Abstract
Whereas genomic data are universally machine-readable, data arising from imaging, multiplex biochemistry, flow cytometry and other cell- and tissue-based assays usually reside in loosely organized files of poorly documented provenance. This arises because the relational databases used in genomic research are difficult to adapt to rapidly evolving experimental designs, data formats and analytic algorithms. Here we describe an adaptive approach to managing experimental data based on semantically-typed data hypercubes (SDCubes) that combine Hierarchical Data Format 5 (HDF5) and Extensible Markup Language (XML) file types. We demonstrate the application of SDCube-based storage using ImageRail, a software package for high-throughput microscopy. Experimental design and its day-to-day evolution, not rigid standards, determine how ImageRail data are organized in SDCubes. We apply ImageRail to the collection and analysis of drug dose-response landscapes in human cell lines at the single-cell level.
INTRODUCTION
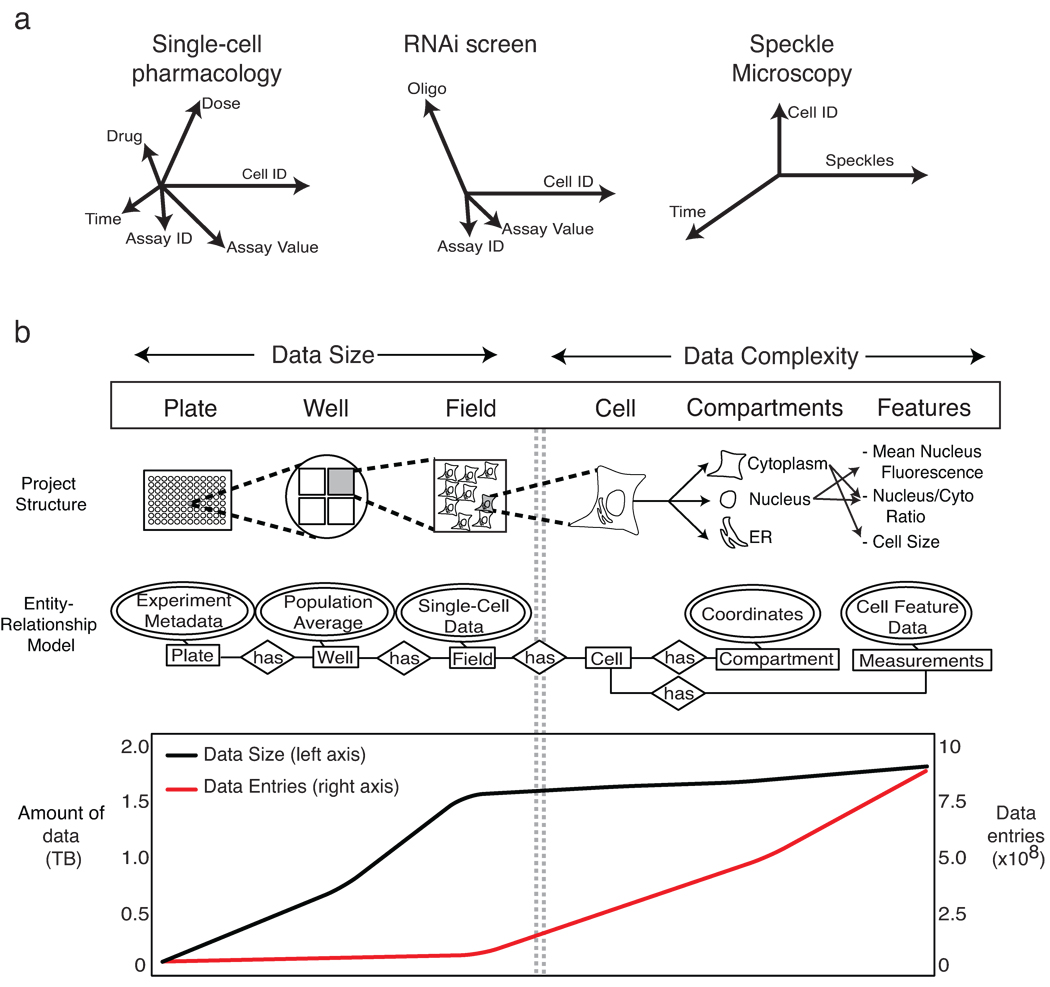
It is widely accepted that biomedical data should be machine-readable and web-accessible. Relational database management systems (RDBMS)1,2 have proven highly effective with sequence data that are string-based, invariant in organization and interpretable without knowledge of the experiments, instruments or algorithms used to gather them. It has proven more difficult to manage data arising from complex biochemical measurements, imaging, flow cytometry and phenotypic assays of cells and tissues. The interpretation of these data, which are often unstructured (e.g., images), is critically dependent on experimental context, and this context changes frequently. The difficulty in developing satisfactory database solutions for “high-content” data is widely ascribed to insufficient standardization or poor implementation3, but we believe the problem is more fundamental: it reflects the impossibility of fully specifying a priori complex experimental designs. Flexible and creative design is the essence of good experimental science, and since design determines data structure (the number of time points, repeats, conditions, etc.), structures frequently change (Fig. 1a). To accommodate these changes, database schema must be reconfigured frequently, a complex and time-consuming task. Thus, most experimental data reside in unlinked, loosely annotated spreadsheets that are easily fragmented or lost4,5. When data scope and complexity demand a more capable repository, a new database is often created ad hoc.
