UniFrac is a β-diversity measure that uses phylogenetic information to compare environmental samples. UniFrac, coupled with standard multivariate statistical techniques including principal coordinates analysis (PCoA), identifies factors explaining differences among microbial communities. A recent simulation study concluded that UniFrac is unsuitable as a distance metric and should not be used for multivariate analysis (Schloss, 2008). We counter this argument by reassessing the data that led to this conclusion and by providing a mathematical proof showing that UniFrac is a distance metric. However, we confirm with actual sequence data that UniFrac values can be influenced by the number of sequences/sample, and recommend sequence jackknifing (that is, determining how often the cluster results are recovered using random subsets of the data) to avoid this issue.
UniFrac (Lozupone and Knight, 2005) has been applied in >150 research publications seeking to understand relationships among microbial communities in systems ranging from human disease to general ecology. UniFrac measures the difference between two collections of sequences (for example, 16S rRNA molecules sequenced from different microbial samples) as the amount of evolutionary history that is unique to either of the two, which is measured as the fraction of branch length in a phylogenetic tree that leads to descendents of one sample or the other but not both. There is also a weighted version that directly accounts for differences in relative abundances that can produce different but complementary results (Lozupone et al., 2007). UniFrac can be used to test if the phylogenetic lineages between samples are significantly different, or to cluster many samples using multivariate statistical techniques. UniFrac's widespread application is facilitated by its user-friendly web interface that handles large data sets from next-generation sequencing technologies (Hamady et al., 2010). UniFrac is also implemented in the QIIME (Caporaso et al., 2010) and mothur (Schloss et al., 2009) microbial community sequence analysis pipelines.
Sequence-based studies of 10–1000 microbial samples are now common due to advances in sequencing technologies, and increase the value of interpreting UniFrac distances using multivariate statistics, such as PCoA and hierarchical clustering. Multivariate analyses with UniFrac frequently recover biologically meaningful patterns from complex data sets, such as differences in the fecal bacteria of dogs fed with different diets (Middelbos et al., 2010) or between wet and dry soils (Castro et al., 2010).
Thus, UniFrac performs well on real data. Why might theoretical simulations disagree? In recent simulations, it was concluded that UniFrac could be used to determine whether communities were significantly different, but should not be used as a distance metric for multivariate statistical analyses. In these simulations, communities with ‘known' diversity and overlap were estimated by drawing ellipses around random points in two-dimensional space, where larger ellipses represent more diverse communities and overlap between ellipses represents the shared community membership (Schloss, 2008). We argue that even these unrealistic two-dimensional simulations demonstrate UniFrac's effectiveness.
The recommendation against using UniFrac as a distance measure is based on two assertions: (1) it does not exhibit a consistent linear correlation with the fraction of overlap between communities and (2) its values are sensitive to sampling. The first assertion is unsupported by the simulations, where community overlap correlates well with unweighted (r=0.97) and weighted (r=0.90) UniFrac measures (Figure 1; Schloss, 2008). The second assertion is based on resampling the data with 50–1000 individuals/community under three community overlap conditions: 0%, 80% and 100%. As the sequences/sample increased, mean UniFrac values stayed constant and standard deviations decreased. In contrast, mean-weighted UniFrac values decreased with sampling, particularly in the communities with 0% overlap (Schloss, 2008).
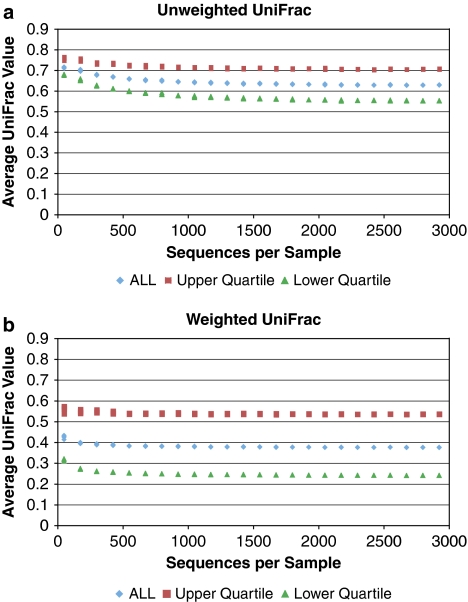
Figure 1.
