Abstract
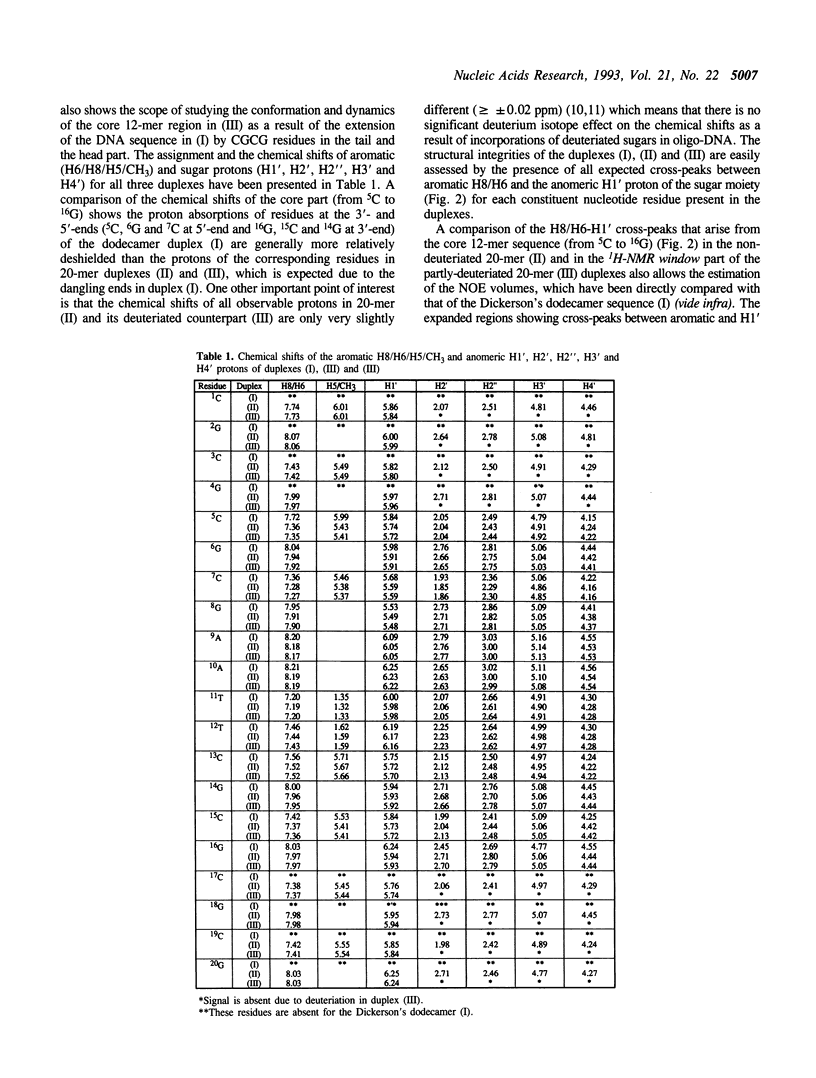
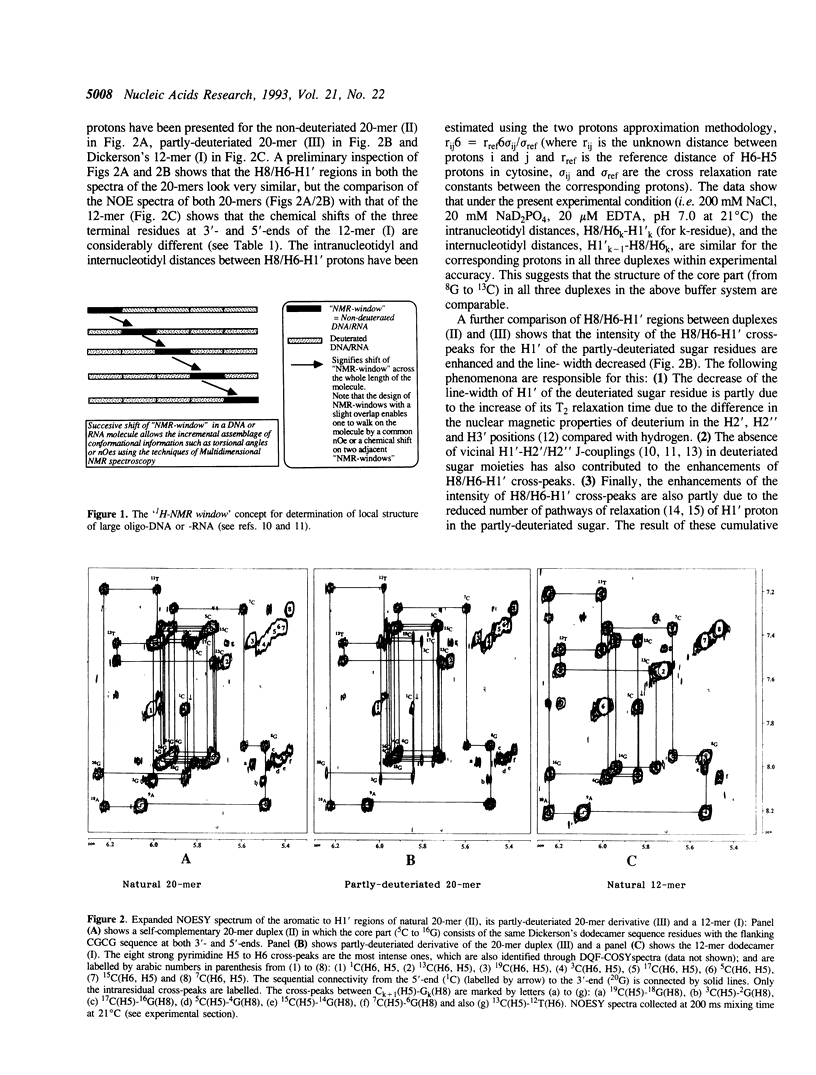
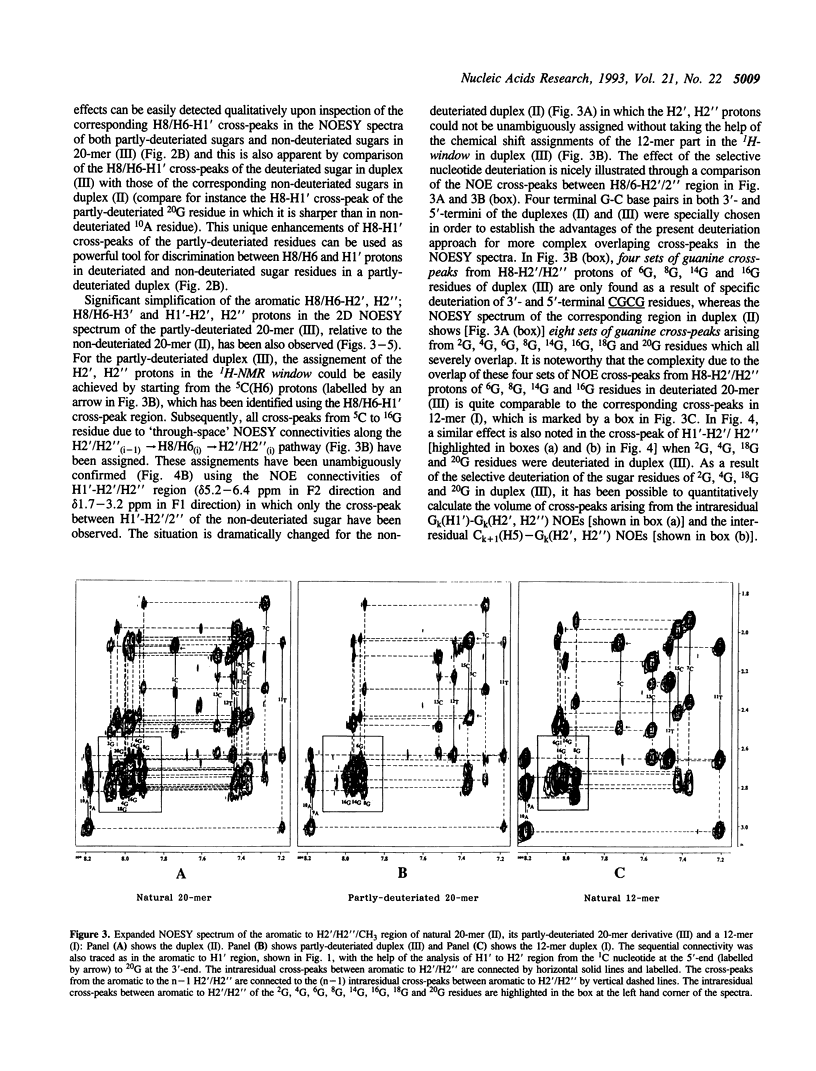
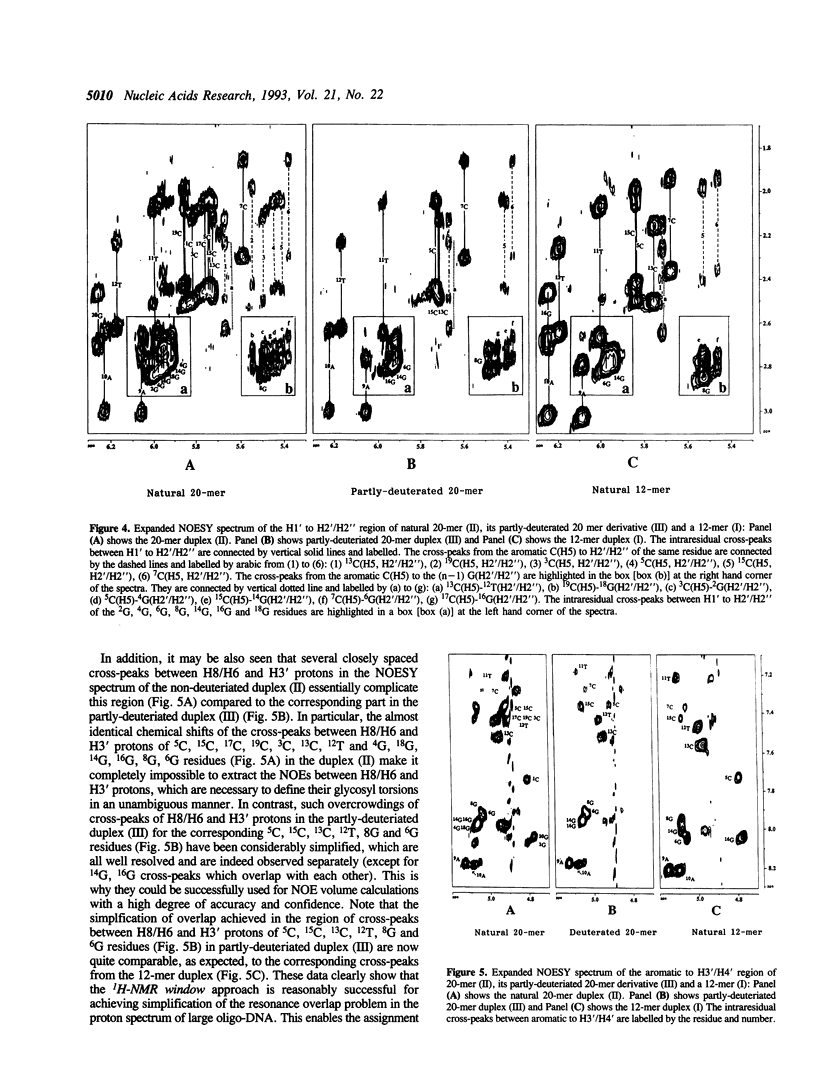
The concept of the 1H-NMR window has been developed and examined through a comparative study of NOESY spectra of a self-complementary Dickerson's dodecamer (I) [5'd(5C6G7C8G9A10A11T12T13C-14G15C16G)2(3')], a self-complementary 20-mer (II) [(5'd(1C2G3C4G5C6G7C8G9A10A11T12T13C14G15C16G17C18G19C20G)2(3')] in which the core part consists of the same Dickerson's dodecamer sequence with the flanking CGCG residues at both 3' and 5'-ends, and the partly-deuteriated (shown by underlined CGCG residues at both 3' and 5'-ends) analogous duplex (III) [5'd(1C2G3C4G5C6G7C8G9A10A11T12T13C14G15C16G17C18G19C20G)2(3')] in which the core 5C to 16G part (i.e. 1H-NMR window) consists of the natural Dickerson's dodecamer sequence. A comparison of their NOESY spectra clearly demonstrates that the severe overlap of