ–PIASy association, and the regulation of PIASy activity most likely depends on PKCζ-mediated increase in PKCζ–PIASy association but not phosphorylation. We also found that PKCζ binds to the RING domain of PIASy, which contains its catalytic site and may alter the structure and enzymatic activity of PIASy. Our findings suggest that d-flow–mediated PKCζ–PIASy association is critical for p53 SUMOylation to induce the endothelial p53 nuclear export and apoptosis.
Materials and methods
Animals
C57BL/6 and p53−/− mice with a C57BL/6 background (P53N4-M) were purchased from Taconic. All mice were maintained under pathogen-free conditions at the Aab Cardiovascular Research Institute at the University of Rochester. All animal procedures were performed with the approval of the University Committee on Animal Resources at the University of Rochester.
Plasmid and adenovirus vector construction
The plasmid encoding human HA-SUMO3 was a gift from R.T. Hay (University of Manchester, Manchester, England, UK; Tatham et al., 2001). The pcDNA3-Myc-PIASy and pcDNA-HA-PKCζ constructs were gifts from M. Takahashi (Nagoya University, Chikusa-ku, Nagoya, Japan; Matsuura et al., 2005) and J.-W. Soh (Columbia University, New York, NY; Soh et al., 1999), respectively. The p53-luciferase (p53-luc) reporter (Addgene plasmid 16442; B. Vogelstein, Johns Hopkins University School of Medicine, Baltimore, MD; el-Deiry et al., 1993) and pcDNA-Flag-p53 (Addgene plasmid 10838; T. Roberts, Harvard Medical School, Boston, MA; el-Deiry et al., 1993; Gjoerup et al., 2001) were obtained from the nonprofit Addgene plasmid repository. The Gal4 wild-type and truncated forms of PKCζ were created by inserting a KpnI–XbaI fragment generated by PCR into the pBIND vector (Promega). V16-PIASy was created by inserting human PIASy isolated from pcDNA3-PIASy into BamH1 and EcoRV1 sites of the pACT vector. Truncated forms of PIASy were constructed by inserting fragments EcoRI–XhoI or BamHI–XhoI generated by PCR into the pGEX-KG or pCMV-Taq2B vector, respectively. All constructs were verified by DNA sequencing.
Antibodies, siRNA, adenovirus, and reagents
Rabbit and mouse anti-PKCζ (C-20 [SC-216] and A-3 [SC-17781]), rabbit and mouse anti-p53 (FL-393 [SC-6243] and DO-1 [SC-126]), rabbit and mouse anti–Bcl-2 (N-19 [SC-492] and C-2 [SC-7382]), rabbit and mouse anti-HA (Y-11 [SC-805] and F-7 [SC-7392]), and anti-myc (A-14; SC-789) were purchased from Santa Cruz Biotechnology, Inc. The phospho-specific antibodies for p-PKCζ were purchased from Cell Signaling Technology (Thr 410; 9378) and Abcam (Thr 560; ab62372). The rabbit and mouse anti-SUMO2/3 were purchased from Abgent (AP1224a) and MBL International (M114-3), respectively. Anti-PIASy was purchased from Abcam (ab58416) and Sigma-Aldrich (P0104). Rat anti–VE-cadherin was purchased from BD (555289). ON-TARGETplus SMARTpool for human PKCζ siRNA was purchased from Thermo Fisher Scientific (L-003526-00), and a nonspecific control siRNA obtained from Invitrogen (12935-122) was used as a negative control. The Ad-DN-PKCζ was purchased from Cell Biolabs. The Ad-DN-PKCζ was generated by mutating the ATP binding site (K281M) from human PKCζ (NCBI Protein database accession no. NP_002735; Diaz-Meco et al., 1993). GST-fused active recombinant human PKCζ was purchased from Stressgen (PPK-468). Peroxynitrite was purchased form EMD. Alexa Fluor 488–conjugated mouse antinitrotyrosine was purchased from Millipore (16–226).
