Abstract
Genetic and fossil evidence supports a single, recent (<200,000 yr) origin of modern Homo sapiens in Africa, followed by later population divergence and dispersal across the globe (the “Out of Africa” model). However, there is less agreement on the exact nature of this migration event and dispersal of populations relative to one another. We use the empirically observed genetic correlation structure (or linkage disequilibrium) between 242,000 genome-wide single nucleotide polymorphisms (SNPs) in 17 global populations to reconstruct two key parameters of human evolution: effective population size (Ne) and population divergence times (T). A linkage disequilibrium (LD)–based approach allows changes in human population size to be traced over time and reveals a substantial reduction in Ne accompanying the “Out of Africa” exodus as well as the dramatic re-expansion of non-Africans as they spread across the globe. Secondly, two parallel estimates of population divergence times provide clear evidence of population dispersal patterns “Out of Africa” and subsequent dispersal of proto-European and proto-East Asian populations. Estimates of divergence times between European–African and East Asian–African populations are inconsistent with its simplest manifestation: a single dispersal from the continent followed by a split into Western and Eastern Eurasian branches. Rather, population divergence times are consistent with substantial ancient gene flow to the proto-European population after its divergence with proto-East Asians, suggesting distinct, early dispersals of modern H. sapiens from Africa. We use simulated genetic polymorphism data to demonstrate the validity of our conclusions against alternative population demographic scenarios.
Genetic and fossil evidence largely supports a single, recent (<200,000 yr) origin of modern Homo sapiens in Africa followed by a later dispersal to the rest of the world (“Out of Africa” model) (Stringer and Andrews 1988; Ingman et al. 2000; Stringer 2002; Cavalli-Sforza and Feldman 2003; Relethford 2008; Tattersall 2009). Although it is clear that humans were thriving across the Old World, from western Europe to southeast Asia, by ∼35,000 yr ago (Goebel 2007), the process of population dispersal “Out of Africa” and subsequently across Eurasia is less clear (Forster 2004; Mellars 2006a,b). Traditionally, the “Out of Africa” event is thought to have occurred in a single wave, although the precise nature of these human migration events has been difficult to discern. The process of global colonization by humans in the past will have played an important role in shaping current patterns of genetic diversity and will partially explain geographic variation in genetic susceptibility to certain diseases (Tishkoff and Verrelli 2003; Novembre and Di Rienzo 2009). The recent availability of high-density genetic information allows us to infer relationships between human populations and, through this, gain an understanding of past demographic events (Sved et al. 2008).
Linkage disequilibrium (LD) is the nonrandom association of alleles between genetic loci (Hill and Robertson 1968). Understanding patterns of LD has been crucial in designing and implementing genome-wide association studies that rely on the ability of a subset of genotyped markers, typically single nucleotide polymorphisms (SNPs), to effectively “tag” other genetic variation, including the causative variants underlying diseases (Donnelly 2008; McCarthy et al. 2008). The extent and strength of LD between any two markers depend on intrinsic cellular factors like recombination, mutation, and gene conversion rates. However, LD patterns are also shaped by extrinsic aspects of the human past, such as effective population size (Ne), migration (admixture), and selection. Through population genetics theory, the extant LD structure can be used to reconstruct these past events (Ardlie et al. 2002).
Ne is a measure of the number of independent breeding individuals in a population and is normally much less than the actual census size (Nc) since real populations depart from idealized theoretical models and their assumptions (Charlesworth 2009). Human Ne is consistently estimated to be about 10,000 (Takahata 1993). Estimates are traditionally calculated from DNA sequence diversity and represent an average Ne over many past generations. However, population size is likely to have varied considerably over human history. Patterns of LD contain information about these changes (Hayes et al. 2003; Tenesa et al. 2007). Finite Ne causes genetic drift – random fluctuations in allele or haplotype frequencies—which leads to increased LD, but this LD decays due to recombination. The greater the recombination rate between a pair of genetic markers, the more quickly LD decays. Consequently, LD between markers that are far apart reflects recent Ne, while LD between markers close together is more affected by ancient Ne (Hill and Robertson 1968; Hayes et al. 2003).
As well as offering the unique ability to monitor fluctuating population size across time, genetic polymorphism data can be used in two parallel ways to date the time (T) since two populations diverged from one another. Under neutral evolutionary theory, the level of genetic differentiation between two populations (measured by variation in allele frequencies using the FST statistic) is determined by genetic drift (Holsinger and Weir 2009). The extent of genetic drift is, in turn, dependent on effective population size and the time since divergence; for example, the impact of genetic drift will be greatest between two populations that have had a small population size and been separated for long periods. Normally, Ne and T are confounded and difficult to tease apart (Hartl and Clark 2007). However, LD data within populations could be used to estimate Ne, while the allele frequencies within and between populations can be used to calculate FST, making an estimate of T possible. These methods present a unique opportunity to independently estimate both historic Ne and the time when two populations diverged. In addition, T may also be derived from the similarity in LD patterns between populations. Immediately after their separation, the two populations are expected to show a perfect correlation in LD structure, but this will gradually decay in a manner dependent on recombination distance and time, allowing us to date the separation point (de Roos et al. 2008; Sved et al. 2008).
We explored human LD patterns using approximately 242,000 SNPs across the genome in 17 population samples from across the globe. We used these to reconstruct two key parameters of human evolution: effective population size (Ne) and population divergence times (T), and through these track the emergence and dispersal of our species “Out of Africa” and beyond. In addition, we used simulated genetic data to evaluate the performance of parameter estimators across a range of population demographic models.
