Abstract
The majority of sporadic carcinomas suffer from a kind of genetic instability in which chromosome number changes occur together with segmental defects. This means that changes involving intact chromosomes accompany breakage-induced alterations. Whereas the causes of aneuploidy are described in detail, the origins of chromosome breakage in sporadic carcinomas remain disputed. The three main pathways of chromosomal instability (CIN) proposed until now (random breakage, telomere fusion and centromere fission) are largely based on animal models and in vitro experiments, and recent studies revealed several discrepancies between animal models and human cancer. Here, we discuss how the experimental systems translate to human carcinomas and compare the theoretical breakage products to data from patient material and cancer cell lines. The majority of chromosomal defects in human carcinomas comprises pericentromeric breaks that are captured by healthy telomeres, and only a minor proportion of chromosome fusions can be attributed to telomere erosion or random breakage. Centromere fission, not telomere erosion, is therefore the most probably trigger of CIN and early carcinogenesis. Similar centromere–telomere fusions might drive a subset of congenital defects and evolutionary chromosome changes.
Introduction
Molecular analysis of tumor samples has led to the subdivision of carcinomas in two classes, each with a specific type of genetic instability. Most solid tumors undergo numerical chromosome alterations, termed aneuploidy, together with gross structural changes such as translocations or deletions. This combination of genetic defects is termed chromosomal instability (CIN) and is found in ∼85% of non-hereditary carcinomas (1,2). Approximately 15% of sporadic carcinomas show a different type of genetic instability termed microsatellite instability (MIN). The alterations responsible for MIN accrue in a small number of genes involved in mismatch repair and bring about a mutator phenotype (3,4). Because of its mutagenic effect on key regulators of cell proliferation (5), the relationship between MIN and cancer is generally accepted. The link between CIN and cancer, however, remains a matter of dispute, notwithstanding the large number of tumors that show this kind of genetic defect.
A better understanding of CIN has come from the finding that aneuploidy arises together with segmental chromosome changes, such as translocations, deletions and amplifications (6,7). Whereas aneuploidy strictly refers to the missegregation of intact chromosomes, segmental changes involve breakage and fusion. Aneuploidy and segmental changes have been recognized individually for a long time; abnormal chromosome numbers were suggested as a cause of cancer nearly a century ago (8), and chromosomes in cancer cells were shown to undergo structural changes when banding techniques became available (9). Only recently, however, aneuploidy and chromosome breakage were suggested to be part of a single phenotype (10).
Our current knowledge concerning the initial steps leading to CIN is largely based on experimental approximations. Even though experimental models can describe one or more phenomena related to cancer, they only reproduce individual aspects, are based on induced phenotypes, and have given problems when extrapolating to human carcinogenesis. The complex etiology of CIN has sometimes led to the idea that instability is caused by a combination of two defects; multiple defects would justify its description by a combination of models. The classical opinion is that spindle errors result in aneuploidy, whereas telomere erosion or random breakage causes segmental alterations. A novel hypothesis, however, indicates that mitotic spindle defects might cause both aneuploidy and chromosome breakage, opening the possibility of a single origin for the full spectrum of genetic alterations in CIN tumors (11,12). Here, we will compare three pathways of DNA breakage, assess if they faithfully describe chromosomal defects in human carcinomas and discuss the role of centromeres and telomeres in the initial phases of CIN.
Aneuploidy alone is not enough
Among the genetic alterations in CIN tumors, aneuploidy is understood in more detail; most carcinomas show variations in chromosome number that arise from continuous losses and gains of entire chromosomes during mitosis (13). Aneuploidy can be reproduced in animal models through the inactivation of genes that control the spindle assembly checkpoint (14,15) but frequently leads to an embryonic lethal phenotype (14,16). In contrast, haploinsufficiency of these checkpoint genes is compatible with life but induces tumor development (17,18). A complete loss of spindle checkpoint control probably causes a high rate of aneuploidy that compromises embryogenesis and masks the tumor development phenotype. Haploinsufficiency or mutations that inhibit apoptosis rescue the embryonic lethality and expose the carcinogenic effects (14,18). A question left unanswered in these studies is whether spindle checkpoint mutants undergo genetic alterations other than aneuploidy.
The simple presence of extra chromosomes does not seem to lead to CIN, and aneuploidy itself slows down cell proliferation (19). Experiments in which MIN cells are released from nocodazole-blocked mitosis yield a mixed population of diploid and aneuploid cells that reverts to a diploid state after a few passages (20). Similar observations have been made in animal models; autosomal trisomy is usually associated with fetal or early postnatal death, although an extra copy of a small chromosome is tolerated (21–23). The reduced viability of aneuploid cells seems at odds with the behavior of CIN tumors, as neither uncontrolled growth nor DNA breakage are explained by numerical chromosome changes alone. Once a certain degree of instability has been reached, aneuploidy can promote amplification of growth-promoting mutations (2,24). This collaboration between mutations and aneuploidy is evident in a combined Bub1/p53 haploinsuffient background (25); as the Bub1 insufficiency generates aneuploidy, lymphomas can acquire two copies of the mutated p53 allele from a heterozygous background. The capacity of gene dosage to maintain a normal chromosome complement is illustrated by the concomitant loss of the wild-type p53 locus. In this way, murine tumors with chromosome segregation errors gain a growth advantage through p53 inactivation but avoid the gene dosage effects of a third chromosome 11. Early carcinogenesis is thus governed by the balance between copy number changes of oncogenes or tumor suppressor genes on one side and overall gene dosage effects on the other side. Given the high level of aneuploidy in advanced carcinomas, tumor development might at some stage involve acquired tolerance for gene dosage effects (20,26).