Cell culture and transfection of the PKCζ and PIASy siRNAs
HUVECs were obtained from collagenase-digested umbilical cord veins (Takahashi and Berk, 1996) and collected in M200 medium supplemented with low serum growth supplement (Cascade Biological) and 5% fetal calf serum (Invitrogen). HUVECs were cultured on 0.2% gelatin-precoated dishes. For transient expression experiments, transfection of the PKCζ SMARTpool siRNA or PIASy siRNA was performed using Lipofectamine 2000 (Invitrogen) following protocols provided by the manufacturer. The Stealth RNAi Negative Control Med GC (Invitrogen), which has no homology to the vertebrate transcriptome, was used as a negative control. The cells were harvested 48 h after siRNA transfection, and protein expressions were monitored by immunoblotting with antibodies against PKCζ, PIASy, or tubulin. The target and control sequences for PIASy siRNAs were 5′-CAAGACAGGUGGAGUUGAU-3′ and 5′-UACCGUCUCCACUUGAUCG-3′.
Flow apparatus
To expose a large number of cells to flow, we used a cone and plate flow apparatus identical to the one used previously (Reinhart-King et al., 2008). Confluent HUVECs cultured in 100-mm dishes were exposed to s-flow in a flow apparatus placed in a cell culture incubator with 5% CO2 and at 37°C for long-term experiments (shear stress = 20 dyn/cm2). To expose cells to d-flow, we used a cone with radial grooves. Shear stress induced by d-flow cannot be calculated, but we rotated the cone with 50 rpm that gives 5 dyn/cm2 of laminar shear stress. We optimized and chose this speed because ECs maintained nonelongated cell shapes with this speed.
Analysis of apoptosis
In accordance with recently published guidelines, apoptosis was quantified by multiple methodologically unrelated criteria (Galluzzi et al., 2009). To induce apoptosis, HUVECs were treated with 100 µM ONOO− or d-flow. Phosphatidylserine externalization was quantified by staining cells with annexin V–Alexa Fluor 568 (Roche), and late-stage apoptosis was quantified morphologically by counting cells showing evidence of positive staining for TUNEL (Ding et al., 2000). TUNEL analyses were performed according to the manufacturer’s instructions, and the cells were counterstained with DAPI (Sigma-Aldrich) to identify nuclei. For each experiment in vitro, a total of 150–200 cells were counted from randomly selected fields of view, and TUNEL-positive cells were expressed as a percentage of total cells counted. Samples were examined using a 40× lens under a fluorescence microscope (BX51; Olympus) capable of imaging two distinct channels without adjustment of the microscope stage position. 10 random fields per sample were examined. Apoptosis was also quantified by Western blot assessment of the cleavage of the caspase 3 using the cleaved caspase 3 antibody. All measurements were performed blinded, and at least three independent experiments were performed.
Mammalian two-hybrid analysis and transfection of cells
Cells were plated in 12-well plates at 5 × 104 cells/well. The mammalian two-hybrid assay was performed as described previously (Woo et al., 2006). In brief, cells were transfected in Opti-MEM (Invitrogen) with Lipofectamine mixture containing the pG5-luc vector and various pBIND and pACT plasmids (Promega) for 4 h. Cells were washed, and fresh DME supplemented with 10% fetal bovine serum was added. The pBIND vector contains the yeast GAL4–DNA-binding domain upstream of a multiple cloning region, and the pACT vector contains the herpes simplex virus VP16 activation domain upstream of a multiple cloning region. Various PKCζ mutants and PIASy were cloned into the pBIND and pACT vector, respectively. Because pBIND also contains the Renilla luciferase gene, the expression and transfection efficiencies were normalized with the Renilla luciferase activity. Cells were collected 36 h after transfection unless indicated otherwise, and the luciferase activity was assayed with the Dual-Luciferase kit (Promega) using a luminometer (TD-20/20; Turner Designs). Transfections were performed in triplicate, and each experiment was repeated at least three times.
GST pull-down assay
HeLa cells (106 cells) were plated in 100-mm dishes and transfected in Opti-MEM with Lipofectamine mixture containing the Flag-tagged PIASy-truncated mutants for 4 h. Cells were washed and cultured for 24 h in fresh complete growth medium. Cell lysates were made by incubating cells in 0.5 ml radioimmunoprecipitation assay lysis buffer for 30 min at 4°C. After centrifugation for 20 min at 12,000 rpm, the whole-cell lysates were incubated with 100 ng of recombinant GST-fused PKCζ, anti-FLAG, and IgG Sepharose beads with constant mixing for 2–5 h at 4°C. The beads were then washed three times with lysis buffer, resuspended in SDS-PAGE sample buffer, and boiled for 8 min. Samples were separated on a 15% SDS-PAGE and analyzed by Western blotting using anti-PKCζ.