Figure 1.
Challenges in management of multi-dimensional data. (a) The schematic illustrates that experimental design is the key determinant of data dimensionality. Different experimental protocols result in experiments that yield data with different numbers and types of dimensions; in the case of single-cell pharmacology, this includes the selection of drug, dose, time and type of assay and number of cells analyzed (represented by axes whose length represents magnitude). (b) High-throughput immunofluorescence experiments managed by ImageRail have a hierarchy describable in an entity-relationship model, as illustrated in the schematic (above). The graph (below) shows the amount of data in Terabytes (black line) during successive steps in the image processing workflow as compared to the number of individual data entries (red line).
As an illustrative problem in biological data management, we focus here on high-throughput, high-content microscopy6,7. Microscopy presents two distinct data management challenges. One is the sheer size of the data, which can exceed many terabytes per month. The second involves the difficulties of working with numerical data extracted by image analysis, which can include a large number of data types that have complex relationships to each other (e.g., the boundaries and intensities of cells or compartments and computed features such as nuclear translocation; Fig. 1b)8. For example, a typical genome-wide RNAi screen might generate ~7 × 105 images (~1.3 terabytes of data); analysis would increase the size only modestly (by ~100 megabytes), but the number of data entries would increase from ~106 images to >109 features (Supplementary Fig. 1). Conventional spreadsheets and comma-separated value (CSV) files perform poorly with 109 data entities, and relational databases impose the organizational costs described above.
In this paper we propose a potential solution to the challenge of managing high-dimensionality biomedical data based on the use of semantically-typed data hypercubes (SDCubes) in which binary data are stored in Hierarchical Data Format 5 (HDF5; http://www.hdfgroup.org/HDF5/) and metadata and data ontologies are stored in Extensible Markup Language (XML; http://www.w3.org/standards/xml). We have created a new open-source Java library, the SDCube Programming Library (Supplementary Software 1, http://www.semanticbiology.com/software/sdcube) that can create SDCubes with appropriate dimensionality, encode the data model in a machine-readable XML ontology, and reformat SDCubes as needed when experiments change (Fig. 2a). To illustrate the use of SDCubes, we have created a second program ImageRail (Supplementary Software 2, http://www.semanticbiology.com/software/imagerail) for high-content microscopy that (i) segments images of cells grown in 96- and 384-well plates to extract features such as cell shape or nuclear fluorescence, (ii) stores experimental metadata and results of image analysis in SDCubes, (iii) computes sets of cellular features from the image (e.g., fluorescence and localization metrics), and (iv) displays metadata, images and analysis in various formats9. By using SDCubes, ImageRail is able to organize data according to the design of an experiment and its day-to-day evolution rather than an inflexible, predetermined schema. We use these tools to characterize the responses of tumor cells to therapeutic small molecules and show that the apparent IC50 for receptor inhibitors varies with ligand dose, that cell-to-cell variability is maximal as ligands and drugs approach concentrations likely to be encountered in vivo, and that variance impacts the shape of dose-response curves. Our results suggest that monitoring variance will be broadly useful in pre-clinical pharmacology. Moreover, because flow cytometry and multiplex biochemistry have similar workflows to imaging4,10, ImageRail and the SDCube Programming Library represent starting points for managing diverse experimental data.
Figure 2.
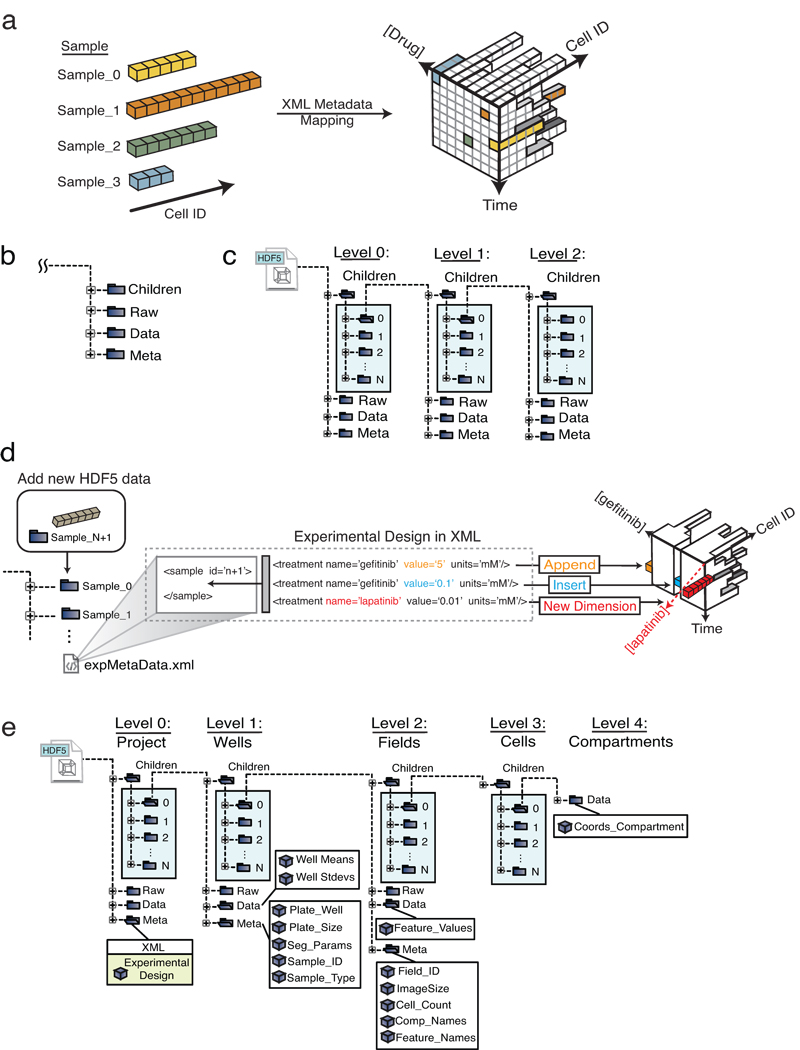
