Abstract
Shewanella oneidensis MR-1 is capable of forming highly structured surface-attached communities. By DNase I treatment, we demonstrated that extracellular DNA (eDNA) serves as a structural component in all stages of biofilm formation under static and hydrodynamic conditions. We determined whether eDNA is released through cell lysis mediated by the three prophages LambdaSo, MuSo1 and MuSo2 that are harbored in the genome of S. oneidensis MR-1. Mutant analyses and infection studies revealed that all three prophages may individually lead to cell lysis. However, only LambdaSo and MuSo2 form infectious phage particles. Phage release and cell lysis already occur during early stages of static incubation. A mutant devoid of the prophages was significantly less prone to lysis in pure culture. In addition, the phage-less mutant was severely impaired in biofilm formation through all stages of development, and three-dimensional growth occurred independently of eDNA as a structural component. Thus, we suggest that in S. oneidensis MR-1 prophage-mediated lysis results in the release of crucial biofilm-promoting factors, in particular eDNA.
Keywords: Shewanella, biofilm, eDNA, lysis, phage
Introduction
Shewanella oneidensis MR-1 belongs to the Gram-negative γ-proteobacteria and is characterized by an enormous respiratory versatility, which allows this species to use an impressive variety of organic and inorganic compounds as alternative terminal electron acceptors when growing under anaerobic conditions (Myers and Nealson, 1988; Venkateswaran et al., 1999). The group of alternative electron acceptors includes metal ions such as Fe(III) and Mn(IV), which are highly abundant in soils and sediments. In addition, a number of radionucleotide oxides can be reduced (Myers and Nealson, 1988; Nealson and Scott, 2003; Icopini et al., 2009). Thus, bacteria such as Shewanella significantly impact biogeochemical cycling processes and are of particular interest with regard to bioremediation processes (Heidelberg et al., 2002; Nealson et al., 2002; Lovley et al., 2004; Ward et al., 2004; Hau and Gralnick, 2007). It has been hypothesized that direct interaction of Shewanella cells with, or close proximity to, an appropriate surface facilitates the deposition of electrons (Das and Caccavo, 2000; Gorby et al., 2006; McLean et al., 2010). In fact, Shewanella species have been demonstrated to adhere to various surfaces and form biofilms (Bagge et al., 2001; Thormann et al., 2004, 2005, 2006; Teal et al., 2006; McLean et al., 2008a; Zhang et al., 2010).
Previous work has demonstrated that, under aerobic hydrodynamic conditions, S. oneidensis biofilm development proceeds via initial attachment of single cells, subsequent surface coverage and finally in the formation of pronounced three-dimensional structures (Thormann et al., 2004; Teal et al., 2006; McLean et al., 2008a). Generally, formation of these structures is thought to rely on the release of extracellular polymeric substances, such as proteins, polysaccharides, lipids or extracellular DNA (eDNA) (Sutherland, 2001; Branda et al., 2005; Flemming et al., 2007). The identity and composition of the extracellular components that are critical for S. oneidensis MR-1 community architecture is still unknown. Mutant analyses have identified several factors whose absence leads to aberrant biofilm formation. One of these factors is type IV pili, which were demonstrated to be required for initial surface attachment and are also implicated in mediating tight cell–cell interactions (Thormann et al., 2004; McLean et al., 2008b; Saville et al., 2010). A similar role has been attributed to the outer membrane protein AggA, which is thought to represent a component of a protein transporter system similar to the Pseudomonas fluorescens Lap system (De Vriendt et al., 2005; De Windt et al., 2006; McLean et al., 2008b; Theunissen et al., 2009). In addition, a four-gene locus, termed mxdABCD, was identified to be critical for three-dimensional growth of the community and is assumed to be involved in the production and/or maintenance of the extracellular matrix (Thormann et al., 2006; Saville et al., 2010). MxdB most likely is a glycosyl transferase, suggesting that mxd might contribute to matrix formation by synthesis of a polysaccharide.
It has long been established that proteins and polysaccharides have an important structural role in bacterial biofilms. In contrast, the significance of eDNA for cellular attachment and structural integrity has more recently been recognized for an increasing number of Gram-negative and Gram-positive species (Whitchurch et al., 2002; Steinberger and Holden, 2005; Allesen-Holm et al., 2006; Moscoso et al., 2006; Jurcisek and Bakaletz, 2007; Qin et al., 2007; Izano et al., 2008; Thomas et al., 2008; Heijstra et al., 2009; Vilain et al., 2009; Harmsen et al., 2010; Lappann et al., 2010). Release of DNA in bacterial biofilms has mainly been attributed to the lysis of a cellular subpopulation, mediated by the activity of autolysis systems (Allesen-Holm et al., 2006; Rice et al., 2007; Thomas et al., 2008, 2009; Mann et al., 2009). The analysis of aggregates formed by S. oneidensis MR-1 in planktonic cultures indicated the presence of proteins, α--mannose or α--glucose containing exopolysaccharides, and substantial amounts of eDNA (McLean et al., 2008b). This finding prompted us to determine whether eDNA is also a critical component of S. oneidensis biofilms.
In this study, we demonstrate that eDNA is, in fact, an important factor through all stages of S. oneidensis biofilm development. Based on mutant studies, we postulate that, in S. oneidensis MR-1, eDNA originates from lysis through induction of three genome-encoded prophages. Our study strongly indicates that phage-mediated lysis during early stages of biofilm formation may have beneficial consequences for subsequent structure formation.
Materials and methods
Growth conditions and media
Bacterial strains used in this study are summarized in Table 1. Escherichia coli strains were routinely grown in LB medium at 37 °C. For strain WM3064, 2,6-diamino-pimelic acid was added to the medium to a final concentration of 300 μ. S. oneidensis strains were routinely grown at 30 °C in LB. For solidification, agar was added to a final concentration of 1.5% (w/v).
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| DH5α λpir | Φ80dlacZΔM15 Δ(lacZYA-argF) U196 recA1 hsdR17 deoR thi-l supE44 gyrA96 relA1/ëpir | Miller and Mekalanos (1988) |
| WM3064 | ThrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | W Metacalf, University of Illinois, Urbana |
| Shewanella oneidensis | ||
| MR-1 | Shewanella oneidensis MR-1 wild type | Venkateswaran et al. (1999) |
| S198 | MR-1, tagged with eGfp in a mini-Tn7 construct, Cmr | This work |
| S176 | MR-1, tagged with Cfp in a mini-Tn7 construct, Cmr | This work |
| S199 | MR-1, tagged with Yfp in a mini-Tn7 construct, Cmr | This work |
| S1407 | MR-1 ΔMuSo1, deletion of gene cluster SO0641–SO0685 | This work |
| S1412 | MR-1 ΔMuSo1, tagged with eGfp in a mini-Tn7 construct, Cmr | This work |
| S1381 | MR-1 ΔMuSo2, deletion of gene cluster SO2651–SO2704 | This work |
| S1392 | MR-1 ΔMuSo2, tagged with eGfp in a mini-Tn7 construct, Cmr | This work |
| S1387 | MR-1 ΔLambdaSo, deletion of gene cluster SO2939–SO3013 | This work |
| S1393 | MR-1 ΔLambdaSo, tagged with eGfp in a mini-Tn7 construct, Cmr | This work |
| S1421 | MR-1 ΔLambdaSo ΔMuSo1 | This work |
| S1426 | MR-1 ΔLambdaSo ΔMuSo2 | This work |
| S1422 | MR-1 ΔMuSo1 ΔMuSo2 | This work |
| S1419 | MR-1 ΔLambdaSo ΔMuSo2 ΔMuSo1 | This work |
| S1461 | MR-1 ΔlambdaSo ΔMuSo2 ΔMuSo1, tagged with eGfp in a mini-Tn7 construct, Cmr | This work |
| Plasmids | ||
| pNTPS-138-R6K | Ori-R6K sacB, suicide plasmid for generating in-frame deletions, Kmr | Lassak et al. (2010) |
| pNTPS-R6K-dMu1 | Fragment for in-frame deletion of the gene region SO0641–SO0685 in pNTPS-R6K | This work |
| pNTPS-R6K-dMu2 | Fragment for in-frame deletion of the gene region SO2651–SO2704 in pNTPS-R6K | This work |
| pNTPS-R6K-dlambda | Fragment for in-frame deletion of the gene region SO2939–SO3013 in pNTPS-R6K | This work |
| pME6031 | repA oriVpVS1 oriVp15A oriT, Tcr, broad-host-range plasmid | Heeb et al. (2000) |
| pME6031-Pmot-lacZ | motAB promoter fused to lacZ, Tcr | This work |
| pUC18-R6KT-miniTn7T | ori-R6K, Tn7 recognition sites, Apr | Choi et al. (2005) |
| pTNS2 | ori-R6K; encodes the TnsABC+D specific transposition pathway, Apr | Choi et al. (2005) |
| pBK-miniTn7-gfp3 | ori-ColE1, gfp3, Apr, Cmr, Gmr | Lambertsen et al. (2005) |
| miniTn7(Gm)PA1/04//03-eyfp-a | ori-ColE1, eyfp, Apr, Cmr, Gmr | Lambertsen et al. (2005) |
| miniTn7(Gm)PA1/04//03-ecfp-a | ori-ColE1, ecfp, Apr, Cmr, Gmr | Lambertsen et al. (2005) |
| pUC18-R6KT-miniTn7T-egfp | NotI-egfp-Cmr-NotI fragment from pBK-miniTn7-gfp3 in pUC18-R6KT-miniTn7T | This work |
| pUC18-R6KT-miniTn7T-ecfp | NotI-ecfp-Cmr-NotI fragment from miniTn7(Gm)PA1/04//03-ecfp-a in pUC18-R6KT-miniTn7T | This work |
| pUC18-R6KT-miniTn7T-eyfp | NotI-eyfp-Cmr-NotI fragment from miniTn7(Gm)PA1/04//03-eyfp-a in pUC18-R6KT-miniTn7T | This work |
| pASK-IBA3plus | expression vector, Apr, Strep-TagII epitope | IBA GmbH, Göttingen, Germany |
| pASK-SO_2963 | SO_2963 in pASK-IBA3plus | This work |
| pASK-SO_2685 | SO_2685 in pASK-IBA3plus | This work |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Tcr, tetracyclin resistance.
Media were supplemented with 6 μg ml−1 chloramphenicol, 30 μg ml−1 kanamycin and/or 2 μg ml−1 tetracycline, where necessary. Biofilms of S. oneidensis were cultivated in LM medium (Paulick et al., 2009) without antibiotics containing 0.5 m lactate. DNase I (Serva Electrophoresis GmbH, Heidelberg, Germany) was used at a concentration of 30 μg ml−1 in medium supplemented with 5 m MgCl2. DDAO (7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one)) was used at a concentration of 4 μ to stain eDNA in biofilms grown under hydrodynamic conditions.
Vector and strain constructions
DNA manipulations were performed according to standard protocols (Sambrook et al., 1989) or following the manufacturer's instructions. Kits for the isolation of chromosomal DNA, the isolation of plasmids and the purification of polymerase chain reaction (PCR) products were purchased from HISS Diagnostics GmbH (Freiburg, Germany). Enzymes were purchased from New England Biolabs (Frankfurt, Germany) and Fermentas (St Leon-Rot, Germany). Strains and plasmids used in this study are summarized in Table 1.
In-frame deletion mutants of S. oneidensis MR-1 were constructed essentially as reported earlier (Lassak et al., 2010) using the suicide vector pNTPS-138-R6K and appropriate primer pairs, as summarized in Supplementary Table 1.
For biofilm studies, S. oneidensis MR-1 strains constitutively expressing gfp were constructed by using a modified Tn7 delivery system (see Supplementary Material). To construct pME-Pmot-lacZ, lacZ was PCR amplified from pBAD-lacZ and cloned into the HindIII/XhoI sites of the broad-host-range vector pME6031. To generate a translational fusion of the S. oneidensis MR-1 motAB promoter region to lacZ, the corresponding region was amplified from S. oneidensis MR-1 chromosomal DNA and cloned into the BamHI/HindIII sites. The resulting vector was introduced into S. oneidensis MR-1 by electroporation (Myers and Myers, 1997).
Cultivation of S. oneidensis MR-1 biofilms
Static conditions
Biofilm cultivation in polystyrene microtitre plates (Sarstedt, Newton, NC, USA) was carried out essentially as previously described (Thormann et al., 2004). Briefly, overnight cultures of S. oneidensis MR-1 strains grown in LM medium were diluted 1:35 in LM medium. The diluted cultures were transferred to wells of polystyrene microtitre plates (170 μl per well) and incubated for the desired time at 30 °C. When required, DNase I (30 μg ml−1) was added to the wells at different time points (0, 4 and 24 h). To serve as control, DNase I was heat inactivated by a 10-min incubation at 65 °C. Prior to processing, the density of the planktonic population in the wells was determined at 600 nm. Afterwards, 10 μl crystal violet (0.5% (w/v)) were added to the wells, followed by incubation for 10 min. The wells were then washed with 200 μl distilled water to remove loosely attached biomass. Subsequently, the crystal violet retained by the cells was redissolved in 200 μl ethanol (96% (w/v)), and the absorbance was determined at 570 nm using an Infinite M200 plate reader (Tecan, Männedorf, Switzerland). The relative amount of surface attachment was normalized to that of the wild type. The assay was repeated in at least three independent experiments.
Hydrodynamic conditions
Biofilms were cultivated at room temperature in LM medium in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm. Microscope cover slips (Roth, Germany) were used as a colonization surface, glued onto the channels with silicone (Sista-Henkel, Germany) and left to dry for 24 h at room temperature prior to use. Assembly, sterilization and inoculation of the flow system were performed essentially as previously described (Thormann et al., 2004). Analyses were carried out in triplicate in at least two independent experiments. For treatment with DNase I, the enzyme was added to the inflow medium reservoir at a concentration of 30 μg ml−1. For DDAO staining, the flow was arrested briefly, and DDAO was added to the medium in the bubble trap and the upstream tubing. This process took no longer than 1 min, and control channels, in which the medium flow was stopped in parallel, ensured that the short arrest did not affect biofilm development. The biofilm cells were incubated with the dye for 1 h. Microscopic visualization using an inverted CLSM was performed at defined spots close to the inflow before and after the treatment.
Microscopy and image acquisition
Microscopic visualization of biofilms and image acquisition were conducted using an inverted Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) equipped with × 10/0.3. Plan-Neofluar and × 63/1.2 W C-Apochromate objectives. For displaying biofilm images, CLSM images were processed using the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop. For the quantification of the surface coverage, the image of the confocal plane displaying the cell layer attached to surface was selected. The amount of surface-attached biomass was determined by the amount of green pixels (cells) in relation to that of the background (black) using Adobe Photoshop. For each data point, at least four individual images from at least two independent experiments were analyzed.