Immunohistochemistry and confocal microscopy
To determine whether the p53 localization is regulated by PKCζ activation, HUVECs grown in 6-well plates were transduced with Ad-LacZ or Ad-DN-PKCζ for 24 h. After treatment with 100 µM ONOO− for 4 h, the cells were quickly washed two times with cold PBS, fixed with 4% paraformaldehyde in PBS for 15 min, and then permeabilized with 0.2% Triton X-100 in PBS for 10 min. Cells were incubated with blocking buffer (5% goat serum and 0.1% NP-40 in PBS) for 60 min to block nonspecific binding and incubated with anti-p53 (1:200 dilution in 2% goat serum and 0.1% NP-40 in PBS) overnight at 4°C. The cells were washed three times with PBS and incubated with Alexa Fluor 546–labeled anti–rabbit IgG or Alexa Fluor 488–labeled anti–rabbit IgG1 (1:2,000 dilution; Invitrogen) for 1 h at room temperature. The cells were counterstained with DAPI to identify nuclei. All of the images were collected using an epifluorescence microscope (BX51) equipped with a charge-coupled device camera (Spot; Diagnostic Instruments, Inc.) and an Acroplan water 60× W lens.
To determine the colocalization of p53 with Bcl-2 or PKCζ with PIASy, HUVECs were stimulated with d-flow for 3 h. After fixation, permeabilization, and 10% goat serum in PBS with 0.1% NP-40 for 1 h, the cells were incubated with mouse anti-p53 and rabbit anti–Bcl-2 or mouse anti-PKCζ and rabbit anti-PIASy (1:200) antibodies in 2% goat serum with PBS overnight at 4°C. The cells were washed three times with PBS and incubated with Alexa Fluor 546–labeled anti–mouse IgG and Alexa Fluor 488–labeled anti–rabbit IgG1 (1:2,000 dilution) for 1 h at room temperature. The samples were analyzed using a laser-scanning confocal microscope (FV1000; Olympus) equipped with a Plapon 60× 1.42 NA oil lens objective. Quantification of the overlay image was performed using the Photoshop (CS; Adobe) program.
Immunoprecipitation (SUMO assay) and Western blot analysis
Cells were collected in PBS containing 10 mM N-ethylmaleimide, and cell extracts were prepared in modified radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM dithiothreitol, 1:200-diluted protease inhibitor cocktail [Sigma-Aldrich], 1 mM PMSF, 10 mM N-ethylmaleimide, and 0.1 mM iodoacetamide). Immunoprecipitation with a mouse monoclonal anti-Flag, HA, or p53 was performed as described previously (Woo et al., 2006). Bound proteins were released in 2× SDS sample buffer, resolved by SDS-PAGE, transferred onto an enhanced chemiluminescence nitrocellulose membrane (Hybond), and visualized by using the enhanced chemiluminescence detection reagents (GE Healthcare) according to the manufacturer’s instructions. p53 SUMOylation was detected by immunoprecipitation with anti-p53 followed by Western blotting with anti-SUMO2/3. Results were normalized to the lowest phosphorylation level within each set of experiments, and statistical significance was determined by comparing the mean level of the control group to each of the experimental data points.