Chromosome breakage can limit gene dosage effects
Experimental creation of a broken—also termed reactive—chromosome end induces a phenomenon termed the breakage-fusion-bridge (BFB) cycle and leads to extensive genome remodeling (27). In contrast to aneuploidy, which refers to numerical changes of entire chromosomes, BFB can modulate the copy number of chromosome segments in which oncogenes or tumor suppressor genes are flanked by a limited amount of DNA. The limitations imposed by gene dosage are illustrated by high-level amplifications and homozygous deletions (28); these genetic defects are usually restricted to a few megabases surrounding the gene that confers the phenotype. This means that segmental alterations, at least theoretically, can contribute to tumorigenesis but have fewer gene dosage effects than aneuploidy. Since aneuploidy alone cannot account for segmental alterations, BFB is now generally accepted as a mechanism that explains genetic plasticity in CIN tumors (2).
Whereas mitotic spindle errors are generally accepted as the leading cause of aneuploidy (29), the origins of segmental alterations in CIN tumors are still poorly understood. All models that include segmental alterations rely on BFB, however, as breakage is essential to obtain different copy numbers for segments of a single chromosome. The original study on the BFB cycle illustrated how breakage is propagated (27) but relied on breaks induced by recombination and thus might reflect an artificial situation. The detection of multiple copies of the n-Myc gene in anaphase bridges (30) directly links BFB to gene amplification in human cancer and indicates at a role for both breakage and fusion in the amplification mechanism (Figure 1).
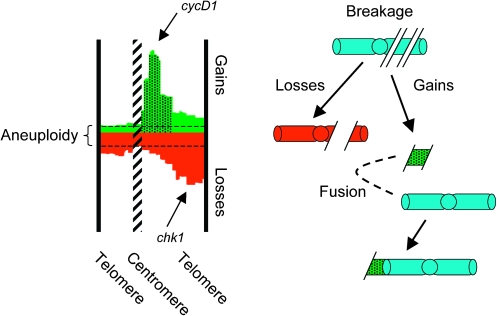
Fig. 1.
Copy number alterations involve chromosome breaks. As aneuploidy strictly refers to numerical changes of whole chromosomes, segmental gains and losses require that part of a chromosome has obtained a different copy number from the remainder of the same chromosome. The example shows chromosome 11 from Figure 3. Segmental gains and losses create a growth advantage by uncoupling the copy number of oncogenes (e.g. cycD1) and tumor suppressor genes (e.g. chk1) from general gene dosage. Frequently, the gains and losses over a single chromosome are complementary, as segments without gains are not copy number neutral but normally show losses. Intrachromosomal segment borders that delineate copy number alterations correspond to unprotected (reactive) ends that are functionally equivalent to breaks.
Since BFB was first described, three routes have been proposed to start breakage: Random breaks due to external factors, and the site-specific telomere erosion and centromere fission (11,31,32). Since the hypotheses concerning chromosome breakage are based on experimental models, the comparison with data from patient material provides an essential verification.
External factors, random breakage and fragile sites
Since chromosomal breaks and translocations were considered random until recently (33), several mechanisms of random breakage have been proposed as causes of BFB. The common theme in models that assume random breakage is a continuous basal rate of break formation due to external factors such as cosmic radiation or reactive oxygen species (31). Normally, these breaks are repaired by one of the multiple repair mechanisms in mammalian cells, but an increased rate of break formation or defects in the repair machinery might offset this equilibrium and lead to the random accumulation of breaks that initiate BFB (31). Whereas radiation-induced damage persists longer in cells with reduced repair capacity to (34), most CIN tumors efficiently repair DNA damage (35) or show augmented break repair activity (36,37). Even models that lack a single repair system maintain genetic stability unless given a break-inducing treatment (38,39). Although a few random breaks might escape detection on the timescale of a human life, this hypothesis is difficult if not impossible to prove experimentally; since late passage human fibroblasts show increased levels of spontaneous but not of induced DNA damage (40), repair-independent mechanisms probably generate DNA damage in aging cells.
One mechanism that could generate repair-independent DNA damage is breakage of stalled replication forks. According to this theory, termed the fragile site hypothesis, our genome has various sites with extensive flexibility and sequence repeats that are believed to be more difficult to replicate in fast-growing cells. The study of fragile sites includes folate deprivation, which results in repair deficiency, or treatment with the DNA polymerase inhibitor aphidicolin (41) conditions that unlikely represent early carcinogenesis. When not induced experimentally, breakage at fragile sites seems to correlate with a loss of function in cell cycle checkpoint and repair proteins (42), suggesting a hereditary component normally absent from sporadic carcinomas. An important argument against random breakage and fragile sites comes from human cancer itself; large-scale analyses of tumor samples show that chromosome breaks in CIN tumors are non-random and show little or no preference for fragile sites (43). Even fragile sites that coincide with tumor suppressor genes (44), the breakage of which might affect carcinogenesis directly, represent only a small fraction of breakpoints in sporadic carcinomas (28). Instead, tumor samples and cancer cell lines show a striking preference for gains and losses of whole arms (43). