Abstract
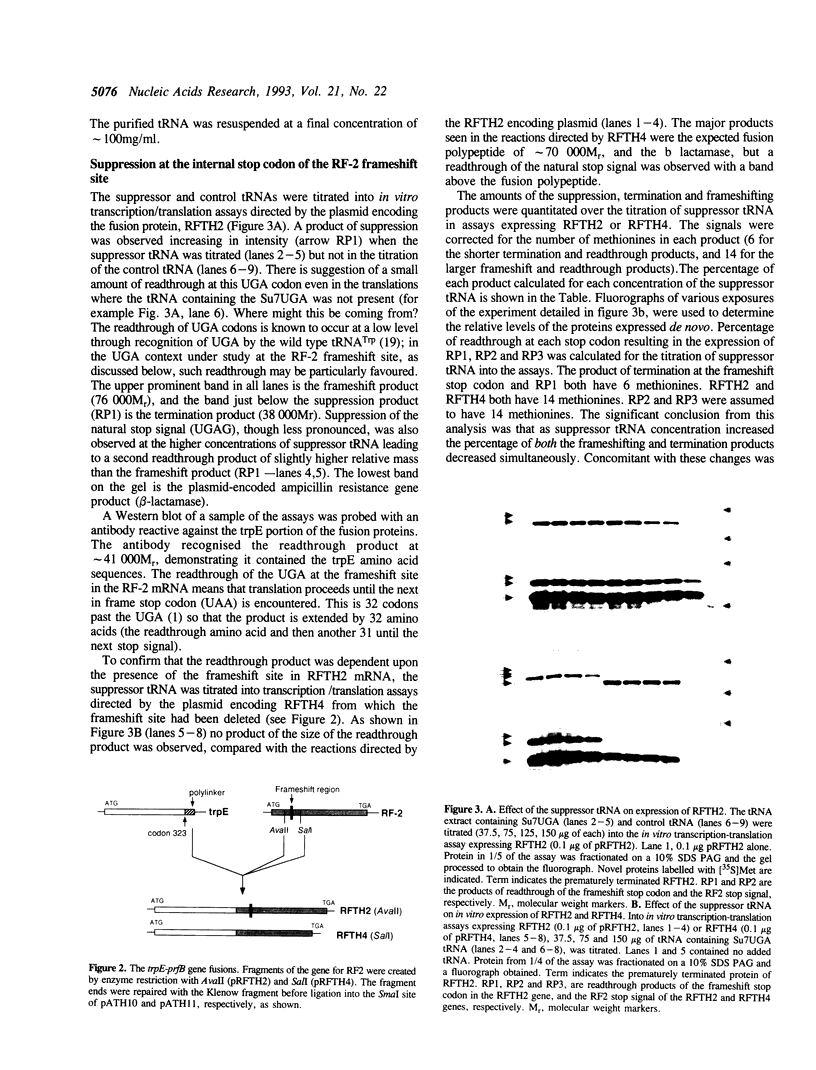
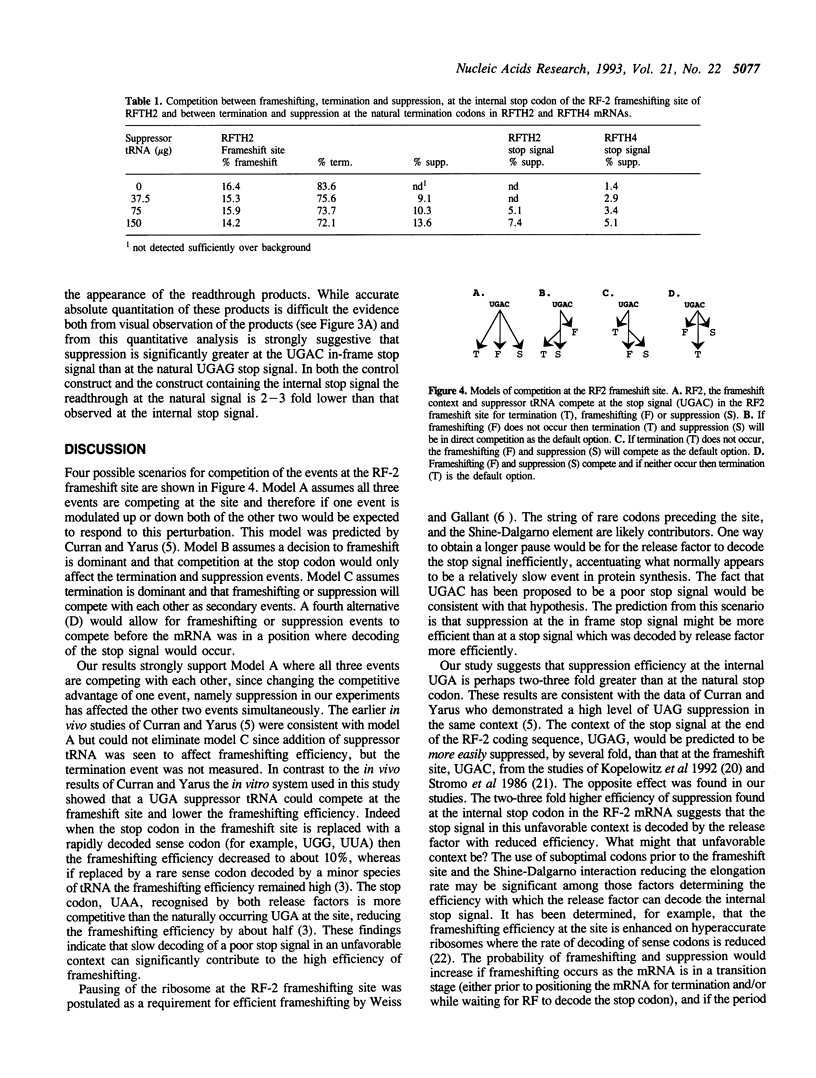
Competition between frameshifting, termination, and suppression at the frameshifting site in the release factor-2 (RF-2) mRNA was determined in vitro using a coupled transcription-translation system by adding a UGA suppressor tRNA. The expression system was programmed with a plasmid containing a trpE-prfB fusion gene so that each of the products of the competing events could be measured. With increasing concentrations of suppressor tRNA the readthrough product increased at the expense of both the termination and the frameshifting product indicating all three processes are in direct competition. The readthrough at the internal UGA termination codon was greater than that at the natural UGA termination codon at the end of the coding sequence. The results suggest that this enhanced suppression may reflect slower decoding of the internal stop codon by the release factor giving suppression a competitive advantage. The internal UGAC stop signal at the frameshift site has been proposed to be a relatively poor signal, but in addition the release factor may be less able to recognise the signal with the mRNA in such a constrained state. Consequently, the frameshifting event itself will be more competitive with termination in vivo because of this longer pause as the release factor is decoding the stop signal.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack K. A., Wade H. E. The sedimentation behaviour of ribonuclease-active and -inactive ribosomes from bacteria. Biochem J. 1965 Sep;96(3):671–680. doi: 10.1042/bj0960671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988 Sep 