En face immunohistochemistry
The s-flow and d-flow areas within the aorta were identified based on the published and generally accepted anatomical locations where such flow patterns are known to occur (Iiyama et al., 1999; Hajra et al., 2000; Jongstra-Bilen et al., 2006). For example, a typical s-flow area is located in the greater curvature area and is marked as an LP region for lesion formation (Hajra et al., 2000), which is also known as a high wall shear stress area. A d-flow area is the lesser curvature area (HP region; Iiyama et al., 1999; Hajra et al., 2000; Jongstra-Bilen et al., 2006), which is also indicated as a low wall shear stress area. In the Fig. 10 legend, we indicated these two areas in the aortic arch. EC shape outlined by anti–VE-cadherin staining was also used to identify s-flow areas (elongated cell shape) and d-flow areas (irregular cell shape). It has been reported that PECAM-1 (CD31) staining, another marker of ECs, was localized to endothelial junctions in the HP region, whereas in the LP region, it is more diffuse. Therefore, the PECAM-1 staining at EC borders was much stronger in the HP region than in the LP region (Hajra et al., 2000). We found the similar tendency of VE-cadherin staining in the mouse aorta. Because these data were very consistent in every mouse, we do not think that this is caused by the damage that specifically occurred in the LP region. C57BL/6 wild-type mice (Taconic) were fed standard chow. Animals of 6–8 wk of age were euthanized by CO2 inhalation. The arterial tree was perfused via the left ventricle with saline containing 40 USPU/ml heparin followed by 4% paraformaldehyde in PBS for 10 min. After adipose tissues were removed, aortas were cut open longitudinally and permeabilized with PBS containing 0.1% Triton X-100 and blocked by TBS containing 10% goat serum and 2.5% Tween 20 for 30 min. Aortas were incubated with 10 µg/ml rabbit anti-p53 (FL-393; Santa Cruz Biotechnology, Inc.; rabbit IgG was used as a control) and 7 µg/ml rat anti–VE-cadherin (an EC marker; BD) in the blocking solution overnight. Specificity of anti-p53 staining was tested by staining with anti-p53 preabsorbed with recombinant p53 (Santa Cruz Biotechnology, Inc.) as shown in Fig. S4. After a PBS rinse, anti–rabbit IgG and anti–rat IgG (1:1,000 dilution; Alexa Fluor 546 and 488, respectively; Invitrogen) were applied for 1 h at room temperature. Nuclei were stained using TO-PRO3 (Invitrogen). For the detection of apoptosis, we used two different methods. To perform an in situ TUNEL assay, aortas immunostained with anti–VE-cadherin (with Alexa Fluor 546 anti–rabbit IgG) were incubated in the TUNEL reaction mixture (Roche) for 1 h at 37°C according to the manufacturer’s instructions (mixture without the enzyme TdT as a control). For annexin V labeling, we injected annexin V–Alexa Fluor 568 into aortas via the left ventricle after perfusion with saline containing 40 USPU/ml heparin followed by 4% paraformaldehyde in PBS for 10 min. Then, these aortas were immunostained with anti–VE-cadherin as described in the previous paragraph. Images were acquired using a confocal laser-scanning microscope (Fluoview 300; Olympus) equipped with krypton/argon/HeNe laser lines and 20× 0.70 NA, 40× 1.0 NA, and 60× 1.4 NA objectives. For quantification of phospho- and total PKCζ expression level, 10–15 optical sections were collected at 0.3–0.5-µm increments so that z stacks of ∼4-µm-thick cell blocks from the luminal surface were obtained. Images were collected using the same confocal settings. For quantification of EC apoptosis, aortas of 7-wk-old wild-type mice (n = 7 each) were prepared. Images were acquired as described in the previous paragraph. To analyze p53 subcellular localization, we took z-stack clip images of 50 optical sections collected at 0.1-µm increments from the luminal surface. We radially extended the edge of the nucleus by the length of the nuclear radius at each point along the edge of the nucleus and delineated an area for each cell. We defined the signal level in this area as total because practically all anti-p53 staining signals (>80–90%) were included in this area. Staining signal levels in the cytoplasm and the nucleus were determined. Confocal images were obtained from s-flow and d-flow areas as shown in Fig. 10.
Statistical analysis
Data are reported as means ± SD. Statistical analysis was performed with the Prism program version 2.00 (GraphPad Software). Differences were analyzed with a one-way or a two-way repeated-measure analysis of variance as appropriate followed by Dunnett’s correction for multiple comparisons. P < 0.05 is indicated by an asterisk, and P < 0.01 is indicated by a double asterisk.
Online supplemental material
Fig. S1 shows that ONOO− increases p53 SUMOylation and p53–Bcl-2 binding. Fig. S2 shows d-flow–induced p53 nuclear export analyzed by immunostaining. Fig. S3 shows whether PKCζ phosphorylates PIASy in vitro by in vitro kinase assay. Fig. S4 shows apoptosis in d-flow and s-flow area of the mouse aortic arch endothelia analyzed by TUNEL and annexin V staining. Fig. S5 shows specificity of anti-p53 staining and nitrotyrosine staining in mouse aortic arch endothelia. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010051/DC1.