Measurements of β-galactosidase activity in culture supernatants
The activity of extracellular β-galactosidase in statically grown biofilms was determined as previously described (Steinmoen et al., 2002). Overnight cultures of S. oneidensis MR-1 strains grown in LM medium were diluted to an OD600 nm of 0.05. The diluted cultures were transferred into Petri dishes and incubated at room temperature for 1, 4 and 24 h. As a control, the supernatant of a 24-h culture incubated at room temperature was used. To obtain cell-free supernatant, the samples were centrifuged at 2 500 × g for 5 min and subsequently filtered (0.2 μm filter). β-Galactosidase assays on supernatants were carried out in Eppendorf tubes at 30 °C according to standard protocols (Miller, 1972). The β-galactosidase activity was normalized to the overall amount of protein in the sample as quantified with the BCA protein assay reagent (Thermo Scientific, Schwerte, Germany).
Isolation of eDNA from biofilm supernatants
S. oneidensis MR-1 biofilms were grown in Petri dishes as described above. After 1, 4 and 24 h of incubation, 500 μl of the supernatant was collected, centrifuged (13 000g, 3 min) and passed through a membrane filter (0.45 μm) to remove all cell material. The supernatant was transferred to a new eppendorf tube. NaCl was added to a concentration of 0.25 M, and the eDNA was precipitated by adding 2:1 volume of ethanol (96% (w/v)). The precipitated eDNA was dissolved in TE (10 mM Tris (pH 8.0); 1 mM EDTA) buffer, and the DNA concentration was determined by spectrophotometry using a NanoDrop ND-1000 (Peqlab, Erlangen, Germany).
Quantification of eDNA in culture supernatants
Sterile supernatants of statically grown S. oneidensis MR-1 biofilms were used in a DNA release assay modified after Hamilton et al. (2001). The supernatants were collected after 1, 4 and 24 h of incubation and filter sterilized. A measure of 100 μl of a 1:200 dilution of PicoGreen fluorescent dye (Molecular Probes; Invitrogen, Darmstadt, Germany) was added to 100 μl of biofilm supernatant. DNA release was immediately measured fluorometrically at an excitation wavelength of 485 nm and an emission wavelength of 535 nm using a Tecan Infinite M200 reader (Tecan). Concentration of eDNA was then calculated using DNA reference standards prepared in culture medium.
Quantitative RT–PCR
Isolated eDNA was used as template for quantitative RT–PCR (Real Time 7300 PCR Machine, Applied Biosystems, Darmstadt, Germany) using the Sybr Green detection system (Applied Biosystems). The signals were standardized to recA, with the CT (cycle threshold) determined automatically by the Real Time 7300 PCR software (Applied Biosystems), and the total number of cycles was set to 40. Samples were assayed in duplicate. The efficiency of each primer pair was determined using four different concentrations of S. oneidensis MR-1 chromosomal DNA (10 ng l−1, 1.0 ng l−1, 0.1 ng l−1 and 0.01 ng l−1) as a template in quantitative PCRs. 0.1 ng of isolated eDNA was used to quantify the ratio of phage DNA to chromosomal DNA.
Immunoblot analyses
Rabbit polyclonal antibodies raised against the heterologously produced phage proteins SO_2685 and SO_2963 (see Supplementary Material) were generated by Eurogentec (Seraing, Belgium).
S. oneidensis MR-1 protein lysates for western blot analyses were prepared from statically grown biofilm cells harvested via scraping. Cells corresponding to an OD600 nm of 0.25 were harvested by centrifugation, resuspended in 10 μl sample buffer, heated at 99 °C for 5 min and stored at −20 °C. For immunoblot analysis, 10 μl of the sample was resolved by denaturing SDS–polyacrylamide gel electrophoresis on 13% acrylamide gels. Subsequently, proteins were transferred to polyvinylidene difluoride membrane by semidry transfer. For detection of the proteins, polyclonal antibodies against SO_2685 and SO_2963 were used at a dilution of 1:5000 and 1:20 000, respectively. Secondary G-horseradish peroxidase-conjugated antibody anti-rabbit immunoglobulin (Thermo Fisher Scientific, Schwerte, Germany) was used at a dilution of 1:20 000, and signals were detected using the SuperSignal West Pico Chemoluminescent Substrate (Thermo Fisher Scientific) followed by exposure to autoradiography film. Representative immunoblot patterns are shown, but similar patterns were obtained from at least two biological replicates.
Determination of phages's lytic activity in biofilm supernatants
To analyze the lytic activity of phage particles in S. oneidensis MR-1 biofilms, the supernatants of 24 h statically grown biofilms or 30 h planktonic cultures were collected, centrifuged and filter sterilized using a 0.45-μm filter (Sarstedt). In parallel, S. oneidensis MR-1 cells of the appropriate strain were cultivated overnight, freshly diluted and grown to an OD600 nm of 1. A volume of 200 μl of the cell suspension was mixed with 40 ml LB soft agar (0.3% (w/v)) and poured into Petri dishes. Cell-free supernatants were spotted onto the cooled soft agar plates. The plates were incubated overnight at 30 °C and checked for plaque formation.
Determination of relative living cell number in biofilms
To determine the relative living cell number in S. oneidensis MR-1 biofilms, cells were grown statically in Petri dishes and collected at certain time points via scraping. The total cell number of each fraction was determined using a Thoma counting chamber (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany). In parallel, aliquots were plated onto LB agar to quantify the number of colony forming units.
Results
eDNA is an important structural component of S. oneidensis biofilms
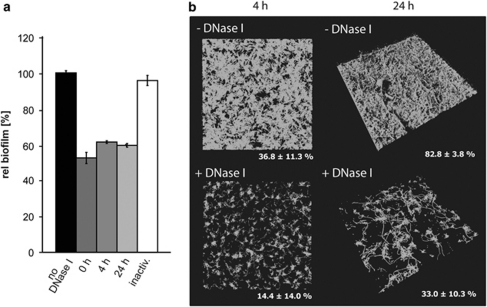
To determine whether eDNA serves as a structural component in the formation of surface-attached communities by S. oneidensis MR-1, we applied DNase I to biofilms in different developmental stages. Biofilms of S. oneidensis MR-1 were grown either in static microtitre dishes or in the hydrodynamic flow chamber system. At appropriate time points, DNase I was added to the cultures in concentration that was previously determined not to interfere with the bacterial growth rate (data not shown). Addition of DNase I to static biofilm cultures of S. oneidensis MR-1 resulted in dissolution of the community within 15–30 min to 52–62% of the mass determined for untreated biofilms, independent of the developmental stage (Figure 1). In contrast, addition of heat-inactivated DNase I had no effect on the biofilm structure. Communities grown in the hydrodynamic flow chamber system also released large amounts of biomass within 60 minutes after exposure to DNase I, and the surface coverage of a 24-h biofilm decreased from 83% to 33%. Dispersal from the biofilm mainly occurred from the less structured areas, whereas the more densely packed three-dimensional structures that started to form after 24 h were more resistant to DNase I treatment and rarely dissolved. Treatment of cells subsequent to initial attachment released the majority of cells directly from the surface, and cultures pretreated with DNase I prior to incubation were significantly deficient in initiating surface contact (Figure 1).
Figure 1.