Results
We examined LD patterns in a range of global populations (Fig. 1; Supplemental Table 1) using the correlation (rLD) and squared correlation (rLD2) in genotype frequencies between approximately 242,000 autosomal SNPs (Weir 2008; Sved 2009). Approximately 5.4 million pairwise LD observations were made between loci separated by genetic distances of between 0.005 and 0.25 cM. rLD2 values were binned into distance categories, averaged, and related to Ne as E(rLD2) ≈ 1/(2 + 4Nec), where c is the genetic distance between loci in morgans (Tenesa et al. 2007). rLD2 values in a particular recombination distance (c) bin give an estimate of Ne t generations in the past as t ≈ 1/(2c) (Hayes et al. 2003). The range of genetic distances in the data can potentially offer a view of Ne from approximately 5000 to 200 generations ago or 125–5 thousand years ago (KYA), assuming a generation time of 25 yr.
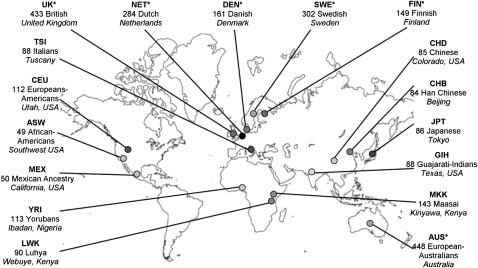
Figure 1.
Populations, codes, and sample sizes. The sampling location is indicated in italics. GenomEUtwin populations are marked *; otherwise samples are from HapMap 3. See also Supplemental Table 1.
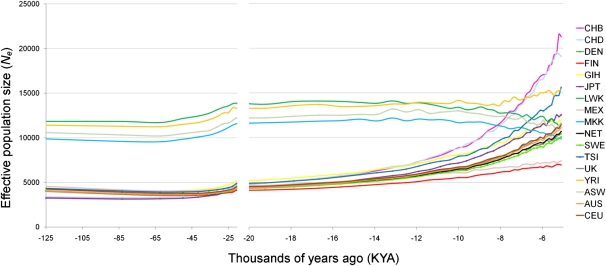
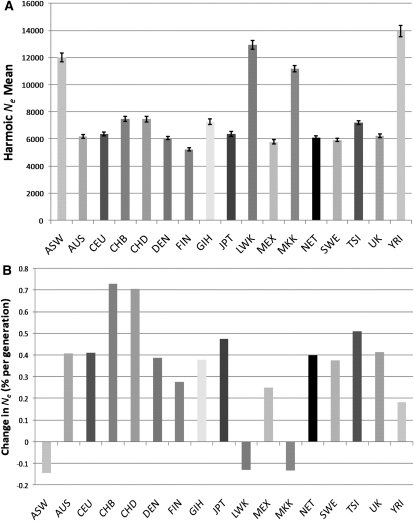
There is considerable variation in Ne estimates across both time (genetic distance categories) and space (geographic populations) (Fig. 2). As long-term Ne behaves as a harmonic mean over all recombination distance classes, it provides an average value for each population. These range from 13,900 in Yorubans (YRI) to 5200 in the Finns (FIN) (Fig. 3A), consistent with Ne estimates by different approaches and methods (Yu et al. 2004; Conrad et al. 2006). Prior to the divergence of African and non-African populations, their Ne should be the same, but we do not observe this convergence. The recombination distance-to-time relationship holds best when a population has been constant in size or grown linearly (Hayes et al. 2003). If non-African populations experienced a drastic “Out of Africa” bottleneck, this would inflate rLD2 even for markers a very small distance apart and so the Ne before the bottleneck would be severely underestimated. Averaging Ne estimates between African and non-African populations over the notional 125–25-KYA period suggests that initial human migration resulted in a substantial reduction in Ne (Fig. 3A). The African population, in contrast, has remained relatively large and stable over most of its history. However, there is evidence for a small increase in the West African Yorubans (YRI) ∼8 KYA, coinciding with declines in the East African Maasai (MKK) and Lubya (LKK) populations at the same time (Figs. 2, 3B).
Figure 2.
Spatial and temporal variation in Ne estimates. Ne was calculated from LD observations in each of 50 recombination distance classes for each population. Note the change in the time axis scale at 20 KYA. The underlying LD structure upon which these Ne estimates are based can be seen in Supplemental Figure 5.
Figure 3.
(A) The harmonic mean of Ne for each population over all recombination distance classes up to 0.25 cM. The Ne illustrated is the average from separate analysis of each autosomal chromosome, which allows the placement of 95% confidence intervals on each estimate as indicated by the error bars. (B) Ne growth rate, calculated as the percentage change in Ne at t = 200 compared to t = 800 averaged over the 600 generations. See Figure 1 for population codes.