Supplementary Material
Acknowledgments
This work is supported by grants from the America Heart Association to Dr. Woo (Postdoctoral Fellowship 0625957T and Scientist Development Grant 0930360N) and to Dr. Le (Postdoctoral Fellowship 4360007) and from the National Institutes of Health to Drs. Abe (HL-064839, HL-077789, and HL-102746) and Berk (HL-064839 and HL-077789). Dr. Abe is a recipient of Established Investigator Awards of the American Heart Association (0740013N).
Footnotes
Abbreviations used in this paper:
- Bcl-2
- B cell lymphoma/leukemia-2
- DN
- dominant negative
- EC
- endothelial cell
- HP
- high probability
- HUVEC
- human umbilical vein EC
- L-NAME
- N-nitro-l-arginine methyl ester
- LP
- low probability
- Mn-TBAP
- Mn (III)tetrakis(4-benzoic acid)porphyrin chloride
- NES
- nuclear export signal
- PIASy
- protein inhibitor of activated STATy
- VE
- vascular endothelial
References
- Akaike M., Che W., Marmarosh N.L., Ohta S., Osawa M., Ding B., Berk B.C., Yan C., Abe J.. 2004. The hinge-helix 1 region of peroxisome proliferator-activated receptor gamma1 (PPARgamma1) mediates interaction with extracellular signal-regulated kinase 5 and PPARgamma1 transcriptional activation: involvement in flow-induced PPARgamma activation in endothelial cells. Mol. Cell. Biol. 24:8691–8704. 10.1128/MCB.24.19.8691-8704.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M.N., Piacenza L., Irigoín F., Peluffo G., Radi R.. 2004. Macrophage-derived peroxynitrite diffusion and toxicity to Trypanosoma cruzi. Arch. Biochem. Biophys. 432:222–232. 10.1016/j.abb.2004.09.015 [DOI] [PubMed] [Google Scholar]
- Bischof O., Schwamborn K., Martin N., Werner A., Sustmann C., Grosschedl R., Dejean A.. 2006. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell. 22:783–794. 10.1016/j.molcel.2006.05.016 [DOI] [PubMed] [Google Scholar]
- Boggon T.J., Eck M.J.. 2004. Structure and regulation of Src family kinases. Oncogene. 23:7918–7927. 10.1038/sj.onc.1208081 [DOI] [PubMed] [Google Scholar]
- Carter S., Bischof O., Dejean A., Vousden K.H.. 2007. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9:428–435. 10.1038/ncb1562 [DOI] [PubMed] [Google Scholar]
- Cheng J., Wang D., Wang Z., Yeh E.T.. 2004. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 24:6021–6028. 10.1128/MCB.24.13.6021-6028.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Meco M.T., Berra E., Municio M.M., Sanz L., Lozano J., Dominguez I., Diaz-Golpe V., Lain de Lera M.T., Alcamí J., Payá C.V., et al. 1993. A dominant negative protein kinase C zeta subspecies blocks NF-kappa B activation. Mol. Cell. Biol. 13:4770–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Price R.L., Goldsmith E.C., Borg T.K., Yan X., Douglas P.S., Weinberg E.O., Bartunek J., Thielen T., Didenko V.V., Lorell B.H.. 2000. Left ventricular hypertrophy in ascending aortic stenosis mice: anoikis and the progression to early failure. Circulation. 101:2854–2862. [DOI] [PubMed] [Google Scholar]
- el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B.. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell. 75:817–825. 10.1016/0092-8674(93)90500-P [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Aaronson S.A., Abrams J., Alnemri E.S., Andrews D.W., Baehrecke E.H., Bazan N.G., Blagosklonny M.V., Blomgren K., Borner C., et al. 2009. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16:1093–1107. 10.1038/cdd.2009.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin G., Abe J.I., Mohan A., Lu W., Yan C., Newby A.C., Rhaman A., Berk B.C.. 2007. Flow antagonizes TNF-alpha signaling in endothelial cells by inhibiting caspase-dependent PKC zeta processing. Circ. Res. 101:97–105. 10.1161/CIRCRESAHA.107.148270 [DOI] [PubMed] [Google Scholar]
- Garner E., Raj K.. 2008. Protective mechanisms of p53-p21-pRb proteins against DNA damage-induced cell death. Cell Cycle. 7:277–282. 10.4161/cc.7.3.5328 [DOI] [PubMed] [Google Scholar]