SDCubes are built from a collection of linked data modules that can encode diverse experimental data with varying requirements. (a) The XML metadata maps the experimental sampling procedure onto the HDF5 data space; data from each cell are represented by colored boxes and different numbers of cells are collected for each condition. (b) The SDCube data module is composed of four HDF5 groups, each storing a different type of data. (c) The Children group in each module can contain additional data modules, generating an arbitrarily complex data tree. (d) A previously defined SDCube can be modified to append a new piece of data to the end of an existing series (orange), insert data into the middle of a series (blue) or add a new type of data that requires addition of a new dimension (red) (in this case, use of lapatinib rather than gefitinib). All three operations are performed by modifying the XML file while recording the data in the appropriate place in the HDF5 file hierarchy. (e) ImageRail uses a five-level SDCube encoding high-throughput fixed-cell imaging data and progressively increasing levels of detail (project, well, field, cell and compartment).
RESULTS
Managing complex and heterogeneous data using SDCubes
HDF5 files can contain both structured and unstructured data, can encode data hierarchically using “groups” (analogous to file system folders), are unlimited in size and can be opened progressively using software libraries that read and write selected slices of data. The latter feature is critical for files that exceed the size of physical memory. To date, HDF5 has been used primarily (if not exclusively) in observational sciences (particularly remote Earth-sensing) involving highly standardized data collection and little or no directed perturbation of the system under study. It has been suggested that HDF5 might be applied to biological imaging11, but no practical implementations exist and HDF5 alone appears to be insufficient to meet the challenges of biological experiments involving complex perturbations such as gene knockdown, drug and ligand dose-response, pulse-chase, etc. SDCubes address this challenge by encoding the design of perturbation-rich experiments in XML and using the design to create HDF5 files of appropriate dimensionality. A two-format solution is needed because XML is ill-suited for storage of large numerical datasets and HDF5 lacks easy integration with “minimum information” standards such as Minimum Information for Biological and Biomedical Investigations (MIBBI)12 and other Web-based ontologies.
The HDF5 component of an SDCube is composed of basic data modules, each of which contains the HDF5 groups Data, Meta, Raw and Children (Fig. 2b). Data contains measured or computed data stored in N-dimensional arrays; Meta contains metadata such as plate address, sample identifiers and the SDCube XML file; and Raw contains original CSV, TIFF, FCS and other primary data as byte arrays. The Children group allows creation of nested data modules, each containing progressively more detailed information (Fig. 2c). The top-level Children group is special in that it is always organized by “sample,” a label identical to “experiment” in the Minimum Information About a Cellular Assay (MIACA) standard12.
The XML component of SDCubes contains four types of information: (i) standard metadata (e.g., investigator and research group); (ii) experimental protocol (e.g., information on cell lines and reagents, in formats conforming to MIBBI standards when possible); (iii) experimental design (e.g., species and other variables in the protocol, such as time or perturbation, that are applicable to each sample); and (iv) the identities of algorithms and free parameters used during conversion of raw data into useful experimental measurements (see Methods for details). Using methods in the SDCube Programming Library (Supplementary Note 1), new samples, dimensions or assays can be appended to or inserted into an existing SDCube simply by modifying the XML file and adding to the Children group at the top level of the HDF5 hierarchy (Fig. 2d). SDCubes are adaptable to a variety of data types (Supplementary Fig. 2) and can combined to aggregate data from other SDCubes or divided up to create subsets of the data..
