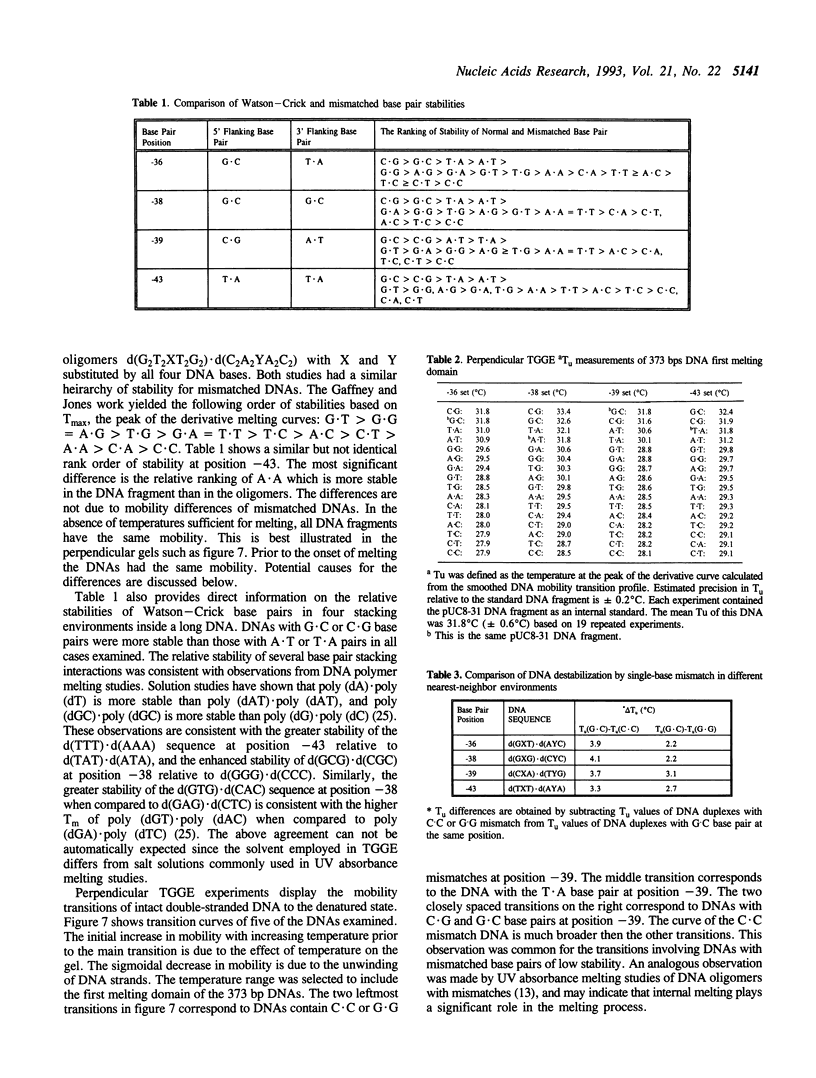
Abstract
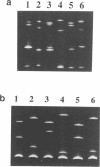
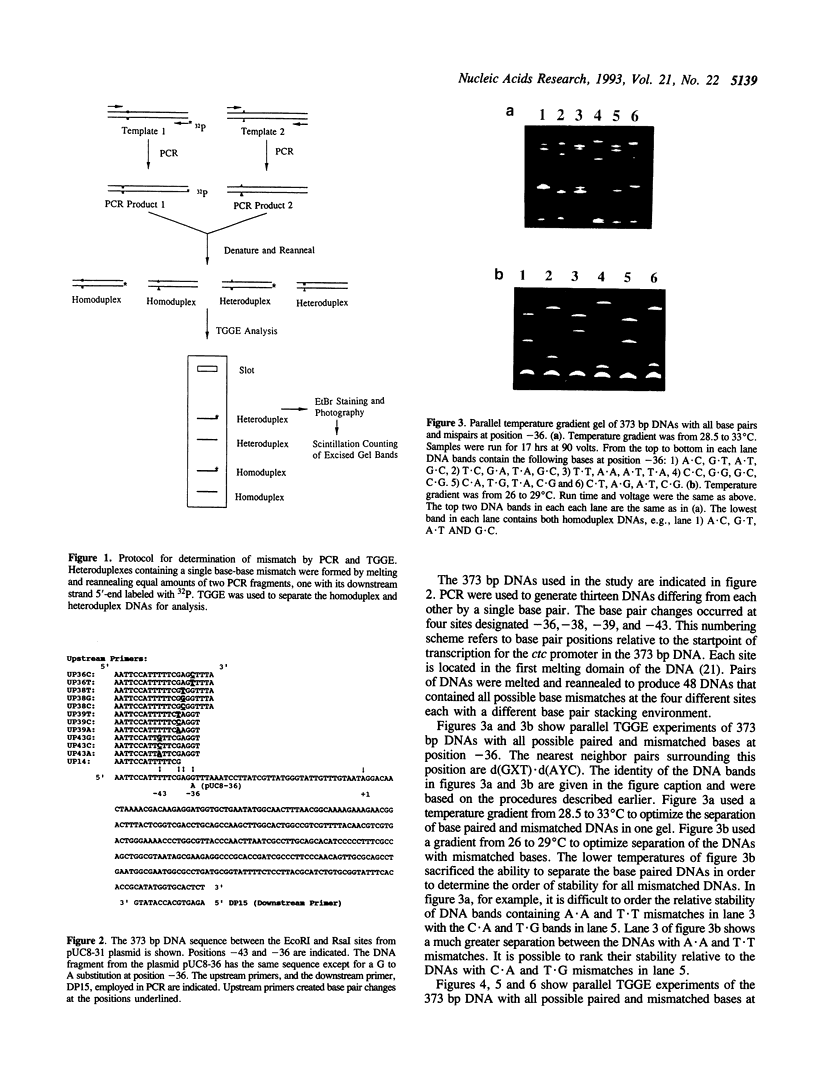
Temperature-gradient gel electrophoresis (TGGE) was employed to determine the thermal stabilities of 48 DNA fragments that differ by single base pair mismatches. The approach provides a rapid way for studying how specific base mismatches effect the stability of a long DNA fragment. Homologous 373 bp DNA fragments differing by single base pair substitutions in their first melting domain were employed. Heteroduplexes were formed by melting and reannealing pairs of DNAs, one of which was 32P-labeled on its 5'-end. Product DNAs were separated based on their thermal stability by parallel and perpendicular temperature-gradient gel electrophoresis. The order of stability was determined for all common base pairs and mismatched bases in four different nearest neighbor environments; d(GXT).d(AYC), d(GXG).d(CYC), d(CXA).d(TYG), and d(TXT).d(AYA) with X,Y = A, T, C, or G. DNA fragments containing a single mismatch were destabilized by 1 to 5 degrees C with respect to homologous DNAs with complete Watson-Crick base pairing. Both the bases at the mismatch site and neighboring stacking interactions influence the destabilization caused by a mismatch. G.T, G.G and G.A mismatches were always among the most stable mismatches for all nearest neighbor environments examined. Purine.purine mismatches were generally more stable than pyrimidine.pyrimidine mispairs. Our results are in very good agreement with data where available from solution studies of short DNA oligomers.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams E. S., Murdaugh S. E., Lerman L. S. Comprehensive detection of single base changes in human genomic DNA using denaturing gradient gel electrophoresis and a GC clamp. Genomics. 1990 Aug;7(4):463–475. doi: 10.1016/0888-7543(90)90188-z. [DOI] [PubMed] [Google Scholar]