- Garner E., Martinon F., Tschopp J., Beard P., Raj K.. 2007. Cells with defective p53-p21-pRb pathway are susceptible to apoptosis induced by p84N5 via caspase-6. Cancer Res. 67:7631–7637. 10.1158/0008-5472.CAN-07-0334 [DOI] [PubMed] [Google Scholar]
- Ghosh A., Stewart D., Matlashewski G.. 2004. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell. Biol. 24:7987–7997. 10.1128/MCB.24.18.7987-7997.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoerup O., Zaveri D., Roberts T.M.. 2001. Induction of p53-independent apoptosis by simian virus 40 small t antigen. J. Virol. 75:9142–9155. 10.1128/JVI.75.19.9142-9155.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra L., Evans A.I., Chen M., Hyduk S.J., Collins T., Cybulsky M.I.. 2000. The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc. Natl. Acad. Sci. USA. 97:9052–9057. 10.1073/pnas.97.16.9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgarth R.S., Murphy L.A., Skaggs H.S., Wilkerson D.C., Xing H., Sarge K.D.. 2004. Regulation and function of SUMO modification. J. Biol. Chem. 279:53899–53902. 10.1074/jbc.R400021200 [DOI] [PubMed] [Google Scholar]
- Hsiai T.K., Hwang J., Barr M.L., Correa A., Hamilton R., Alavi M., Rouhanizadeh M., Cadenas E., Hazen S.L.. 2007. Hemodynamics influences vascular peroxynitrite formation: Implication for low-density lipoprotein apo-B-100 nitration. Free Radic. Biol. Med. 42:519–529. 10.1016/j.freeradbiomed.2006.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.L., Li S., Shyy J.Y., Chien S.. 1999. Sustained JNK activation induces endothelial apoptosis: studies with colchicine and shear stress. Am. J. Physiol. 277:H1593–H1599. [DOI] [PubMed] [Google Scholar]
- Iiyama K., Hajra L., Iiyama M., Li H., DiChiara M., Medoff B.D., Cybulsky M.I.. 1999. Patterns of vascular cell adhesion molecule-1 and intercellular adhesion molecule-1 expression in rabbit and mouse atherosclerotic lesions and at sites predisposed to lesion formation. Circ. Res. 85:199–207. [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H., Zhu L., Beckman J.S.. 1992. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 298:446–451. 10.1016/0003-9861(92)90433-W [DOI] [PubMed] [Google Scholar]
- Jongstra-Bilen J., Haidari M., Zhu S.N., Chen M., Guha D., Cybulsky M.I.. 2006. Low-grade chronic inflammation in regions of the normal mouse arterial intima predisposed to atherosclerosis. J. Exp. Med. 203:2073–2083. 10.1084/jem.20060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S.E., Pietenpol J.A., Thiagalingam S., Seymour A., Kinzler K.W., Vogelstein B.. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 256:827–830. 10.1126/science.1589764 [DOI] [PubMed] [Google Scholar]
- Klotz L.O., Schieke S.M., Sies H., Holbrook N.J.. 2000. Peroxynitrite activates the phosphoinositide 3-kinase/Akt pathway in human skin primary fibroblasts. Biochem. J. 352:219–225. 10.1042/0264-6021:3520219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek S.S., Derry J., Tyner A.L., Shen Z., Gudkov A.V.. 2001. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene. 20:2587–2599. 10.1038/sj.onc.1204362 [DOI] [PubMed] [Google Scholar]
- Levrand S., Pesse B., Feihl F., Waeber B., Pacher P., Rolli J., Schaller M.D., Liaudet L.. 2005. Peroxynitrite is a potent inhibitor of NF-kappaB activation triggered by inflammatory stimuli in cardiac and endothelial cell lines. J. Biol. Chem. 280:34878–34887. 10.1074/jbc.M501977200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hahn C.N., Parsons M., Drew J., Vadas M.A., Gamble J.R.. 2004. Role of protein kinase Czeta in thrombin-induced endothelial permeability changes: inhibition by angiopoietin-1. Blood. 104:1716–1724. 10.1182/blood-2003-11-3744 [DOI] [PubMed] [Google Scholar]