proton resonances in the larger DNA duplex (II) has been successfully reduced in the partly-deuterated duplex (III) as a result of specific incorporations of the sugar-deuteriated nucleotide residues in the latter [stereospecific > 97 atom % 2H enrichment at H2', H2'' and H3' sites, approximately 85 atom % 2H enrichment at H4' and approximately 20 atom % 2H enrichment at H1' (see refs. 10 and 11) in the 20-mer duplex (III)]. These simplifications of the resonance overlap by the deuteriation approach have enabled unequivocal chemical shift assignments and extraction of the quantitative NOE data in the 1H-NMR window part of duplex (III). A comparison of the 12-nucleotide long 1H-NMR window in (III) with that of the 12-mer duplex (I) also shows the scope of studying the changes in conformation and dynamics of the core 12-mer region in (III) which result from the increase of molecular weight due to the DNA chain extension. It is noteworthy that such a study is clearly impossible for the natural 20-mer (II) because of the inherent problem of the overlap of resonances.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Földesi A., Nilson F. P., Glemarec C., Gioeli C., Chattopadhyaya J. NMR spectroscopic properties (1H at 500 MHz) of deuterated* ribonucleotide-dimers ApU*, GpC*, partially deuterated 2'-deoxyribonucleotide-dimers d(TpA*), d(ApT*), d(GpC*) and their comparison with natural counterparts (1H-NMR window). J Biochem Biophys Methods. 1993 Feb;26(1):1–26. doi: 10.1016/0165-022x(93)90018-j. [DOI] [PubMed] [Google Scholar]
- Majumdar A., Hosur R. V. A frequency-time domain 3D COSY-J technique for measurement of 1H-31P coupling constants in oligonucleotides. J Biomol NMR. 1991 Jul;1(2):205–208. doi: 10.1007/BF01877231. [DOI] [PubMed] [Google Scholar]
- Maltseva T. V., Yamakage S. I., Agback P., Chattopadhyaya J. Direct estimation of base-pair exchange kinetics in oligo-DNA by a combination of NOESY and ROESY experiments. Nucleic Acids Res. 1993 Sep 11;21(18):4288–4295. doi: 10.1093/nar/21.18.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus D., Wagner G., Vasák M., Kägi J. H., Wüthrich K. Systematic application of high-resolution, phase-sensitive two-dimensional 1H-NMR techniques for the identification of the amino-acid-proton spin systems in proteins. Rabbit metallothionein-2. Eur J Biochem. 1985 Sep 2;151(2):257–273. doi: 10.1111/j.1432-1033.1985.tb09096.x. [DOI] [PubMed] [Google Scholar]
- Oda Y., Nakamura H., Yamazaki T., Nagayama K., Yoshida M., Kanaya S., Ikehara M. 1H NMR studies of deuterated ribonuclease HI selectively labeled with protonated amino acids. J Biomol NMR. 1992 Mar;2(2):137–147. doi: 10.1007/BF01875525. [DOI] [PubMed] [Google Scholar]
- Pachter R., Arrowsmith C. H., Jardetzky O. The effect of selective deuteration on magnetization transfer in larger proteins. J Biomol NMR. 1992 Mar;2(2):183–194. doi: 10.1007/BF01875529. [DOI] [PubMed] [Google Scholar]