From ∼25 KYA, all non-African populations start to expand, and distinct growth trajectories become apparent moving toward the present, reflecting the emergence of each population as a separate entity (Fig. 2). The Chinese population shows the strongest recent population growth (∼0.7% per generation, averaged over the period 20–5 KYA) (Fig. 3B), leaving them with the highest most recent Ne estimate of ∼21,000. Italians (TSI) show a significantly higher recent (<12.5 KYA) Ne than Northern/Western Europeans (10,500 vs. an average of 8200 in the CEU, NET, SWE, DEN, and UK populations; two-tailed t-test, p < 1 × 10−5), likely the consequence of bottlenecks associated with the depopulation and recolonization of Northern Europe before and after the last glacial maximum (LGM; 30–18 KYA) (Reich et al. 2001). Growth accelerates moving forward in time, with the average rate about threefold higher in the period 8–5 KYA than 20–8 KYA, presumably representing the impact of agricultural innovations on population density.
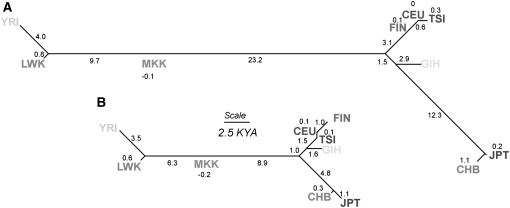
As well as offering the ability to monitor fluctuating population size across time and space, LD can be used in two parallel ways to explicitly date population divergence times (T). Under the neutral theory, the level of population differentiation (measured by FST) is determined by genetic drift (Holsinger and Weir 2009). The extent of genetic drift is dependent on Ne and T as FST ≈ T/(2Ne) (Nei 1987). Therefore, we estimate T in generations by 2NeFST (we refer to this estimator of T as TF). Normally, Ne and T are confounded together, but here we can estimate Ne from the LD structure and use SNP allele frequencies to calculate FST. We initially relied on average Ne from recombination distance classes up to 0.10 cM so as to obtain a long-term estimate of Ne. The matrix of interpopulation TF values (Supplemental Table 2) is summarized in a neighbor-joining (NJ) phylogenetic tree, for a subset of the nine most differentiated populations (Fig. 4A). This provides a clear picture of the historical relationship between populations with three broad groupings apparent: Africans, East Asians, and Europeans. Early human dispersal patterns can be inferred through estimates of T between these three main groups. The average TF estimate between these African and European populations is ∼36 KYA, ∼44 KYA for Africans and East Asians, and ∼20 KYA between East Asians and Europeans. If Ne estimates derived over 0.25 cM are used, then average TF between African and European or Asian populations increases to ∼48 KYA and ∼66 KYA, respectively, and ∼34 KYA for East Asian–European comparisons (Supplemental Table 2). The estimates of non-African population foundation are in the range of those calculated from other genetic loci and methods: 40–50 KYA and 52–60 KYA from Y-chromosome (Thomson et al. 2000) and mitochondrial (mt) DNA (Ingman et al. 2000) sequence, respectively, and 37–57 KYA from autosomal STRs (Zhivotovsky et al. 2003). They also accord well with fossil and archaeological evidence that modern Homo sapiens had reached Western Europe and East Asia by at least 30–40 KYA (Stringer 2002).
Figure 4.
Unrooted neighbor-joining (NJ) networks illustrating (A) TF and (B) TLD divergence times. TF and TLD are estimated using information from genetic distance classes ≤0.10 cM. Branch lengths are proportional to divergence times in thousands of years ago (KYA) with actual values reported along each branch or over the population name where the branch length is too small for labeling. To maintain the additivity of distances along the tree, the NJ method can introduce negative branch lengths by chance. Divergence times between six Northern European (NET, UK, AUS, DEN, SWE, and CEU) and two Chinese (CHD and CHB) population samples were very small (less than 10 generations) for both TF and TLD, suggesting that they are very similar populations in relative global terms. We therefore included only one representative of each set of populations (CEU and CHB for Northwest Europe and China, respectively) in the construction of the NJ tree. We also did not include the Mexican (MEX) and African-Americans (ASW) samples in NJ construction since they are known to be a product of recent admixture between different continental parental populations (see Supplemental Fig. 4). Neighbor-joining trees were generated using the PHYLIP package (version 3.68 available at http://evolution.genetics.washington.edu/phylip.html).
As an empirical control, T may also be estimated from LD alone (TLD). Immediately after divergence, two populations will have a perfect correlation in LD structure, but this will gradually decay in a manner dependent on recombination distance and time but approximately independent of Ne (de Roos et al. 2008; Sved et al. 2008). The NJ tree topology based on an interpopulation matrix of TLD values (Supplemental Table 2), calculated from pairs of SNPs ≤0.10 cM apart (Fig. 4B), is similar to that based on TF, and there is a strong correlation between the rank order of divergence times between TF and TLD (r = 0.97). Notwithstanding this, TLD values are generally substantially less than TF, and, accordingly, the TLD NJ tree is generally deflated in scale by ∼50% (Fig. 4B). As with TF, the average Europe–Africa TLD is significantly smaller than Africa–East Asia (∼17 KYA and ∼20 KYA, respectively; p = 9.7 × 10−5, two-tailed t-test), while European–East Asia divergence was placed at ∼9 KYA (Supplemental Table 3). TLD estimates based on the correlation in LD structure up to 0.25 cM are 15 KYA, 17 KYA, and 11 KYA, respectively, for the three intercontinental comparisons. (See Supplemental Table 2.) TLD is expected to underestimate the absolute divergence times of African, European, and Asian populations compared to TF, probably largely due to fixation bias and migration (Supplemental Figs. 1, 2; Sved et al. 2008; see Methods).
Estimates of TF and TLD between European, East Asian, and African populations suggest a more complex “Out of Africa” dispersal pattern than that proposed by a single-wave “Out of Africa” model. Under a single-wave “Out of Africa” model, the divergence times of Africa versus Europe and Africa versus Asia are expected to be roughly similar. However, the Europe–Africa TF is significantly smaller (more recent) than that from the East Asia–Africa comparison (p < 1 × 10−17) (see Supplemental Table 3).
Simulations
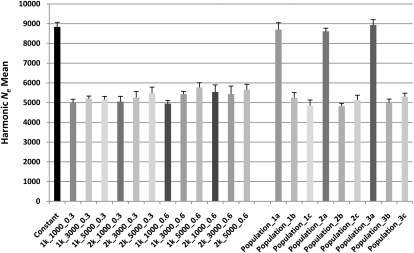
From the genetic data of simulated population scenarios, we use LD patterns among loci separated by genetic distances of between 0.005 and 0.25 cM, representing approximately 200 to 5000 generations ago, to assess the behavior of the estimators Ne, TLD, and TF. The extent of the bottleneck (r) has little effect on estimates of Ne at generations both before and after the simulated bottleneck event (Fig. 5; Supplemental Fig. 3). The population maintained at a constant Ne = 10,000 (Constant) has a consistent estimate of historic Ne, although there is a downward bias of ∼10%. Populations with simulated bottlenecks show inflated levels of LD even at markers close together, leading to Ne severely underestimated at generations prior to the bottleneck, consistent with estimates of Ne in non-African populations. Simulated population growth is clearly apparent in estimates of Ne in generations after the bottlenecks, demonstrating the ability of the estimator to identify differences in the extent of growth among populations.
Figure 5.
The harmonic mean of Ne for each simulated population scenario over all genetic distances between 0.05 and 0.25 cM. The Ne illustrated is the mean from 100,000 independent simulations, with standard errors shown with error bars. The population characteristics are described in Methods and Supplemental Figure 7.
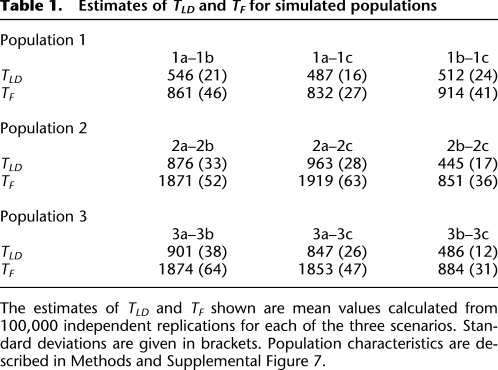
To obtain a clearer evaluation of human population divergence patterns, we used the LD and allele frequency information from simulated population scenarios mimicking “Out of Africa” events to evaluate the performance of TF and TLD estimators (Fig. 5; Table 1). Under scenarios following a standard single-wave “Out of Africa” (e.g., Population_2), estimates of TF for Population_2a–Population_2b and Population_2a–Population_2c are very similar (Table 1), showing that TF is capable of accurately estimating true divergence times under a single-wave “Out of Africa” model. Estimates of TF and TLD between populations are not strongly influenced by the extent of bottlenecks, consistent with estimations of Ne shown from single population scenarios (Fig. 5). Naturally, caution is needed when interpreting results from simulations that follow simple population scenarios; however, our results clearly show that the estimators TF and TLD are robust to differences in the extent of population bottleneck events and are able to accurately estimate divergence times for populations relative to one another.
Table 1.
Estimates of TLD and TF for simulated populations
The estimates of TLD and TF shown are mean values calculated from 100,000 independent replications for each of the three scenarios. Standard deviations are given in brackets. Population characteristics are described in Methods and Supplemental Figure 7.
Discussion
We used LD patterns in 17 population samples to estimate Ne and to date population divergence times, with estimator performance evaluated using simulated genetic data. As well as allowing the incorporation of information from across the entire genome simultaneously, LD-based estimations of Ne have an advantage of allowing us to track changes in population size across time, as represented by different recombination distances; and space, through a global spread of populations. The results capture the substantial “bottleneck” that accompanied the emergence of modern humans from Africa and the subsequent re-expansion.