- Liaudet L., Vassalli G., Pacher P.. 2009. Role of peroxynitrite in the redox regulation of cell signal transduction pathways. Front. Biosci. 14:4809–4814. 10.2741/3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Hsu P.P., Chen B.P., Yuan S., Usami S., Shyy J.Y., Li Y.S., Chien S.. 2000. Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc. Natl. Acad. Sci. USA. 97:9385–9389. 10.1073/pnas.170282597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magid R., Davies P.F.. 2005. Endothelial protein kinase C isoform identity and differential activity of PKCzeta in an athero-susceptible region of porcine aorta. Circ. Res. 97:443–449. 10.1161/01.RES.0000179767.37838.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T., Shimono Y., Kawai K., Murakami H., Urano T., Niwa Y., Goto H., Takahashi M.. 2005. PIAS proteins are involved in the SUMO-1 modification, intracellular translocation and transcriptional repressive activity of RET finger protein. Exp. Cell Res. 308:65–77. 10.1016/j.yexcr.2005.04.022 [DOI] [PubMed] [Google Scholar]
- Mercer J., Figg N., Stoneman V., Braganza D., Bennett M.R.. 2005. Endogenous p53 protects vascular smooth muscle cells from apoptosis and reduces atherosclerosis in ApoE knockout mice. Circ. Res. 96:667–674. 10.1161/01.RES.0000161069.15577.ca [DOI] [PubMed] [Google Scholar]
- Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U.M.. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 11:577–590. 10.1016/S1097-2765(03)00050-9 [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F., Slee E.A., Lu X.. 2008. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 9:702–712. 10.1038/nrm2451 [DOI] [PubMed] [Google Scholar]
- Newton A.C. 2001. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 101:2353–2364. 10.1021/cr0002801 [DOI] [PubMed] [Google Scholar]
- O’Keefe K., Li H., Zhang Y.. 2003. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 23:6396–6405. 10.1128/MCB.23.18.6396-6405.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuswamy P., Ostermeier E., Schröttle A., Chen J., Huang P.L., Ertl G., Nieswandt B., Kuhlencordt P.J.. 2009. Oxidative stress and compartment of gene expression determine proatherosclerotic effects of inducible nitric oxide synthase. Am. J. Pathol. 174:2400–2410. 10.2353/ajpath.2009.080730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart-King C.A., Fujiwara K., Berk B.C.. 2008. Physiologic stress-mediated signaling in the endothelium. Methods Enzymol. 443:25–44. 10.1016/S0076-6879(08)02002-8 [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr 2005. Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 331:1–14. 10.1016/j.bbrc.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R.. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088–3103. 10.1101/gad.944801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.H., Utama B., Wang J., Raveendran M., Senthil D., Waldman W.J., Belcher J.D., Vercellotti G., Martin D., Mitchelle B.M., Wang X.L.. 2004. Human cytomegalovirus causes endothelial injury through the ataxia telangiectasia mutant and p53 DNA damage signaling pathways. Circ. Res. 94:1310–1317. 10.1161/01.RES.0000129180.13992.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L., Wang Z., Smith J.B.. 2003. Caspase processing activates atypical protein kinase C zeta by relieving autoinhibition and destabilizes the protein. Biochem. J. 375:663–671. 10.1042/BJ20030926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh J.W., Lee E.H., Prywes R., Weinstein I.B.. 1999. Novel roles of specific isoforms of protein kinase C in activation of the c-fos serum response element. Mol. Cell. Biol. 19:1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P., Xie Z., Wu Y., Xu J., Dong Y., Zou M.H.. 2008. Protein kinase Czeta-dependent LKB1 serine 428 phosphorylation increases LKB1 nucleus export and apoptosis in endothelial cells. J. Biol. Chem. 283:12446–12455. 