Effect of DNase I on S. oneidensis MR-1 biofilms grown under static and hydrodynamic conditions. (a) Wild-type cells were incubated in microtitre plates and DNase I was added directly (0), or after 4 and 24 h of incubation. As a control, a 24-h biofilm was treated with heat-inactivated DNase I. The values are means of three replicates, and the s.d. is displayed by error bars. (b) Biofilms formed by Gfp-tagged wild-type cells were cultivated under hydrodynamic conditions. After 4 and 24 h of incubation, DNase I was added to the medium. The biofilms were irrigated with DNase I-containing medium for 2 h prior to analysis by CLSM (lower panel). The lateral edge of a micrograph is 250 μm. The numbers display the average surface coverage of at least four individual scans from at least two independent experiments.
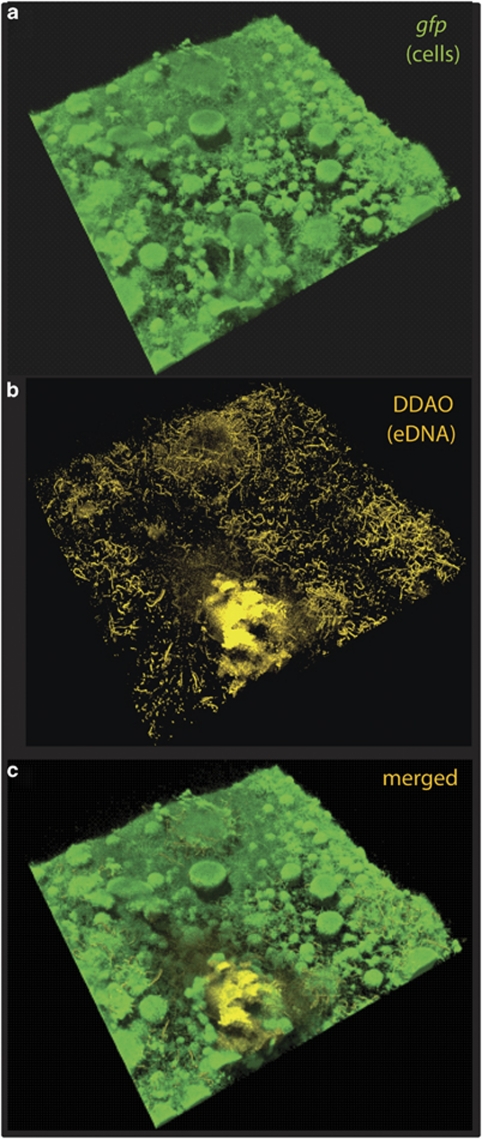
To visualize eDNA, biofilms grown under hydrodynamic conditions were treated with cell membrane-impermeable DNA stain DDAO. eDNA appeared as a faint haze with string-like structures pervading the community (Figure 2). Notably, elevated amounts of eDNA were observed in the towering three-dimensional structures, even though these were more resistant against DNase I treatment. We determined an eDNA concentration of up to 3.5 ng μl−1 in the supernatant of static S. oneidensis MR-1 cultures after 24 h of incubation. However, eDNA directly associated with cells might have been lost during sample preparation, thus, local DNA concentrations within the community may be much higher. Isolated eDNA from cell-free supernatants of static S. oneidensis MR-1 biofilms was used as template in PCR reactions with different primer pairs that covered distant loci of the genome. All reactions gave rise to distinct products (Supplementary Figure S1), indicating that the DNA isolated from the supernatant represents the full chromosome. We further determined whether the addition of eDNA stimulates initial attachment or biofilm formation of S. oneidensis MR-1. To that end, the culture medium was supplemented with purified chromosomal DNA or herring sperm DNA. However, no stimulation of initial attachment occurred with either type of DNA, and further biofilm development was not affected (data not shown).
Figure 2.
Extracellular DNA is a structural component of the matrix in S. oneidensis MR-1 biofilms. Shown are projections of a 72-h-old biofilm of Gfp-tagged S. oneidensis cells grown under hydrodynamic conditions that was subjected to DDAO staining. The upper image (a) displays the Gfp-tagged cells, the middle image (b, yellow) reveals the spatial distribution of extracellular DNA within the biofilm. The lower image (c) displays a merged projection of both upper images. The lateral edge of the micrograph is 250 μm.
These data provide evidence that eDNA is a significant factor through all stages of S. oneidensis MR-1 biofilm formation and enhances initial surface attachment. However, as addition of DNase I never resulted in complete biofilm dissolution, additional structural components, such as proteins or polysaccharides, must be important for biofilm development. Moreover, we conclude that auxiliary factors may be required for the eDNA to mediate cell–cell and cell–surface interactions.
A role for the LambdaSo and MuSo prophages in cell lysis
A number of bacterial species was demonstrated to release eDNA by lysis of a subpopulation of the cells. So far, potential autolysis systems have not been characterized in Shewanella species. However, according to the genome data, S. oneidensis MR-1 harbors three prophages, designated LambdaSo (SO_2939-SO_3013; 50.84 kbp), MuSo1 (SO_0641-SO_0683; 34.55 kbp) and MuSo2 (SO_2652-SO_2704; 34.53 kbp) (Heidelberg et al., 2002). All three prophages have previously been shown to be upregulated upon exposure to environmental stresses such as UV and ionizing radiation (Qiu et al., 2005, 2006). Consistent with this finding, cell death upon UV radiation has been attributed to lysis mainly caused by LambdaSo as demonstrated by the occurrence of phage particles in the supernatant (Qiu et al., 2005). We therefore hypothesized that one or more of the prophages may be involved in cell lysis and eDNA release during S. oneidensis MR-1 biofilm formation. To identify a potential role of the prophages in cell lysis, we generated S. oneidensis MR-1 mutants that harbored a single prophage (ΔλΔMu2, ΔλΔMu1 and ΔMu1ΔMu2, respectively) and a mutant in which all three prophages were deleted (ΔΔΔ). To that end, the full prophage-encoding sequence was removed by double homologous recombination. By PCR and Southern blot analyses, we confirmed that no additional integrated or non-integrated copy of the phage genome remained (data not shown). As large gene regions including potential integration sites for the phages were deleted, these mutants could not be complemented by ectopic expression or reintegration of the deleted fragment. Therefore, we tested strains that had restored the original genotype after the second recombination step of the deletion procedure. These strains phenotypically equaled the corresponding predecessor strains, and we concluded that the phenotypes associated with the loss of the prophage genomes were not due to spontaneous mutations.
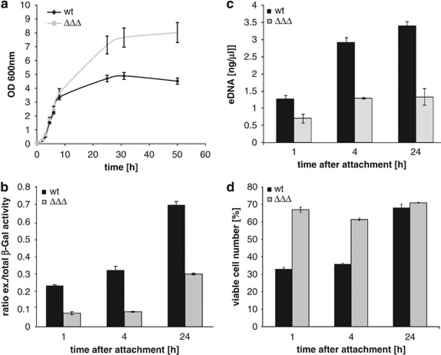
We then determined whether the presence of the prophages influenced cell physiology during growth in complex media. The mutant devoid of all prophages reached a significantly higher final optical density than the wild type (7.4 versus 4.9 after 31 h; Figure 3), indicating that the prophages may induce cellular lysis under appropriate conditions. To further demonstrate that a prophage-less mutant is less prone to autolysis than the wild type under static biofilm growth conditions, β-galactosidase was constitutively produced from a plasmid in the mutant and the wild-type strain. To that end, the strains were grown in static cultures, and the extent of cellular lysis was assessed by measuring the activity of β-galactosidase in the cell-free supernatant relative to that of the cells after 1, 4 and 24 h (Figure 3). A ratio of 0.7 was measured for the wild type after 24 h, whereas that of the phage-less mutant was significantly lower (0.3). As a second indicator for cell lysis, we determined the concentration of DNA in the medium supernatant. Up to 3.4 ng ml−1 was measured for the wild type after 24 h, compared with 1.34 ng ml−1 released by the mutant strain. In parallel, the relative number of colony forming units was determined. The relative amount of colony forming units of the phage-less mutant was almost twice as high as that of wild-type cells after 1 h (67–33%) and 4 h (61–36%). After 24 h, the viable cell number of the mutant and wild type was similar (∼70%). Taken together, the prophage-less mutant released significantly lower amounts of β-galactosidase and DNA at all time points and, after 1 and 4 h of incubation, the relative amount of colony forming units was significantly higher than for the wild type. From these results, we concluded that loss of the prophages results in decreased cell lysis and increased cell viability.