While Ne shows evidence of the expected bottleneck effect under the “Out of Africa” model, estimates of population divergence times are inconsistent with its simplest manifestation: a single dispersal from the continent followed by a split into Western and Eastern Eurasian branches. Under this scenario, the divergence times of these two groups relative to Africa would be expected to be similar. Both TF and TLD, two T estimators calculated by different means from the same data, consistently demonstrate a significantly more recent relationship between Europe and Africa than between East Asia and Africa. Using simulated populations, we show that under the single-wave “Out of Africa” model, TF and TLD estimate very similar divergence times between the two diverged populations and the ancestral population. Thus, the pattern of TF and TLD among human populations appears at odds with the standard single-wave “Out of Africa” model. Previous studies have noted the relationship of higher genetic distances (Keinan et al. 2007), lower levels of diversity (Ramachandran et al. 2005), and longer-range LD (Jakobsson et al. 2008) with increasing geographic distance from East Africa. A likely explanation for the pattern of population divergence seen among human populations is that they are the result of serial founder effects, and consequent greater genetic drift, as repeated subsets of the total population gradually advanced across the globe. Differences in the nature and number of founder events could potentially explain a discrepancy in some divergence time estimators since East Asia is obviously geographically more distant from Africa than Europe. However, our estimate of TF incorporates independent population-specific estimates of Ne that will reflect the history of genetic drift, while the parallel TLD estimator is approximately independent of Ne. Simulations demonstrate that the extent of bottlenecks and rates of growth are not likely to lead to the patterns of TF and TLD observed among human populations (Fig. 5; Table 1). An alternative explanation for the difference, consistent with previous modeling of human dispersal (Schaffner et al. 2005; Gutenkunst et al. 2009), is a greater migration rate between Africa and Europe, although it is less clear from these previous studies when this gene flow occurred and what form it took. Our results begin to shed light on these latter questions. The ratios of average Europe–Africa to East Asia–Africa divergence times are 0.82 and 0.89 by TF and TLD, respectively, using LD information over pairs of SNPs ≤0.1 cM apart. However, a deviation from 1 in TLD estimates over smaller distances (≤0.05 cM) corresponding to a more ancient timeframe (>25 KYA) is still apparent (0.89). If recent gene flow occurred between populations, estimates of TLD are expected to be more greatly biased over larger distances, leading to inconsistencies in TLD estimates across distance classes. Our estimators of TF and TLD are calculated under assumptions of no gene flow between populations subsequent to divergence. If such gene flow exists, it will bias the estimates of divergence times downward (Sved et al. 2008). While the exact bias is difficult to estimate (Sved et al. 2008), it appears that post-divergence migration rates from Africa to Europe would need to be approximately constant because we observe consistent ratios of TF and TLD at different genetic distances. Thus, the observations are suggestive that greater migration to Europe from sub-Saharan African has been a long-term phenomenon.
Y-chromosome and mtDNA lineages are generally highly differentiated between continents, making them powerful genetic markers of intercontinental migration. Most of the lineages that are characteristic of sub-Saharan Africa are absent in Europe (and vice versa) (Cavalli-Sforza and Feldman 2003; Underhill and Kivisild 2007). However, the coalescent time and geographic distribution of the Y-chromosome E3b (E-M215) haplogroup points to a late Pleistocene migration from Eastern Africa to Western Eurasia via the Nile Valley and Sinai Peninsula ∼20–25 KYA (Cruciani et al. 2004, 2007; Luis et al. 2004). However, these Y chromosomes are concentrated in southern Europe (Cruciani et al. 2004), whereas the smaller average divergence times between Europe and Africa relative to East Asia and Africa are still readily apparent across each individual northern European sample population (Supplemental Table 2). This suggests that the discrepancy has, at least partially, an even earlier and more pervasive origin, being established prior to the appearance, and consequent migration tagging ability, of the current range of mtDNA and Y-chromosome haplogroups.
Genetic and fossil evidence strongly supports the “Out of Africa” model (Stringer and Andrews 1988; Stringer 2002; Cavalli-Sforza and Feldman 2003; Relethford 2008). However, how, where, and when modern humans emerged from Africa is the subject of considerable debate (Lahr and Foley 1998; Mellars 2006a). mtDNA diversity supports an origin in East Africa via the “Southern route,” across the Red Sea to Arabia, ∼65 KYA, splitting shortly afterward into proto-West and Eastern Eurasian branches. The latter branch departed relatively rapidly for Eastern Eurasia and Australia, while the former moved north to Europe through the Levant at a later stage (Quintana-Murci et al. 1999; Kivisild et al. 2003; Macaulay et al. 2005). Our results, which look at divergence times in West and East Eurasian populations simultaneously, point to a more complex “Out of Africa” scenario. Firstly, they suggest a substantial gap between African/Eurasian and West/East Eurasian divergence (∼20 KYA from TF estimates), indicating an appreciable pause between leaving Africa and departure for East Eurasia. Secondly, they support further early gene flow to the remaining proto-West Eurasian population from Africa after Eurasian divergence, perhaps as a second smaller dispersal (Mellars 2006a).
Caution is warranted in trying to condense human population history into clean population splits and migration events. Our estimates of population effective size and divergence times from LD and allele frequency differences incorporate information from the entire autosomal genome and contribute to novel inference on the complex emergence of modern H. sapiens out of the African evolutionary cradle and their subsequent colonization of the globe.
Methods
Populations, samples, and genotypes
The data set consisted of 11 global HapMap 3 (Release 2A) and six GenomEUtwin Northern European population samples (Fig. 1; Supplemental Table 1; McEvoy et al. 2009) genotyped for approximately 282,000 overlapping autosomal SNPs in both population sets. Individuals or SNPs with >10% missing data were excluded from the analysis. We used Principal Component Analysis (PCA), as implemented in the EIGENSTRAT package (Patterson et al. 2006), to give an overview of the major population structure. Principal component 1 (PC1) separates the Africans from the other populations, while PC2 does likewise for East Asians (Supplemental Fig. 4). As the African-American (ASW) and Mexican (MEX) populations are clearly recently admixed groups, they were not included (or results not considered) in some analyses.
Linkage disequilibrium patterns
The estimates of Ne and divergence time (T) are based on the recombination or genetic distance between SNPs (Supplemental Fig. 5). A total of 271,497 SNPs were assigned a genetic map position using HapMap2 (Release #22) recombination data. Since rLD2 values are influenced by allele frequencies (Wray 2005), it is important to consider the impact of ascertainment bias (Clark et al. 2005). Many SNPs were discovered or included on the genotyping platform because of their frequency in one (typically European) population (Clark et al. 2005). Previous studies of LD using similar data argued that as we are effectively examining polymorphic haplotypes, the impact of ascertainment bias that affects single loci is greatly reduced (Jakobsson et al. 2008; see also Conrad et al. 2006). Secondly, we restricted our analysis to those SNPs (241,997) that were segregating (polymorphic) in all populations to further mitigate the impact of any bias (Reich et al. 2001; International HapMap 3 Consortium 2010). We observed a strong correlation in LD patterns based on fully resequenced ENCODE3 data and that based on SNP genotypes in the same individuals and populations (Supplemental Fig. 6).
For each pair of SNPs separated by <0.25 cM, for each population separately, we described linkage disequilibrium (LD) levels by the correlation (rLD) and squared correlation (rLD2) in genotype frequencies. Compared to alternative measures based on haplotypic or phased data, such correlation methods yield similar results but are computationally very efficient for whole genome data (Jakobsson et al. 2008; Weir 2008; Sved 2009). rLD can be positive or negative. A total of approximately 5.4 million pairwise LD observations (rLD and rLD2 values) in all 17 populations were made, and these were binned into one of 50 recombination distance categories with incremental upper boundaries of 0.005 cM up to 0.25 cM. We did not include the first category (LD observations in which pairs of SNPs were separated by <0.005 cM) in our analysis since these may have been particularly affected by gene conversion, for which our methods do not account (Tenesa et al. 2007).
Ne estimation
The expected relationship between average rLD2 values in each recombination distance category and Ne is approximately E(rLD2) ≈ 1/(α + 4Nec), where α = 2, accounting for the impact of mutation, and c is the recombination distance between loci in morgans (Sved 1971; Hill 1975; Weir and Hill 1980; McVean 2002). Ne can thus be estimated for each population in each distance category as Ne = 1/(4c) * [(1/r LD2) − 2]. As experimental sampling introduces chance LD, all individual r LD2 values were adjusted: r LD2 − (1/n), where n is the sample size, prior to the calculation of Ne (Weir and Hill 1980; Tenesa et al. 2007). If a population has been constant in size or has grown linearly, then E(r LD2) ≈ 1/(2 + 4Ne(t)c) is approximately true for the Ne t generations ago, where t = 1/(2c) (Hayes et al. 2003). Therefore, LD patterns over smaller recombination distances provide information about Ne from more distant times in the past, while those between markers that are separated by greater distances are informative about recent Ne. Both because some populations might deviate from the assumed growth characteristics and LD patterns are affected by a variety of other factors, the relationship t = 1/(2c) should be regarded as an approximate but useful indication of timeframes (de Roos et al. 2008).
Divergence time TF
LD can be used in two parallel ways to date population divergence (T). The first of these, which we termed TF, is calculated from information on population differentiation (FST) and Ne. FST is the proportion of the variance in allele frequencies that is found between groups (Holsinger and Weir 2009). In the absence of selection (neutral evolution) and migration, FST between two populations is essentially governed by genetic drift or random fluctuations in allele frequencies. The impact of genetic drift is largely determined by Ne (genetic drift is stronger in smaller populations) in both populations and T (the effects of genetic drift accumulate over time) as FST ≈ T/2Ne for small T/(2Ne) (Nei 1987). Smaller Ne or larger T leads to greater FST values. Therefore, we can, in theory, estimate TF in generations by 2NeFST.
FST was calculated for each SNP individually, between pairs of populations, as described by Weir (1996) under the random population model for diploid loci, and these were averaged to obtain a single pairwise population measure. Ne was calculated as the average of the harmonic means between the two populations in question over the relevant recombination distance categories. We primarily relied on distances categories ≤0.1 cM [prior to 500 generations or 12.5 KYA assuming t = (2c) and a generation time of 25 yr] to estimate interpopulation Ne, since this gives a better longer-term indication of the parameter than including the many more recombination distance classes up to 0.25 cM.
Divergence time TLD
T can also be estimated from the similarity in LD structure between populations (TLD). Directly after a split in an ancestral population, the rLD between any given pair of SNPs will be the same in both daughter populations and therefore the correlation of rLD values in population A with those in population B across all pairs of SNPs (rpop) will be 1.0. The expected value of rLD within a population decays with time due to recombination at a rate c per generation. Consequently, rpop will gradually decay in a manner dependent on recombination distance and time but approximately independent of Ne (Sved et al. 2008). For each pair of populations, rpop was computed from observed rLD values between markers (which can be positive or negative) in each recombination distance category. During the calculation of the rpop correlation, the variances in rLD values for each population were adjusted by (1/n) to account for experimental sample size, where n is the sample size of each population in the pairing under consideration. The expected decay in rpop after t generations over different recombination distances (c) of rpop = e−2ct forms the basis of the divergence time estimate TLD. The ln(rpop) is thus expected to follow a linear decrease as a function of distance c with a slope of −2TLD (de Roos et al. 2008). To ensure comparability with TF, we mainly relied on estimates of TLD over distance classes up to and including 0.1 cM.
Relationship between TF and TLD
Coalescent simulations (Sved et al. 2008) suggest that LD methods like TLD will carry an inherent downward bias in T estimates, due to the impact of fixation and migration. Fixation bias occurs when an allele at either locus in a pair in either population reaches a frequency of 1, making it impossible to measure LD (by r LD2, for instance) in one or other of the populations and determine the correlation (rpop) between populations. Therefore, observed LD is only from a subset of loci that would have been segregating in the ancestral population, increasing apparent LD similarity across populations and consequently decreasing TLD estimates. Fixation in one population or another is expected to be more common, and a more significant problem, for populations that have a small Ne relative to their divergence time (or a higher ratio of T/Ne) since the impact of genetic drift is greatest in small populations over longer periods. We observe a significant relationship between the ratio of TLD/Ne (both parameters estimated from LD over 0.1 cM recombination distance span) and the discrepancy between TF/TLD (measured as the ratio of TF/TLD) (see Supplemental Figs. 1, 2), consistent with the operation of fixation bias. The simulations also demonstrated that migration can substantially affect divergence time estimates based on LD since it increases similarity between populations, decreasing apparent divergence times. A migration rate of 0.01% (1 in 10,000 individuals) per generation can deliver a downward bias of 50% in TLD (Sved et al. 2008).
Confidence intervals
To determine the statistical significance of differences in our Ne and T estimates (e.g., the divergence time between Europe and Africa and East Asia and Africa), we used an approach based on observed variation in the estimators across chromosomes. Each estimator (Ne, TF using Ne calculated from recombination distance classes up to 0.1 cM or 0.25 cM, and TLD using the interpopulation correlation [rpop] in LD structure over 0.1 cM) was calculated separately for chromosomes 1 to 22 allowing the standard error of mean differences to be derived. Ninety-five percent confidence intervals can then be placed around any statistic and/or a standard t-test used to assess the significance of a specific difference. We were not able to use this approach with TLD estimates based on pairs of SNPs separated by the full range of distance categories up to 0.25 cM since the number of LD observations in some of the greater distance categories for smaller chromosomes was low, leading to stochastic negative rpop correlations, which render calculation of TLD from these data subsets impossible.
Simulations
A neutral coalescent approach was used to simulate genetic polymorphism data under the infinite sites model of mutation (Hudson 2002). Biological processes such as recombination, changes of Ne, population splits, and growth rates can be modeled using scaled parameters, allowing for flexible population demographic scenarios to be simulated. Simulations were used to evaluate the validity of conclusions drawn from the human populations by assessing the performance of the estimators Ne, TLD, and TF under a range of population demographic scenarios. Multiple 1-Mb segments were simulated for the following population scenarios.
Single population models
SNP level data were generated for populations with an original Ne = 10,000. At t (1000, 2000) generations ago, a bottleneck event was simulated. At the bottleneck, Ne was reduced by r (90, 70, and 50) percent. After the bottleneck event, population growth was g (0.3, 0.6) percent per generation. In addition, a population with a constant Ne = 10,000 and no bottleneck or growth was simulated. A total of 13 population scenarios were simulated; 12 representing the combinations of t, r, and g and a constant Ne population. For each scenario, a total of 100,000 independent 1-Mb chromosome segments were simulated.
Three population models
A series of more complex population demographic patterns was simulated, whereby a single ancestral population undergoes bottleneck events, splitting into three distinct populations. For each of the three scenarios described below, 100,000 independent 1-Mb chromosome segments were simulated. The relationships between populations at t = 0 are evaluated using TLD and TF estimates. A graphical representation of simulated multiple populations are given in Supplemental Figure 7.
Population_1
Ancestral population with an original Ne = 10,000. At t = 1000, the population splits into three populations. Population_1a does not undergo a bottleneck event or any growth rate, leaving Ne remaining at 10,000. Population_1b has a reduction in Ne by r = 90%, followed by population growth of 0.3% per generation. Population_1c has a reduction in Ne by 50%, followed by population growth of 0.3% per generation.
Population_2
Ancestral population with an original Ne = 10,000. At t = 2000, the population splits into two populations. Population_2a's Ne remains constant. Population_2b has a reduction in Ne by 50%, followed by population growth of 0.3% per generation. At t = 1000, population_2c splits from population_2b with a bottleneck event of 50%, followed by population growth of 0.3% per generation.
Population_3
The same simulated population demographics as population_2, with population_3c undergoing a bottleneck event of 90%.
At t = 0, SNP data were extracted (for details, see Supplemental File 1) for samples of 500 individuals randomly selected from each population. For both single- and three-population scenarios, Ne was estimated for t generations ago. Estimators of population divergence times, TLD and TF, were estimated for the three-population scenarios, using methods described above. Values of Ne, TLD, and TF were averaged across the 100,000 replications for each simulated scenario.
Acknowledgments
This work was supported by grants from the Australian National Health and Medical Research Council (grants 389892 and 613601) and the Australian Research Council (grant DP0770096). We thank Nick Martin, Leena Peltonen, Jaako Kaprio, Tim Spector, Kirsten Ohm Kyvik, Dorret Boomsma, and Nancy Pederson for access to the GenomEUtwin genotype data; and Bill Hill and Nick Martin for helpful discussion and comments on the manuscript. For the purpose of QIMR policy, “equal contributions” of B.P.M. and J.E.P. equates to joint first authorship.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.119636.110.
References
- Ardlie KG, Kruglyak L, Seielstad M 2002. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 3: 299–309 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Feldman MW 2003. The application of molecular genetic approaches to the study of human evolution. Nat Genet (Suppl) 33: 266–275 [DOI] [PubMed] [Google Scholar]
- Charlesworth B 2009. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R 2005. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res 15: 1496–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK 2006. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet 38: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Cruciani F, La Fratta R, Santolamazza P, Sellitto D, Pascone R, Moral P, Watson E, Guida V, Colomb EB, Zaharova B, et al. 2004. Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet 74: 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, La Fratta R, Trombetta B, Santolamazza P, Sellitto D, Colomb EB, Dugoujon JM, Crivellaro F, Benincasa T, Pascone R, et al. 2007. Tracing past human male movements in northern/eastern Africa and western Eurasia: New clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol Biol Evol 24: 1300–1311 [DOI] [PubMed] [Google Scholar]
- de Roos AP, Hayes BJ, Spelman RJ, Goddard ME 2008. Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle. Genetics 179: 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P 2008. Progress and challenges in genome-wide association studies in humans. Nature 456: 728–731 [DOI] [PubMed] [Google Scholar]
- Forster P 2004. Ice Ages and the mitochondrial DNA chronology of human dispersals: a review. Philos Trans R Soc Lond B Biol Sci 359: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel T 2007. Anthropology. The missing years for modern humans. Science 315: 194–196 [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5: e1000695 doi: 10.1371/journal.pgen.1000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG 2007. Principles of population genetics, 4th ed Sinauer Associates, Sunderland, MA [Google Scholar]
- Hayes BJ, Visscher PM, McPartlan HC, Goddard ME 2003. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res 13: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG 1975. Linkage disequilibrium among multiple neutral alleles produced by mutation in finite population. Theor Popul Biol 8: 117–126 [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A 1968. Linkage disequilibrium in finite populations. Theor Appl Genet 38: 226–231 [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS 2009. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 10: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Paabo S, Gyllensten U 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708–713 [DOI] [PubMed] [Google Scholar]
- International HapMap 3 Consortium. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451: 998–1003 [DOI] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D 2007. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet 39: 1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, et al. 2003. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72: 313–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr MM, Foley RA 1998. Towards a theory of modern human origins: Geography, demography, and diversity in recent human evolution. Am J Phys Anthropol Suppl 27 1998: 137–176 [DOI] [PubMed] [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioglu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ 2004. The Levant versus the Horn of Africa: Evidence for bidirectional corridors of human migrations. Am J Hum Genet 74: 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, et al. 2005. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308: 1034–1036 [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369 [DOI] [PubMed] [Google Scholar]
- McEvoy BP, Montgomery GW, McRae AF, Ripatti S, Perola M, Spector TD, Cherkas L, Ahmadi KR, Boomsma D, Willemsen G, et al. 2009. Geographical structure and differential natural selection among North European populations. Genome Res 19: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GA 2002. A genealogical interpretation of linkage disequilibrium. Genetics 162: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P 2006a. Going east: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science 313: 796–800 [DOI] [PubMed] [Google Scholar]
- Mellars P 2006b. A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature 439: 931–935 [DOI] [PubMed] [Google Scholar]
- Nei M 1987. Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Novembre J, Di Rienzo A 2009. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet 10: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D 2006. Population structure and eigenanalysis. PLoS Genet 2: e190 doi: 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437–441 [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci 102: 15942–15947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. 2001. Linkage disequilibrium in the human genome. Nature 411: 199–204 [DOI] [PubMed] [Google Scholar]
- Relethford JH 2008. Genetic evidence and the modern human origins debate. Heredity 100: 555–563 [DOI] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res 15: 1576–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C 2002. Modern human origins: progress and prospects. Philos Trans R Soc Lond B Biol Sci 357: 563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer CB, Andrews P 1988. Genetic and fossil evidence for the origin of modern humans. Science 239: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Sved JA 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor Popul Biol 2: 125–141 [DOI] [PubMed] [Google Scholar]
- Sved JA 2009. Correlation measures for linkage disequilibrium within and between populations. Genet Res 91: 183–192 [DOI] [PubMed] [Google Scholar]
- Sved JA, McRae AF, Visscher PM 2008. Divergence between human populations estimated from linkage disequilibrium. Am J Hum Genet 83: 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N 1993. Allelic genealogy and human evolution. Mol Biol Evol 10: 2–22 [DOI] [PubMed] [Google Scholar]
- Tattersall I 2009. Human origins: Out of Africa. Proc Natl Acad Sci 106: 16018–16021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, Visscher PM 2007. Recent human effective population size estimated from linkage disequilibrium. Genome Res 17: 520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW 2000. Recent common ancestry of human Y chromosomes: Evidence from DNA sequence data. Proc Natl Acad Sci 97: 7360–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC 2003. Patterns of human genetic diversity: Implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet 4: 293–340 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Kivisild T 2007. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet 41: 539–564 [DOI] [PubMed] [Google Scholar]
- Weir BS 1996. Population structure. In Genetic data analysis II, pp. 161–200 Sinauer Associates, Sunderland, MA [Google Scholar]
- Weir BS 2008. Linkage disequilibrium and association mapping. Annu Rev Genomics Hum Genet 9: 129–142 [DOI] [PubMed] [Google Scholar]
- Weir BS, Hill WG 1980. Effect of mating structure on variation in linkage disequilibrium. Genetics 95: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR 2005. Allele frequencies and the r2 measure of linkage disequilibrium: Impact on design and interpretation of association studies. Twin Res Hum Genet 8: 87–94 [DOI] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li WH 2004. Nucleotide diversity in gorillas. Genetics 166: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW 2003. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet 72: 1171–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]