Implementing the HDF5-XML SDCube standard in ImageRail
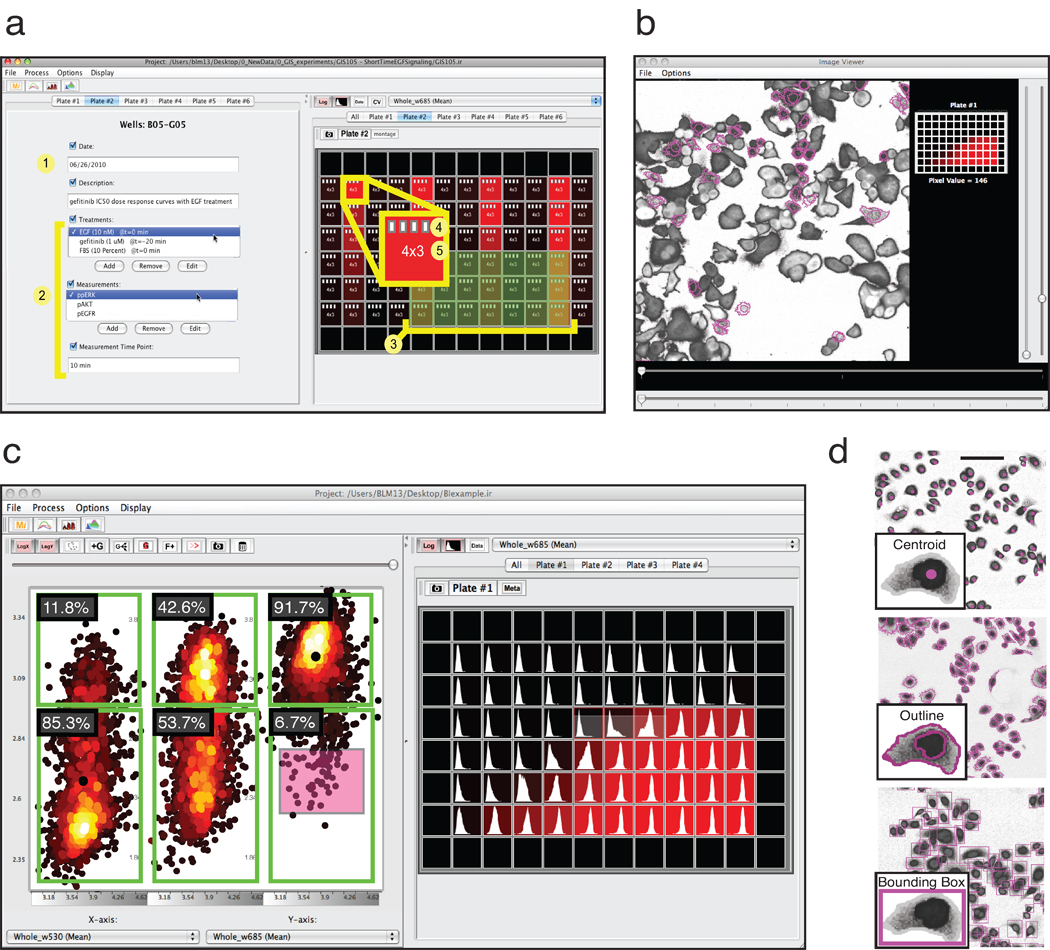
ImageRail is a standalone program for high-throughput image analysis that creates and manipulates SDCubes and serves as a test of the concepts outlined above. ImageRail has four software components. First, formatting tools create and modify SDCubes so that the Children group is formatted to create a five-level data hierarchy comprising project, plate, well, (image) field and cell and (cellular) compartment (conforming to the entity-relationship model in Fig. 1b and 2e). Drop-down lists and a GUI for highlighting wells make it possible to specify which experimental conditions map to which wells, thereby specifying the experimental design and SDCube dimensionality and creating XML annotation (Fig. 3a). Second, image analysis tools create and store segmentation masks based on standard algorithms for cell monolayers, which can be extended using existing software such as ImageJ13 (Fig. 3b). Third, data viewers display raw data and computed features as images, line plots, histograms, scatter plots and multi-well plate views. Scatter plotting includes multi-dimensional gating similar to that used for analysis of flow cytometry data (Fig. 3c). Finally, embedded routines enable dynamic linking of data points to specific image features. Dynamic linking allows users to highlight cells in an image that correspond to selected data points in a scatter plot (Fig. 3b and Fig. 3c), facilitating the identification of outliers and experimental artifacts such as bubbles, tissue culture debris or edge effects (Supplementary Fig. 3). Users choose the level of detail at which to store the link between segmentation and data; at one extreme, pixel-by-pixel information can be stored, but we generally find it more useful to store either the centroid of each cell or a bounding box (Fig. 3d).
Figure 3.
Annotated and simplified screen shots from ImageRail software (also see Supplementary Note 2). (a) (1) General experiment metadata and (2) computable information derived from image analysis across perturbations and measurements are associated with (3) selected wells of a microtiter plate. (4) White document icons represent the number of image fields that have single-cell data stored in the HDF5 file available for analysis, and (5) numbers represent imaged fields and wavelengths. (b) Dynamic linking of extracted data to the source images shows which cells gave rise to which measurements and is implemented using an image viewer and scatter plot (red box in c). (c) Data visualization includes single-cell scatter plots with flow cytometry-style gating (left) and plate heat maps of population averages along with a representation of the underlying single-cell distributions (right). (d) Results of image segmentation can be stored in different ways, including centroid, outline and bounding box. Scale bar = 100µm.
Although the SDCube data group “Raw” can store image data, we are in the process of integrating ImageRail with the Open Microscopy Environment Remote Objects (OMERO) image server14. Thus, ImageRail currently stores TIFF files alongside SDCubes and not within them. OMERO provides powerful tools for processing and organizes images, is used widely in open-source and commercial image management applications14,15 and OME-TIFF has found wide acceptance as a file standard for biological microscopy.