5;203(1):75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Donly B. C., Edgar C. D., Adamski F. M., Tate W. P. Frameshift autoregulation in the gene for Escherichia coli release factor 2: partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 1990 Nov 25;18(22):6517–6522. doi: 10.1093/nar/18.22.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson G., Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988 Sep;52(3):354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Gold L. Translation of the UGA triplet in vitro by tryptophan transfer RNA's. J Mol Biol. 1971 Jun 14;58(2):459–468. doi: 10.1016/0022-2836(71)90363-9. [DOI] [PubMed] [Google Scholar]
- Kopelowitz J., Hampe C., Goldman R., Reches M., Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol. 1992 May 20;225(2):261–269. doi: 10.1016/0022-2836(92)90920-f. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipley J., Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986 Aug 26;14(16):6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Murphy J. P., Gallant J. A. Genetic screen for cloned release factor genes. J Bacteriol. 1984 Apr;158(1):362–364. doi: 10.1128/jb.158.1.362-364.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Gallant J. Mechanism of ribosome frameshifting during translation of the genetic code. 1983 Mar 31-Apr 6Nature. 302(5907):389–393. doi: 10.1038/302389a0. [DOI] [PubMed] [Google Scholar]
- Williams J. M., Donly B. C., Brown C. M., Adamski F. M., Trotman C. N., Tate W. P. Frameshifting in the synthesis of Escherichia coli polypeptide chain release factor two on eukaryotic ribosomes. Eur J Biochem. 1989 Dec 22;186(3):515–521. doi: 10.1111/j.1432-1033.1989.tb15237.x. [DOI] [PubMed] [Google Scholar]