10.1074/jbc.M708208200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Szabó C., Ischiropoulos H., Radi R.. 2007. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov. 6:662–680. 10.1038/nrd2222 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Berk B.C.. 1996. Mitogen-activated protein kinase (ERK1/2) activation by shear stress and adhesion in endothelial cells. Essential role for a herbimycin-sensitive kinase. J. Clin. Invest. 98:2623–2631. 10.1172/JCI119083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura K., Beckman J.S., MacMillan-Crow L.A., Crow J.P.. 1999. Rapid and irreversible inactivation of protein tyrosine phosphatases PTP1B, CD45, and LAR by peroxynitrite. Arch. Biochem. Biophys. 369:197–207. 10.1006/abbi.1999.1374 [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M., Botting C.H., Naismith J.H., Hay R.T.. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368–35374. 10.1074/jbc.M104214200 [DOI] [PubMed] [Google Scholar]
- Tian H., Wittmack E.K., Jorgensen T.J.. 2000. p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 60:679–684. [PubMed] [Google Scholar]
- Traub O., Berk B.C.. 1998. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler. Thromb. Vasc. Biol. 18:677–685. [DOI] [PubMed] [Google Scholar]
- Utama B., Shen Y.H., Mitchell B.M., Makagiansar I.T., Gan Y., Muthuswamy R., Duraisamy S., Martin D., Wang X., Zhang M.X., et al. 2006. Mechanisms for human cytomegalovirus-induced cytoplasmic p53 sequestration in endothelial cells. J. Cell Sci. 119:2457–2467. 10.1242/jcs.02974 [DOI] [PubMed] [Google Scholar]
- van Vlijmen B.J., Gerritsen G., Franken A.L., Boesten L.S., Kockx M.M., Gijbels M.J., Vierboom M.P., van Eck M., van De Water B., van Berkel T.J., Havekes L.M.. 2001. Macrophage p53 deficiency leads to enhanced atherosclerosis in APOE*3-Leiden transgenic mice. Circ. Res. 88:780–786. 10.1161/hh0801.089261 [DOI] [PubMed] [Google Scholar]
- Witty J., Aguilar-Martinez E., Sharrocks A.D.. 2010. SENP1 participates in the dynamic regulation of Elk-1 SUMOylation. Biochem. J. 428:247–254. 10.1042/BJ20091948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won D., Zhu S.N., Chen M., Teichert A.M., Fish J.E., Matouk C.C., Bonert M., Ojha M., Marsden P.A., Cybulsky M.I.. 2007. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am. J. Pathol. 171:1691–1704. 10.2353/ajpath.2007.060860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.H., Massett M.P., Shishido T., Itoh S., Ding B., McClain C., Che W., Vulapalli S.R., Yan C., Abe J.. 2006. ERK5 activation inhibits inflammatory responses via peroxisome proliferator-activated receptor delta (PPARdelta) stimulation. J. Biol. Chem. 281:32164–32174. 10.1074/jbc.M602369200 [DOI] [PubMed] [Google Scholar]
- Woo C.H., Shishido T., McClain C., Lim J.H., Li J.D., Yang J., Yan C., Abe J.I.. 2008. Extracellular signal-regulated kinase 5 SUMOylation antagonizes shear stress-induced antiinflammatory response and endothelial nitric oxide synthase expression in endothelial cells. Circ. Res. 102:538–545. 10.1161/CIRCRESAHA.107.156877 [DOI] [PubMed] [Google Scholar]
- Yeh E.T. 2009. SUMOylation and De-SUMOylation: wrestling with life’s processes. J. Biol. Chem. 284:8223–8227. 10.1074/jbc.R800050200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wang Y.Z., Kagan E., Bonner J.C.. 2000. Peroxynitrite targets the epidermal growth factor receptor, Raf-1, and MEK independently to activate MAPK. J. Biol. Chem. 275:22479–22486. 10.1074/jbc.M910425199 [DOI] [PubMed] [Google Scholar]
- Zhong L.T., Sarafian T., Kane D.J., Charles A.C., Mah S.P., Edwards R.H., Bredesen D.E.. 1993. bcl-2 inhibits death of central neural cells induced by multiple agents. Proc. Natl. Acad. Sci. USA. 90:4533–4537. 10.1073/pnas.90.10.4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.