Figure 3.
The role of prophages in cell lysis in S. oneidensis MR-1 cultures. (a) Growth analysis of batch cultures of wild-type (wt, black) and phage-less mutant ΔLambdaSoΔMuSo2ΔMuSo1 (ΔΔΔ, light gray) in LB medium. (b) Extracellular β-galactosidase activity in cell-free biofilm culture supernatants of the wild-type (black) and a phage-less mutant (light gray) transformed with vector pME-Pmot-lacZ. Cells were grown in Petri dishes and supernatant was collected after 1, 4 and 24 h of incubation. The average and associated s.d. of three replicates are shown. (c) Concentration of eDNA in cell-free supernatants (as obtained for β-galactosidase activity measurements) of the wild-type (black) and the phage-less mutant (ΔΔΔ, light gray). (d) Determination of the relative live cell number in static biofilm cultures of the wild-type (black) and phage-less mutant (light gray) strains. Cells were incubated in Petri dishes and harvested 1, 4 and 24 h after attachment. The bars represent the ratio of colony forming units of each fraction to the total cell number. Average values and s.d. displayed by error bars resulted from three replicates.
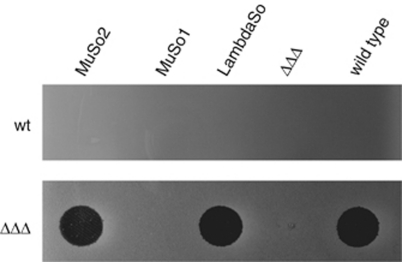
LambdaSo and MuSo2 produce infectious phage particles
To further determine whether all three phages are able to form infectious virus particles during biofilm formation and planktonic growth, cell-free supernatants were harvested from static biofilm cultures of the wild type and the mutants bearing a single prophage that were incubated for 24 h. Supernatants from the planktonic cultures were harvested after 30 h. Small aliquots were spotted onto soft-agar lawns of either the wild type and the prophage-less mutant (ΔΔΔ). Supernatants from mutants harboring only the LambdaSo and the MuSo2 prophage produced plaques in this assay, indicating the production of infectious LambdaSo and MuSo2 virus particles (Figure 4; Supplementary Figure S2). In contrast, supernatants from a MuSo1-harboring strain did not lead to plaque formation, suggesting that this prophage might still exert detrimental effects on its host cell but is not able to assemble a functional virus particle. This finding is in agreement with an earlier prophage gene analysis that reported the absence of several tail genes and insertions in the MuSo1 gene cluster encoding the head subunits (Canchaya et al., 2003). We determined whether MuSo1 is excised during the process of biofilm formation. To that end, PCR was performed using DNA as template that was purified from 24 h biofilms (Supplementary Figure S3). No product was obtained with primer pairs bracketing the MuSo1 locus, strongly indicating that the phage genome is not excised. In contrast to the triple mutant cleared of all the prophages, the wild type was not lysed by supernatants of any mutant or the wild-type strain. Given the higher growth rate of the triple mutant, it cannot be excluded that the mutant strain is in a different physiological stage that renders it more susceptible for phage infection. However, we propose that cell lysis under the conditions tested is rather due to induction of the prophage's lytic cycle and subsequent lysis of the host cell, but not to reinfection and lysis of other prophage-bearing wild-type cells by the released phage particles. This would suggest that the presence of the prophages protects the cells from reinfection.
Figure 4.
Detection of phages in the supernatant of statically grown biofilms of S. oneidensis MR-1. Cell-free supernatants of 24 h-old biofilms formed by mutants exclusively harboring MuSo2 (ΔMu1Δλ, lane 1), MuSo1 (ΔMu2Δλ, lane 2) and LambdaSo (ΔMu1ΔMu2, lane 3), by the phage-less mutant (ΔΔΔ, lane 4) and the by wild type (lane 5) were spotted on soft agar lawns of wild-type cells (upper panel) and phage-less mutant cells (lower panel).
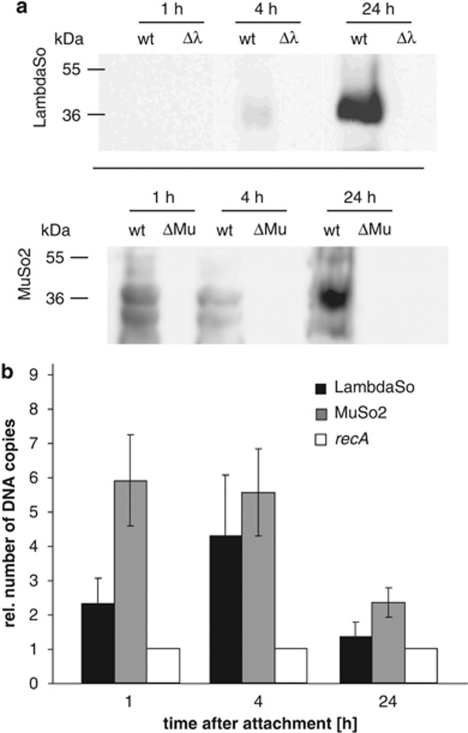
To determine the timing of phage release, supernatants and biofilm cells of static biofilm cultures were tested after 1, 4 and 24 h of incubation for the occurrence of phage proteins (Figure 5) using antibodies raised against the predicted major head subunit of MuSo2 (SO_2685) and the predicted major capsid protein of LambdaSo (SO_2963). Substantial amounts of the MuSo2 major head protein were already detected after 1 h incubation and after prolonged incubation for 24 h. Notably, after 4 h, the concentration of the protein was significantly lower. In contrast, the corresponding protein of LambdaSo was only detected at low concentrations after 4 h of incubation but occurred in substantial amounts in later stages of biofilm formation. Accordingly, quantification of phage DNA in the total eDNA by quantitative RT–PCR revealed an enrichment of MuSo2 DNA during early biofilm formation. An increase in the occurrence of LambdaSo DNA was observed after 4 h of incubation (Figure 5). However, after 24 h, the relative amount of phage DNA dropped, suggesting that, at a certain stage, no more phage particles are produced although the bacterial culture continuous to grow. This is in accordance with the finding that the living cell number of the phage-less mutant and wild type is similar and suggests that induction of the phages and phage-mediated cell lysis decreases between 8 and 24 h. The copy number of MuSo1 DNA equaled that of the control gene recA (data not shown). This is indicating that the phage genome is not replicated and is consistent with the finding that MuSo1 does not produce infectious particles.
Figure 5.