Monitoring cell-to-cell variability in drug responses
It is widely hypothesized that variability in cellular responses to drugs and the presence of drug-resistant cell subpopulations can impact cancer therapy16. One application of ImageRail is to systematize single cell drug-response studies and uncover the origins and significance of variability. Our proof-of-principle studies focused on the impact of changes in the concentration of epidermal growth factor (EGF) on the IC50 of ATP-competitive EGF receptor (EGFR) inhibitors erlotinib and gefitinib17. We assayed inhibition by immunofluorescence microscopy, using antibodies specific for the pT202/pY204-modified form of the downstream kinase ERK1/2 (henceforth ppERK). EGFR mutation and over-expression are implicated in a wide range of tumors18, and erlotinib and gefitinib are used clinically to treat lung, colorectal and other cancers19,20.
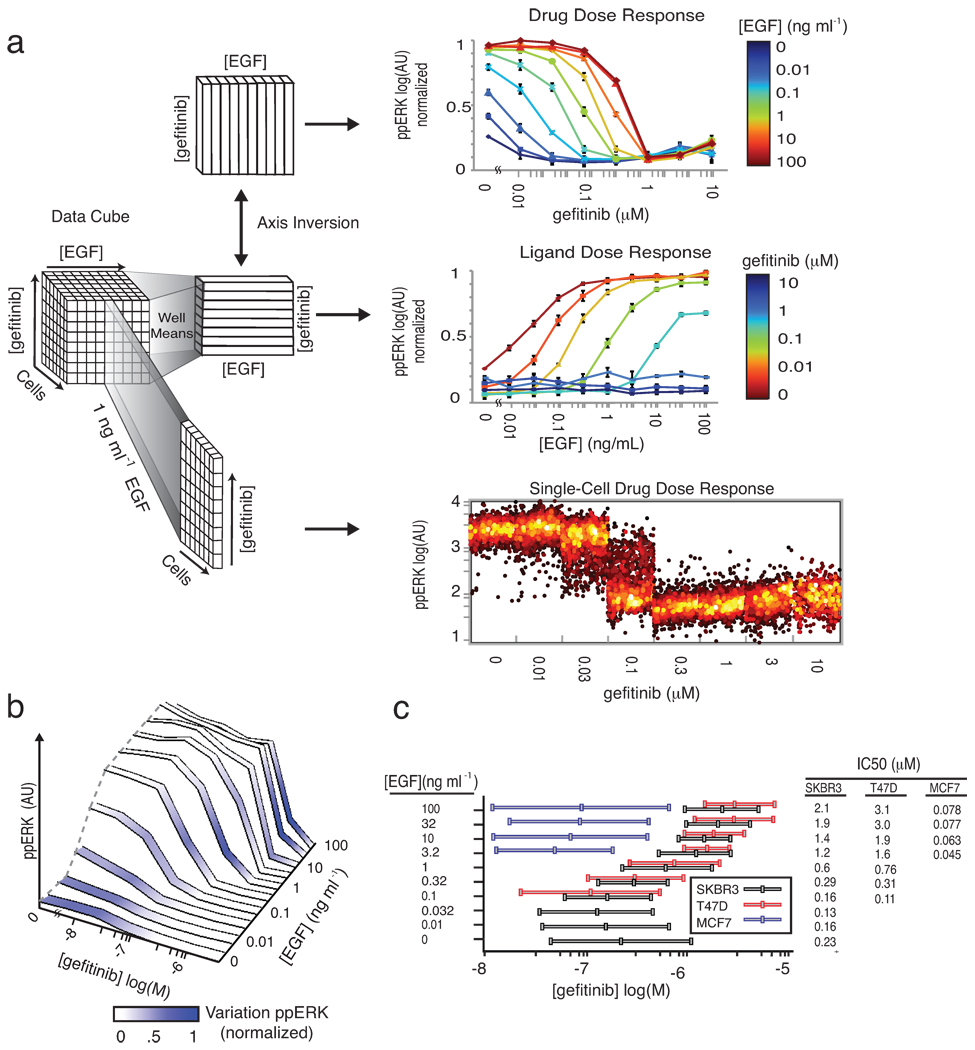
Here, we exposed cells to EGF at 10 doses over a 104 range in combination with gefitinib at 8 doses over a 103 range using a simple adaptive design in which each 96-well plate was subjected to a different and changeable set of treatments and measurements. To enable image segmentation with a standard watershed algorithm, we treated cells with nuclear and cytoplasmic stains (Supplementary Fig. 4). The dataset comprised 160 conditions, 1.4×106 individual cells and an SDCube with 2.8×106 entries (data are available in the supplemental materials in SDCube and CSV formats; a 10-fold larger dataset involving more proteins is shown in Supplementary Fig. 5). By accessing different slices of the cube, we can view data as a series of IC50 curves at differing EGF concentrations ([EGF]), or as a set of EGF dose-response curves at different drug concentrations ([drug]); cell-to-cell variability can also be visualized at any point (Fig. 4a). We observed that average levels of ppERK increased with [EGF] and decreased with [gefitinib], and that the apparent IC50 was sensitive to EGF concentration, varying ~20-fold as exogenous EGF varied from 0 to 100 ng/mL (Fig. 4b). Well-average data computed from images closely matched dose-response data obtained using conventional biochemical assays (Supplementary Fig. 6). The relationship between IC50 and [EGF] varied substantially with cell type (Fig. 4c): whereas IC50 was strongly sensitive to [EGF] in SKBR3 and T47D cells, it was less so in MCF7 cells (Supplementary Fig. 7). Data exploration of this type is intuitively simple, but involves the manipulation of many data entries; because HDF5 successively loads data, there is no limit a priori to the number of entries, and ImageRail has been validated with ~108–109 data points.
Figure 4.
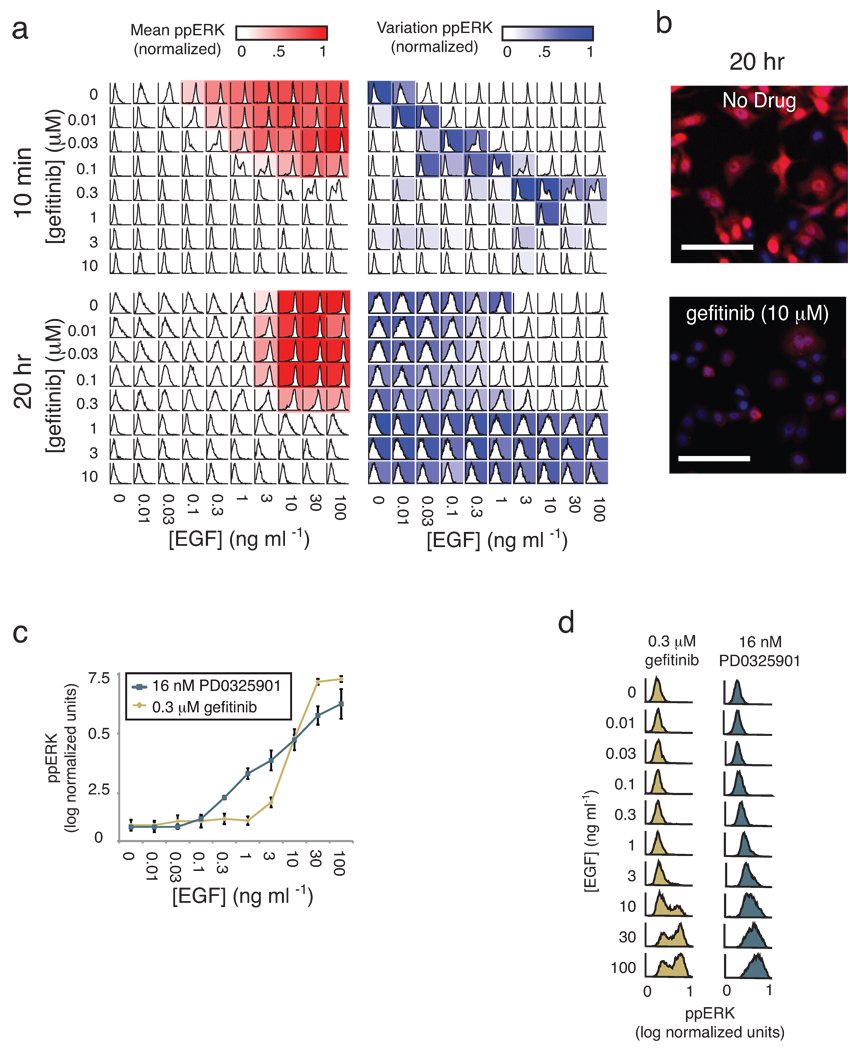
Exploring different dimensions of a multivariate drug and ligand dose-response series using SDCubes. (a) Well-mean values are computed from single-cell data recorded from cultured SKBR3 cells exposed to exogenous EGF for 10 min over a range of concentrations and then stained with antibodies specific for ppERK. Data are plotted to show a series of conventional drug dose-response relationships at different ligand concentrations (top). Inverting the axes allows the same data to be plotted as a ligand dose-response curve at different drug doses (middle). For each mean value in either plot, the underlying single-cell distribution can be visualized as a series of dot-plots (bottom panel shows gefitinib dose-response at 1 ng/mL EGF). (b) The ppERK response surface for SKBR3 cells treated as in (a) and colored according to the degree of cell-to-cell variation; darker blue represents a high coefficient of variation. (c) Whisker plots of gefitinib IC10, IC50 and IC90 values for the inhibition of ERK phosphorylation by gefitinib in SKBR3, T47D and MCF7 cells treated for 10 min with a range of EGF concentrations.
On comparing mean ppERK levels with cell-to-cell variance using plate maps (Fig. 5a), we observed maximum variability at physiologically relevant doses of drug and ligand (estimated to be 0.1–1.5 ng/mL for EGF and 0.4–50 µM for gefitinib21,22). Mean value and variance in response changed over time, such that 20 hours post-EGF/gefitinib treatment, IC50 was less dependent on [EGF] but the variance increased. By linking back to the underlying images, we observed that even in cells exposed to saturating doses of gefitinib (10 µM) for 20 hr, a subpopulation of cells (~1%) exhibited elevated ppERK levels. This implies not only that these cells were drug-resistant but also that ERK signaling could be sustained in the absence of exogenous ligand (a behavior different from that of cells that are simply gefitinib-insensitive; Fig. 5a and Fig. 5b). Thus, single-cell data revealed three interesting features of cellular responses to gefitinib and EGF. First, IC50 varied with the concentration of extracellular ligand, particularly at early time points. Second, the extent of cell-to-cell variability was maximal near intermediate, physiologically-relevant concentrations; conversely, it was masked when drug or ligand were added at high levels. Third, cell-to-cell heterogeneity changed over time, being dominated initially by broad distributions and subsequently by rare cells with sustained signaling. Whether the differences we observe are genetic23, epigenetic24 or stochastic25 in origin is not yet clear, but reversibility implies that some are indeed stochastic, as we have previously demonstrated for TNF-responsive apoptosis-inducing ligand (TRAIL)25.
Figure 5.
Single-cell analysis of drug-ligand dose responses uncovers cell-to-cell heterogeneity. (a) ppERK was measured at 10 min and 20 hr in SKBR3 cells treated with a combination of EGF and gefitinib at the indicated doses. Shown are heat maps of the mean values (red) and coefficients of variation (blue) of the underlying cell population histograms overlaid on representation of a standard 96-well microtiter plate. (b) Selected immunofluorescence images of ppERK (red) and Hoechst (blue) staining of cells 20 hr after exposure first to 10 µM gefitinib and then to 100 ng/mL EGF. Scale bar = 100µm. (c) EGF-induced ppERK dose-response curves in SKBR3 cells pretreated with sub-saturating doses of gefitinib or the MEK inhibitor PD0325901 (MEKi). Error bars represent the standard error of the mean of biological triplicates. (d) Corresponding single-cell distributions for the population mean data shown in c.
The shape of dose-response curves for drugs and ligands often depends on the agent and cell type (Fig. 4c and Supplementary Fig. 7). Gefitinib dose-response of cells exposed to EGF conformed to a sigmoidal shape as expected for simple ligand-receptor binding; however, the dose-response for an inhibitor of MEK kinase (PD0325901)26, an enzyme lying immediately upstream of ERK1/2 kinases in EGFR signaling, was nearly linear over a 103 EGF range (Fig. 5c). At the single cell level, responses to gefitinib were bimodal, with low ppERK levels in some cells and 100-fold higher levels in other cells (Fig. 5d) but responses to PD0325901 were continuous with cells exhibiting a wide range of activities. We conclude that the mean-value dose-response curves for PD0325901 and gefitinib differed in shape because of variability at the single cell level, and speculate that this might be a general explanation for non-sigmoidal dose-response relationships.
DISCUSSION
By creating a lightweight data repository customized to the design of a specific experiment and then storing the design in a machine-readable XML format, the SDCube Programming Library places experimental design foremost in organizing data for storage. The use of XML to encode ontologies simplifies harmonization with existing Web-based standards12, and the use of HDF5 allows progressive access to even very large files. As the design changes or expands, the dimensionality of SDCubes changes as do the metadata tags that point to specific data elements. The result is an approach to data and metadata storage that aim to addresses the competing demands of data integrity and flexibility. Little attention has been paid to computer-readable experimental designs, and only one public specification exists (Minimum Information for Data Analysis in Systems biology; MIDAS)27. However, it is possible to document the format of any hypothesis-driven or systematic experiment in XML, making it straightforward to use Resource Description Framework (RDF) and Web Ontology Language (OWL) to share and analyze experimental designs, a critical step in making the results from complex experiments machine-interpretable in light of their purpose and context.
Although we demonstrate the application of SDCubes to microscopy data using ImageRail, the SDCube format is in principle adaptable to any type of high-dimensional data, and we have created preliminary schemata for multi-color flow cytometry10 and multiplex or array-based biochemical assays28 (see supplementary materials). MATLAB users will recognize that some SDCube functionality is already present in MATLAB, which makes extensive internal use of multi-dimensional data arrays (indeed, MATLAB can read HDF5 files). However, MATLAB files cannot duplicate key features of SDCubes: they cannot be read incrementally, their data models cannot be referenced to external ontologies or parsed using web-based tools, and MATLAB is not open-source—an important consideration for a data standard.
ImageRail is designed to be interoperable with existing open-source image analysis software, including ImageJ, CellProfiler and OME 13–15,29 Interoperability is important to avoid duplication of effort but ImageRail also needs to function as a stand-alone application; hence the inclusion of common segmentation and visualization routines.
The ability of SDCubes and ImageRail to systematize data from complex dose-response experiments has made it possible to implement an efficient scheme for single-cell pharmacology. Exposing tumor cells to growth factors and kinase inhibitors in combination reveals many examples of cell-to-cell variability; some of these are likely to have non-genetic origins, by direct analogy to the variability observed in cellular responses to TRAIL25, T-cell receptor agonists30 and other ligands31. Variability is maximal at doses close to the IC50 of gefitinib or the EC50 of EGF, precisely the doses likely to prevail in real patients. It therefore seems reasonable that application of single-cell pharmacology will help to uncover the basis of fractional killing by anti-cancer drugs and assist in dissecting the origins of intrinsic and acquired drug resistance32.