Rarefaction of data from a study of obese twins (Turnbaugh et al., 2009). This study produced >1 million reads from the V2 region of ribosomal RNA using pyrosequencing. The samples with less than 3000 sequences were first excluded (leaving 112 samples). For five replicate trials, sequences from all 112 samples were subsampled so that each sample had a set number of sequences (between 50 and 2925 with a step size of 125). Pairwise UniFrac values were calculated with both the unweighted (a) and weighted (b) versions for all pairs of samples. To assess the effects of community divergence (the raw UniFrac value) on the sensitivity to sampling, the most similar and most different pairs of samples were identified from the most heavily subsampled data set (2925 sequences per sample) as those in the upper and lower quartile of UniFrac values respectively (calculated separately for unweighted and weighted). The points represent the average UniFrac value at each sample depth for (1) all pairwise comparisons and (2) the pairs that were identified as being in the upper and lower quartiles. Individual points for each of the five replicate trials are plotted, but the values of the replicates were close enough that they are generally on top of each other except for the smallest subsamples.
The decrease in unweighted UniFrac standard deviations with increased sampling indicates that deeper sequencing can help resolve relationships among similar samples because the whole-population value is more reliably estimated. Even at the smallest sample size in the simulations (50 sequences), however, identical communities had the highest UniFrac values and ‘no overlap' the lowest, reinforcing the strong correlation between community overlap and the UniFrac values (Figure 2; Schloss, 2008). We find that more similar communities do require more sequencing to reliably recover their relationships. For instance, in a recent reanalysis of 16S rRNA sequences from a survey of bacterial communities in different human body habitats (for example, the gut, mouth, skin and so on) in which subsampling was used to investigate how many sequences would have been required to recover the results, just 10 sequences were sufficient to show that communities were more similar within individuals than between individuals, but insufficient to reliably detect variability within an individual over time (Kuczynski et al., 2010).
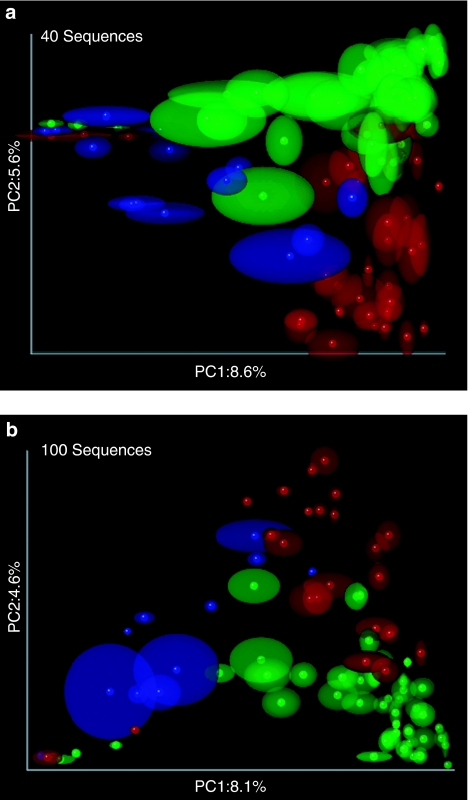
Figure 2.
The results of PCoA jackknifing of the bacteria from the stool of 106 individuals from 60 mammal species reported in Ley et al. (2008) for 100 replicates with unweighted UniFrac and (a) 40 or (b) 100 sequences. The full data set had between 21 and 1060 sequences/sample and the main clustering was explained by diet. This clustering pattern was recaptured with only 40 sequences/sample with the herbivores (green), omnivores (red) and carnivores (blue) largely clustering with each other (samples with less than 40 sequences were excluded from the analysis). In total, 100 sequences/sample show the same trend but with less variability in the point distribution, consistent with the decrease in the standard deviation with sample depth detected in the simulations. These plots were made using QIIME (Caporaso et al., 2010), which supplies a 3D view in which the confidence ellipses for selected PC axes can be viewed dynamically.
In contrast, the reported decrease in the average-weighted UniFrac values with sampling depth is of concern to microbiologists. With uneven sampling, as is common in pooled pyrosequencing because of normalization issues, the communities represented by fewer sequences will appear artificially different. To test whether this effect occurs in real data, we calculated average UniFrac values from subsamples of data from a recent study of obesity and the human gut microbiota (Turnbaugh et al., 2009), and confirm that small samples can inflate weighted UniFrac values (Figure 1). Similar to the simulation results, these effects were most pronounced for the most divergent pairs of samples. Unlike in the simulations, mean-unweighted UniFrac values also decreased with increased sampling. The sampling effect was actually stronger for the unweighted measure in this particular example, with the weighted having inflated values in only the smallest sample sizes (Figure 1).