Phage release during attachment and biofilm formation of S. oneidensis MR-1. (a) Presence of phages LambdaSo (upper panel) and MuSo2 (lower panel) in wild-type cells (wt) and appropriate deletion mutant cells (Δλ and ΔMuSo2). Cells were grown under static conditions and harvested 1, 4 and 24 h after attachment. Cell lysates were subjected to SDS–polyacrylamide gel electrophoresis followed by western blotting and detection with specific antisera against the LambdaSo and MuSo2 phage major head proteins. (b) Determination of the relative DNA copy number of the phages LambdaSo (black) and MuSo2 (light gray) compared with recA (representing the chromosomal DNA (white) in biofilm supernatants). Extracellular DNA was isolated from the supernatant of statically grown biofilms 1, 4 and 24 h after attachment. DNA levels were analyzed via quantitative RT–PCR. Amplification of recA was used as control to determine the number of chromosomal DNA copies. The values are means of three replicates. Error bars display the s.d.
Taken together, we demonstrated that at least two of the three S. oneidensis MR-1 prophages are capable of mediating cell lysis and, thus, may contribute to the release of eDNA in the process of biofilm formation. MuSo2 and LambdaSo are induced during different stages of biofilm development. We therefore speculated that loss of the prophages and subsequent decrease in cell lysis might influence S. oneidensis biofilm formation due to a lack of eDNA.
Mutants lacking the prophages are defective in biofilm formation
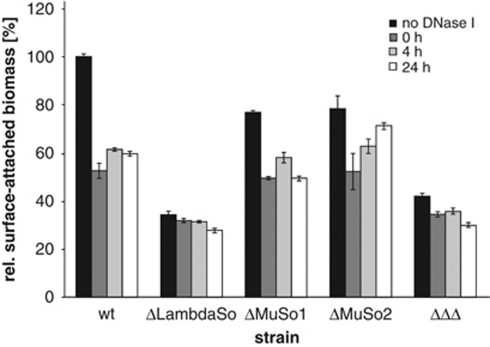
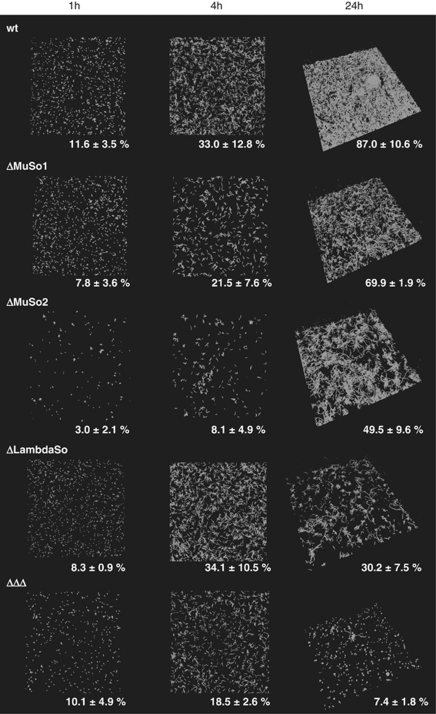
To determine whether the prophages are involved in biofilm formation of S. oneidensis MR-1 by release of eDNA, we tested the biofilm-forming capacities of strains in which one of the three prophages was deleted and that of the triple mutant lacking all three prophages. In the static microtitre plate assay, deletion of the MuSo1 or MuSo2 prophage resulted in a decrease of surface-attached biomass to ∼80% of that of the wild-type level (Figure 6). Addition of DNase I further released biomass to similar levels observed for wild-type biofilms that were equally treated with DNase I (∼50%). A mutant lacking the LambdaSo prophage was drastically impaired in biofilm formation under static conditions (Figure 6). Compared with the wild type, it accumulated only ∼40% surface-associated biomass, which could not be further dispersed by treatment with DNase I. The mutant lacking all three prophages displayed a biofilm phenotype similar to that of the LambdaSo deletion strain. When grown in the hydrodynamic flow chamber system, mutants lacking MuSo1 and, in particular, MuSo2 were delayed in biofilm formation (Figure 7). However, after prolonged incubation for 24 and 48 h, both mutants were able to cover the surface (70% and 50% after 24 h, respectively) and to develop pronounced distinct three-dimensional structures. In contrast, a LambdaSo mutant was not affected in the initial stages of biofilm formation. The mutant, however, was unable to cover the surface (30% after 24 h) and to form the distinct three-dimensional structures after 24 h (Figure 7). The triple-mutant lacking all three prophages displayed an additive biofilm-deficient phenotype. Very small amounts of surface-attached biomass were detected even after 48 h of incubation. The appearance of the biofilm formed by the triple mutant equaled that of wild-type cells treated with DNase I (Figure 1). As has been observed in the static microtitre plate assay, the biofilm structures formed by the triple-prophage mutant could not be dispersed by addition of DNase I (Supplementary Figure S4). In addition, no eDNA was visualized in biofilms formed by the triple mutant by DDAO staining. Notably, biofilm formation of phage mutants was not restored upon addition of herring sperm DNA or S. oneidensis MR-1 chromosomal DNA (data not shown). From the biofilm analysis, we concluded that all three prophages contribute to biofilm formation, most likely by lysis of a subpopulation of cells and release of cytoplasmic factors such as eDNA. The LambdaSo phage appears to have a predominant role during that process, particularly at later stages of biofilm formation.
Figure 6.
Effects of DNase I treatments on statically grown biofilms formed by phage mutants. S. oneidensis MR-1 wild-type and mutant cells were incubated in microtitre plates and DNase I was added after 0, 4 or 24 h of incubation. The biofilm was quantified using a crystal violet assay. The values are means of three replicates, and the s.d. are displayed by error bars.
Figure 7.
Involvement of prophages MuSo1, MuSo2 and LambdaSo in biofilm formation under hydrodynamic conditions. Gfp-tagged S. oneidensis MR-1 wild-type and mutant cells were incubated in flow chambers, and biofilm formation was microscopically analyzed via CLSM after 1 (left panel), 4 (middle panel) and 24 h (right panel) of attachment. Displayed are three-dimensional shadow projections. The numbers represent the average surface coverage. The lateral edge of each micrograph is 250 μm in length.
Discussion
eDNA occurs in significant amounts in terrestrial and aquatic environments where it may serve as important nutrient reservoirs, particularly for nitrogen and phosphorus (Deflaun et al., 1986; Paul et al., 1991; Niemeyer and Gessler, 2002; Dell'Anno and Danovaro, 2005). Accordingly, a previous study showed that Shewanella species are capable of using DNA as source of phosphorus, nitrogen, carbon and energy (Pinchuk et al., 2008). Here we demonstrate that, in addition, eDNA has a major role in surface attachment and development of three-dimensional structures during S. oneidensis MR-1 biofilm formation. First conclusive evidence for eDNA as an important factor in the structural integrity of microbial biofilms was presented for Pseudomonas aeruginosa (Whitchurch et al., 2002; Allesen-Holm et al., 2006). Since that time, there has been emerging evidence from numerous studies on different bacterial species that identify eDNA as a common structural component in biofilm formation, although its exact role still remains unknown. As demonstrated here for S. oneidensis MR-1, eDNA is already involved in early attachment events, similar to what has been reported for other species (Whitchurch et al., 2002; Izano et al., 2008; Vilain et al., 2009; Harmsen et al., 2010; Lappann et al., 2010). Recent studies on Bacillus cereus, Listeria monocytonogenes and Staphylococcus epidermidis suggest that the bacterial cell surface may be decorated with DNA, resulting in acid–base interactions that increase the ability for either cell–cell and cell–surface interactions (Vilain et al., 2009; Das et al., 2010; Harmsen et al., 2010). Furthermore, short DNA fragments smaller than 500 bp added to a DNA-free culture of Listeria monocytogenes were demonstrated to prevent initial adhesion. This lead to the hypothesis that high-molecular-weight DNA bound to a limited number of attachment sites is required to mediate cell–surface interactions (Harmsen et al., 2010). It remains to be shown if a similar mechanism exists in S. oneidensis MR-1. Later multicellular stages of S. oneidensis biofilms were less prone to DNase I-induced dispersal, suggesting that eDNA is not the only structural component of the biofilm matrix. Further, but not complete, detachment of biomass could be achieved by additional exposure to proteases (Gödeke and Thormann, unpublished data). Thus, proteinaceous compounds and exopolysaccharides are likely involved in structural integrity. Notably, addition of DNA to the media did not stimulate biofilm formation of S. oneidensis MR-1 or complement biofilm formation of the phage mutants. We therefore hypothesize that an auxiliary factor, in addition to eDNA, is involved. For L. monocytogenes, it was demonstrated that peptidoglycan is required as an additional prerequisite for DNA-dependent biofilm formation (Harmsen et al., 2010), and a similar factor might be required in S. oneidensis MR-1 biofilms as well.