In many exploratory biological experiments, data collection and analysis are iterative processes undertaken by a limited number of people. In this environment, the high-integrity, multi-user, read-write operations enabled by conventional databases represent unnecessary overhead and SDCubes offer an effective alternative. However, as data become more mature or an experiment nears completion, it will often be advantageous to move key results to a relational database. One way to accomplish this is to create a specialized summary view of an SDCube and then import the summary data into a database. Only data conforming to a pre-existing standard would be accessible in the database, but an SDCube containing all primary data could easily be called using a Uniform Resource Identifier (URI, akin to a Web URL). It is possible that new types of databases will be developed with science in mind (e.g., SciDB; http://www.scidb.org/), but we predict that lightweight, adaptable, file-based data storage will always co-exist with server-based data management, and that sophisticated file formats such as SDCubes will provide a missing link between creative experimentation and machine-interpretable data.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institute of Health grants HG006097, HG005693 and GM68762. We thank G. Danuser, T. Mitchison and M. Eisenstein for help with the manuscript, Applied Precision Inc., C. Brown and K. Teplitz for help with instrumentation and G. Odell and J. Baker for inspiration.
Footnotes
AUTHOR CONTRIBUTIONS
B.L.M, M.P.M. and J.L.M. programmed the software. B.L.M, M.N., J.L.M. and P.K.S. developed the method and wrote the manuscript.
COMPETING FINANCIAL INTEREST
P.K.S. is a founder and stockholder in Glencoe Software, a private company that develops software based on Open Microscopy Environment standards. Glencoe developed the OMERO server mentioned in this article (the software is available under a GNU General Public Library License at http://www.openmicroscopy.org). P.K.S. is also a member of the Board of Directors of Applied Precision Inc., which manufactured the scanning microscope used in this study.
REFERENCES
- 1.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maheswari U, et al. The Diatom EST Database. Nucleic Acids Res. 2005;33:D344–D347. doi: 10.1093/nar/gki121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawley JB. Handbook of Biological Confocal Microscopy. 3rd Edition. New York, NY: Springer Science+Business Media, LLC; 2006. [Google Scholar]
- 4.Gaudet S, et al. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad C, Gerlich DW. Automated microscopy for high-content RNAi screening. J Cell Biol. 2010;188:453–461. doi: 10.1083/jcb.200910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–453. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 8.Snijder B, et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 9.Gehlenborg N, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 10.Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty MT, et al. Unifying Biological Image Formats with HDF5. ACM Queue. 2009 doi: 10.1145/1562764.1562781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor CF, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 14.Moore J, et al. Open tools for storage and management of quantitative image data. Methods Cell Biol. 2008;85:555–570. doi: 10.1016/S0091-679X(08)85024-8. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg IG, et al. The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 17.Ciardiello F, et al. Antitumor effect and potentiation of cytotoxic drugs activity in human cancer cells by ZD-1839 (Iressa), an epidermal growth factor receptor-selective tyrosine kinase inhibitor. Clin Cancer Res. 2000;6:2053–2063. [PubMed] [Google Scholar]
- 18.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 19.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 20.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 21.Blaimauer K, et al. Effects of epidermal growth factor and keratinocyte growth factor on the growth of oropharyngeal keratinocytes in coculture with autologous fibroblasts in a three-dimensional matrix. Cells Tissues Organs. 2006;182:98–105. doi: 10.1159/000093064. [DOI] [PubMed] [Google Scholar]
- 22.McKillop D, et al. Tumor penetration of gefitinib (Iressa), an epidermal growth factor receptor tyrosine kinase inhibitor. Mol Cancer Ther. 2005;4:641–649. doi: 10.1158/1535-7163.MCT-04-0329. [DOI] [PubMed] [Google Scholar]
- 23.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown A, Carlson T, Loi C-M, Graziano M. Pharmacodynamic and toxicokinetic evaluation of the novel MEK inhibitor, PD0325901, in the rat following oral and intravenous administration. Cancer Chemotherapy and Pharmacology. 2007;59:671–679. doi: 10.1007/s00280-006-0323-5. [DOI] [PubMed] [Google Scholar]
- 27.Saez-Rodriguez J, et al. Flexible informatics for linking experimental data to mathematical models via DataRail. Bioinformatics. 2008;24:840–847. doi: 10.1093/bioinformatics/btn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albeck JG, et al. Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol. 2006;7:803–812. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- 29.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 30.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr Opin Chem Biol. 2009;13:556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang R, Niepel M, Mitchison TK, Sorger PK. Dissecting variability in responses to cancer chemotherapy through systems pharmacology. Clin Pharmacol Ther. 2010;88:34–38. doi: 10.1038/clpt.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray-Rust P, Rzepa HS. Chemical markup, XML, and the World Wide Web. 4. CML schema. J Chem Inf Comput Sci. 2003;43:757–772. doi: 10.1021/ci0256541. [DOI] [PubMed] [Google Scholar]
- 34.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 35.Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat Methods. 2006;3:825–831. doi: 10.1038/NMETH931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Curr Opin Chem Biol. 2009;13:398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexopoulos LG, Saez-Rodriguez J, Cosgrove BD, Lauffenburger DA, Sorger PK. Networks inferred from biochemical data reveal profound differences in toll-like receptor and inflammatory signaling between normal and transformed hepatocytes. Mol Cell Proteomics. 2010;9:1849–1865. doi: 10.1074/mcp.M110.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WW, et al. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol. 2009;5:239. doi: 10.1038/msb.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendriks BS, Espelin CW. DataPflex: a MATLAB-based tool for the manipulation and visualization of multidimensional datasets. Bioinformatics. 2010;26:432–433. doi: 10.1093/bioinformatics/btp667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.