- Akiyama M., Maki H., Sekiguchi M., Horiuchi T. A specific role of MutT protein: to prevent dG.dA mispairing in DNA replication. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3949–3952. doi: 10.1073/pnas.86.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F. H., Wolk S., Cruz P., Tinoco I., Jr Structure, dynamics, and thermodynamics of mismatched DNA oligonucleotide duplexes d(CCCAGGG)2 and d(CCCTGGG)2. Biochemistry. 1987 Jun 30;26(13):4068–4075. doi: 10.1021/bi00387a049. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Hess S. T., Nicholson-Tuell J. The influence of nearest neighbors on the rate and pattern of spontaneous point mutations. J Mol Evol. 1992 Mar;34(3):189–200. doi: 10.1007/BF00162968. [DOI] [PubMed] [Google Scholar]
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Leonard G. A., Booth E. D., Kneale G. Influence of pH on the conformation and stability of mismatch base-pairs in DNA. J Mol Biol. 1990 Apr 5;212(3):437–440. doi: 10.1016/0022-2836(90)90320-L. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney B. L., Jones R. A. Thermodynamic comparison of the base pairs formed by the carcinogenic lesion O6-methylguanine with reference both to Watson-Crick pairs and to mismatched pairs. Biochemistry. 1989 Jul 11;28(14):5881–5889. doi: 10.1021/bi00440a026. [DOI] [PubMed] [Google Scholar]
- Gibbs R. A., Nguyen P. N., Caskey C. T. Detection of single DNA base differences by competitive oligonucleotide priming. Nucleic Acids Res. 1989 Apr 11;17(7):2437–2448. doi: 10.1093/nar/17.7.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldberg P., Güttler F. A simple method for identification of point mutations using denaturing gradient gel electrophoresis. Nucleic Acids Res. 1993 May 11;21(9):2261–2262. doi: 10.1093/nar/21.9.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton J. R. Renaturation kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic Acids Res. 1977 Oct;4(10):3537–3555. doi: 10.1093/nar/4.10.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S. H., Kelly P. J., Wartell R. M., Hunter S., Varma V. A. Selecting DNA fragments for mutation detection by temperature gradient gel electrophoresis: application to the p53 gene cDNA. Electrophoresis. 1993 Jul;14(7):561–565. doi: 10.1002/elps.1150140188. [DOI] [PubMed] [Google Scholar]
- Klump H., Burkart W. Calorimetric measurements of the transition enthalpy of DNA in aqueous urea solutions. Biochim Biophys Acta. 1977 Apr 19;475(4):601–604. doi: 10.1016/0005-2787(77)90320-3. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Fischer S. G., Hurley I., Silverstein K., Lumelsky N. Sequence-determined DNA separations. Annu Rev Biophys Bioeng. 1984;13:399–423. doi: 10.1146/annurev.bb.13.060184.002151. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Nishigaki K., Husimi Y., Masuda M., Kaneko K., Tanaka T. Strand dissociation and cooperative melting of double-stranded DNAs detected by denaturant gradient gel electrophoresis. J Biochem. 1984 Mar;95(3):627–635. doi: 10.1093/oxfordjournals.jbchem.a134651. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Roongta V. A., Jones C. R., Gorenstein D. G. Effect of distortions in the deoxyribose phosphate backbone conformation of duplex oligodeoxyribonucleotide dodecamers containing GT, GG, GA, AC, and GU base-pair mismatches on 31P NMR spectra. Biochemistry. 1990 Jun 5;29(22):5245–5258. doi: 10.1021/bi00474a005. [DOI] [PubMed] [Google Scholar]
- Singer B., Chavez F., Goodman M. F., Essigmann J. M., Dosanjh M. K. Effect of 3' flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatti K. M., Moran C. P., Jr Utilization of one promoter by two forms of RNA polymerase from Bacillus subtilis. Nature. 1985 Mar 14;314(6007):190–192. doi: 10.1038/314190a0. [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Hosseini S. H., Moran C. P., Jr Detecting base pair substitutions in DNA fragments by temperature-gradient gel electrophoresis. Nucleic Acids Res. 1990 May 11;18(9):2699–2705. doi: 10.1093/nar/18.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werntges H., Steger G., Riesner D., Fritz H. J. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res. 1986 May 12;14(9):3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]