A main question remaining is how eDNA release is mediated, and different mechanisms for DNA release in bacterial biofilms have been discussed. Species such as Neisseria are capable of active DNA export (Hamilton et al., 2005) and also DNA transport through vesiculation has been suggested (Whitchurch et al., 2002; Allesen-Holm et al., 2006). However, as opposed to active transport, several studies provide evidence that, in biofilms of many bacterial species, eDNA rather originates from the lysis of a cellular subpopulation. In Neisseria, DNA release is thought to be mediated by lytic transglycosylases and N-acetylmuramyl--alanine amidase (Lappann et al., 2010). Other factors implicated in cell lysis are toxin/antitoxin system that have been characterized, for example, in Enterococcus faecalis (Thomas et al., 2008) and Staphylococcus sp. (Qin et al., 2007; Rice et al., 2007; Mann et al., 2009). However, a role for toxin/antitoxin systems in biofilm formation is not necessarily directly linked to cell lysis, as has been demonstrated for E. coli (Kim et al., 2009, 2010). Corresponding systems in Shewanella are yet to be characterized. A putative holin/antiholin autolysis system with homology to the Staphylococcus cid system was identified in S. oneidensis MR-1 (SO_1046-SO_1048) (Bayles, 2007). However, mutant analyses revealed that this system does not have a significant role in S. oneidensis biofilm formation (Gödeke and Thormann, unpublished data). Instead, we identified prophage-mediated cell lysis as a likely mechanism for DNA release.
Bacteriophages are highly abundant in all environments and are thought to outnumber prokaryotes in nature by a factor of 10 (Rohwer and Edwards, 2002; Rice et al., 2009). As opposed to phages that predominantly lyse cells, temperent phages can integrate as prophage into host cell genomes in a way that may benefit both host and prophage (Weinbauer, 2004; Chen et al., 2005). Genome analyses revealed the presence of prophage-like elements in almost all bacterial genomes (Canchaya et al., 2003). By mutant analyses, we demonstrated for the first time that at least two out of three prophages previously identified in S. oneidensis MR-1 are capable of mediating cell lysis, and that two of the prophages, LambdaSo and MuSo2, form infectious phage particles. A mutant lacking all prophages is less prone to cell lysis and biofilm formation of such a mutant occurs independently of eDNA as a structural component. Thus, our study links cell lysis to the release of factors promoting cell–cell and cell–surface attachment, in particular eDNA. Our present results strongly suggest that, in S. oneidensis, phage-mediated cell lysis already affects early stages of biofilm formation, and complements previous reports on the role of prophages in bacterial biofilm formation. The best-studied example in this regard is the role of the filamentous phage Pf4 in P. aeruginosa biofilm formation. Mutants lacking the phage form smaller colonies during the first days of biofilm formation and a potential role of phage-mediated cell lysis in eDNA release has been discussed but has not directly been demonstrated (Allesen-Holm et al., 2006; Rice et al., 2009). At later stages of biofilm development, Pf4 is thought to convert into a superinfective lytic form that causes cell death and hollowing of the structures, and, by that, significantly contributes to seeding dispersal of the community (Webb et al., 2003; Rice et al., 2009). In addition, Pf4 has been linked to phenotypic variations in the dispersed cells, leading to small-colony variants that are characterized by accelerated biofilm formation (Webb et al., 2004; Rice et al., 2009). In contrast to MuSo2 and LambdaSo, MuSo1 does not form functional phage particles under the conditions tested. However, a mutant lacking this phage still had a slightly delayed biofilm phenotype. Notably, cryptic prophages unable to produce infectious phage particles have recently been demonstrated to affect bacterial biofilm formation. In E. coli, the prophage CP4-57 excises its genome from the bacterial chromosome during early biofilm formation. Subsequent loss of the excised prophage results in the induction of genes related to flagella-mediated motility and cell lysis, two factors known to affect biofilm development (Wang et al., 2009). So far, we have no indications that MuSo1 is excised from the chromosome, and further studies will address how MuSo1 might affect cellular functions.
Our results also indicate that the prophages may contribute differentially to biofilm formation: whereas the Mu-like phages affect early steps of development, LambdaSo is the major contributing factor to the formation of three-dimensional structures. Transcription of genes from all three prophages has previously been demonstrated to be upregulated upon stresses such as UVB and UVC irradiation and ionizing radiation (Qiu et al., 2005, 2006). However, the nature of the signals that trigger the prophages to enter the lytic cycle during biofilm formation in S. oneidensis is thus far unknown. Notably, phage genes have been shown to be strongly upregulated in biofilms of several bacterial species (Whiteley et al., 2001; Ren et al., 2004; Domka et al., 2007). It remains to be shown whether phage induction in S. oneidensis MR-1 is a direct response to surface attachment and/or nutrient limitations as indicated by cell lysis occurring in planktonic cultures in late exponential phase and whether the prophages are also involved in the dispersal of S. oneidensis biofilms.