Sensitivity to sampling depth is not unique to UniFrac; simulations have detected the same trend for the Jaccard and Sørenson indices of compositional similarity (Chao et al., 2005), which have been widely applied in the microbiology and ecological literature. Inflated distances at small sample sizes are because of shared rare species being falsely scored as unique because they were detected in one sample but not the other (Chao et al., 2005). Sampling depth effects were particularly strong for diverse assemblages with a high fraction of rare species, which is the most common structure for microbial populations.
We had previously conjectured that sample depth and evenness could affect UniFrac cluster results, and a jackknifing protocol that assesses the robustness of UniFrac hierarchical cluster nodes to these factors is implemented in the UniFrac web interfaces (Hamady et al., 2010). In the jackknifing technique, samples are subsampled evenly for n replicate trials and UniFrac distance matrices are calculated for each replicate. The results are summarized as the frequency that each node in the full-data cluster is supported among the replicates. We have also used sequence jackknifing to visualize the effects of sample depth and evenness on PCoA results (Ley et al., 2008, Lozupone et al., 2007). This functionality is available in QIIME (Caporaso et al., 2010). In PCoA jackknifing, Procrustes analysis is first applied to each PCoA replicate to scale and orient the axes, and the average position of each sample in each principal coordinate axis is plotted, surrounded by an ellipse representing the interquartile range of the distribution of points among the replicates (Figure 2). Jackknifing techniques have often shown support for biologically relevant clustering patterns identified with uneven sampling (Castro et al., 2010, Ley et al., 2008, Lozupone and Knight, 2005, Lozupone et al., 2007, Middelbos et al., 2010).
We emphasize that sensitivity to sampling depth does not make UniFrac unsuitable as a distance metric, and believe there may be confusion in the microbial ecology literature about what a distance metric actually is. In this study, we present a mathematical proof that both weighted and unweighted UniFrac values have the formal requirements of a distance metric (Rudin, 1987): they are always non-negative, symmetric and satisfy the triangle inequality, and identical sequence sets have a value of 0 (Supplementary Methods). The sensitivity to sampling depth indicates that standardizing the number of sequences/sample or jackknifing the sequences is recommended, whether using UniFrac or any other ecological distance/similarity measure. Adapting recently described approaches to correct for these effects for the Jaccard and Sørenson indices (Chao et al., 2005) to UniFrac is also something that we are currently pursuing.
In conclusion, we emphasize that despite potential shortcomings of ecological distance measures, such as UniFrac, in undersampled environments, pairing them with multivariate statistical methods is still a powerful method for analyzing the complex data sets that are now commonly generated in microbial ecology, with many samples and extensive metadata. Their application allows for visualization of which measured variables correlate best with differences between samples. In contrast, the significance tests extensively investigated in the simulations of Schloss (2008) are only described in the context of determining whether two communities differ significantly. These significance tests can only be extended to complex data sets by performing many pairwise tests. This approach lacks power because of the need to correct for multiple comparisons, such as with the Bonferroni correction. Finally, we note that the P-value for a particular pair of samples is also affected by sampling depth, with significant P-values potentially reflecting that certain pairs were sequenced deeper, and not that they are particularly divergent. We hope these conclusions and recommendations will assist microbial ecologists in choosing the correct tools to answer their biological questions.
Acknowledgments
CA Lozupone was funded by the NIH Training grant, 5T15 LM009451-03, Computational Bioscience Program, University of Colorado Denver. Mike Robeson provided valuable feedback on the paper.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol. 2010;76:999–1007. doi: 10.1128/AEM.02874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Chazdon RL, Colwell RK, Shen TJ. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett. 2005;8:148–159. [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Costello EK, Nemergut DR, Zaneveld J, Lauber CL, Knights D, et al. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol. 2010;11:210. doi: 10.1186/gb-2010-11-5-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelbos IS, Vester Boler BM, Qu A, White BA, Swanson KS, Fahey GC., Jr Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One. 2010;5:e9768. doi: 10.1371/journal.pone.0009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin W.1987Real and Complex Analysisvol. 3rd edn.McGraw-Hills: New York [Google Scholar]
- Schloss PD. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2008;2:265–275. doi: 10.1038/ismej.2008.5. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.