Acknowledgments
We are grateful to Martin Thanbichler and Penelope Higgs for critically reading the manuscript and helpful remarks and discussions, and we thank Erhard Bremer for helpful discussions on bacterial phages. We thank Herbert Schweizer for kindly providing the Tn7 delivery vectors that were developed by his group. This work was supported by the Deutsche Forschungsgemeinschaft (DFG TH-831) and the Max-Planck-Gesellschaft.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Bagge D, Hjelm M, Johansen C, Huber I, Gram L. Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl Environ Microbiol. 2001;67:2319–2325. doi: 10.1128/AEM.67.5.2319-2325.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Golding I, Sawai S, Guo L, Cox EC. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005;3:e229. doi: 10.1371/journal.pbio.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Das A, Caccavo F., Jr Dissimilatory Fe(III) oxide reduction by Shewanella alga BrY requires adhesion. Curr Microbiol. 2000;40:344–347. doi: 10.1007/s002849910068. [DOI] [PubMed] [Google Scholar]
- Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol. 2010;76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vriendt K, Theunissen S, Carpentier W, De Smet L, Devreese B, Van Beeumen J. Proteomics of Shewanella oneidensis MR-1 biofilm reveals differentially expressed proteins, including AggA and RibB. Proteomics. 2005;5:1308–1316. doi: 10.1002/pmic.200400989. [DOI] [PubMed] [Google Scholar]
- De Windt W, Gao H, Kromer W, Van Damme P, Dick J, Mast J, et al. AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology. 2006;152:721–729. doi: 10.1099/mic.0.28204-0. [DOI] [PubMed] [Google Scholar]
- Deflaun MF, Paul JH, Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986;52:654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Anno A, Danovaro R. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science. 2005;309:2179. doi: 10.1126/science.1117475. [DOI] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Neu TR, Wozniak DJ. The EPS matrix: the ‘house of biofilm cells'. J Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Hamilton HK, Schwartz KJ, Dillard JP. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J Bacteriol. 2001;183:4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen M, Lappann M, Knøchel S, Molin S. The role of extra-cellular DNA during biofilm formation of Listeria monocytogenes. Appl Environ Microbiol. 2010;76:2271–2279. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in Gram-negative, plant-associated bacteria. Mol Plant-Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- Heijstra BD, Pichler FB, Liang Q, Blaza RG, Turner SJ. Extracellular DNA and type IV pili mediate surface attachment by Acidovorax temperans. Antonie Van Leeuwenhoek. 2009;95:343–349. doi: 10.1007/s10482-009-9320-0. [DOI] [PubMed] [Google Scholar]
- Icopini GA, Lack JG, Hersman LE, Neu MP, Boukhalfa H. Plutonium(V/VI) reduction by metal reducing bacteria Geobacter metallireducens GS-15 and Shewanella oneidensis MR-1. Appl Environ Microbiol. 2009;75:3641–3647. doi: 10.1128/AEM.00022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wang X, Ma Q, Zhang XS, Wood TK. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wang X, Zhang XS, Grigoriu S, Page R, Peti W, et al. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ Microbiol. 2010;12:1105–1121. doi: 10.1111/j.1462-2920.2009.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- Lappann M, Claus H, van Alen T, Harmsen M, Elias J, Molin S, et al. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol Microbiol. 2010;75:1355–1371. doi: 10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- Lassak J, Henche AL, Binnenkade L, Thormann KM. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl Environ Microbiol. 2010;76:3263–3274. doi: 10.1128/AEM.00512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Holmes DE, Nevin KP. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol. 2004;49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Majors PD, Reardon CL, Bilskis CL, Reed SB, Romine MF, et al. Investigations of structure and metabolism within Shewanella oneidensis MR-1 biofilms. J Microbiol Methods. 2008a;74:47–56. doi: 10.1016/j.mimet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- McLean JS, Pinchuk GE, Geydebrekht OV, Bilskis CL, Zakrajsek BA, Hill EA, et al. Oxygen-dependent autoaggregation in Shewanella oneidensis MR-1. Environ Microbiol. 2008b;10:1861–1876. doi: 10.1111/j.1462-2920.2008.01608.x. [DOI] [PubMed] [Google Scholar]
- McLean JS, Wanger G, Gorby YA, Wainstein M, McQuaid J, Ishii SI, et al. Quantification of electron transfer rates to a solid phase electron acceptor through the stages of biofilm formation from single cells to multicellular communities. Environ Sci Technol. 2010;44:2721–2727. doi: 10.1021/es903043p. [DOI] [PubMed] [Google Scholar]
- Miller JH.(ed) (1972A Short Course in Bacterial Genetics Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso M, Garcia E, Lopez R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CR, Myers JM. Replication of plasmids with the p15A origin in Shewanella putrefaciens MR-1. Lett Appl Microbiol. 1997;24:221–225. doi: 10.1046/j.1472-765x.1997.00389.x. [DOI] [PubMed] [Google Scholar]
- Myers CR, Nealson KH. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Belz A, McKee B. Breathing metals as a way of life: geobiology in action. Antonie van Leeuwenhoek. 2002;81:215–222. doi: 10.1023/a:1020518818647. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Scott J.2003Ecophysiology of the genus Shewanella The Prokaryotes: An Evolving Electronic Resource for the Microbial CommunityDworkin M (ed).Springer-NY, LLC New York: USA; 1133–1151. [Google Scholar]
- Niemeyer J, Gessler F. Determination of free DNA in soils. J Plant Nutr Soil Sci. 2002;165:121–124. [Google Scholar]
- Paul JH, Jiang SC, Rose JB. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl Environ Microbiol. 1991;57:2197–2204. doi: 10.1128/aem.57.8.2197-2204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, et al. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol. 2009;71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- Pinchuk GE, Ammons C, Culley DE, Li SM, McLean JS, Romine MF, et al. Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp: ecological and physiological implications for dissimilatory metal reduction. Appl Environ Microbiol. 2008;74:1198–1208. doi: 10.1128/AEM.02026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Daly MJ, Vasilenko A, Omelchenko MV, Gaidamakova EK, Wu L, et al. Transcriptome analysis applied to survival of Shewanella oneidensis MR-1 exposed to ionizing radiation. J Bacteriol. 2006;188:1199–1204. doi: 10.1128/JB.188.3.1199-1204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Sundin GW, Wu L, Zhou J, Tiedje JM. Comparative analysis of differentially expressed genes in Shewanella oneidensis MR-1 following exposure to UVC, UVB, and UVA radiation. J Bacteriol. 2005;187:3556–3564. doi: 10.1128/JB.187.10.3556-3564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Setlow P, Thomas SM, Ye RW, Wood TK. Gene expression in Bacillus subtilis surface biofilms with and without sporulation and the importance of yveR for biofilm maintenance. Biotechnol Bioeng. 2004;86:344–364. doi: 10.1002/bit.20053. [DOI] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook K, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press: Cold Spring Harbor, NY; 1989. [Google Scholar]
- Saville RM, Dieckmann N, Spormann AM. Spatiotemporal activity of the mshA gene system in Shewanella oneidensis MR-1 biofilms. FEMS Microbiol Lett. 2010;308:76–83. doi: 10.1111/j.1574-6968.2010.01995.x. [DOI] [PubMed] [Google Scholar]
- Steinberger RE, Holden PA. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmoen H, Knutsen E, Havarstein LS. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci USA. 2002;99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol. 2006;11:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen S, De Smet L, Dansercoer A, Motte B, Coenye T, Van Beeumen JJ, et al. The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res Microbiol. 2009;161:144–152. doi: 10.1016/j.resmic.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. 2008;190:5690–5698. doi: 10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Saville RM, Shukla S, Pelletier DA, Spormann AM. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J Bacteriol. 2004;186:8096–8104. doi: 10.1128/JB.186.23.8096-8104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Saville RM, Shukla S, Spormann AM. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol. 2005;187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, et al. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Internat J Syst Bacteriol. 1999;2:705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- Vilain S, Pretorius JM, Theron J, Brözel VS. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilm. Appl Environ Microbiol. 2009;75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MJ, Fu QS, Rhoads KR, Yeung CH, Spormann AM, Criddle CS. A derivative of the menaquinone precursor 1,4-dihydroxy-2-naphthoate is involved in the reductive transformation of carbon tetrachloride by aerobically grown Shewanella oneidensis MR-1. Appl Microbiol Biotechnol. 2004;63:571–577. doi: 10.1007/s00253-003-1407-3. [DOI] [PubMed] [Google Scholar]
- Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:22. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Zhang M, Ginn BR, Dichristina TJ, Stack AG. Adhesion of Shewanella oneidensis MR-1 to oron (oxy)(hydr)oxides: microcolony formation and isotherm. Environ Sci Technol. 2010;44:1602–1609. doi: 10.1021/es901793a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.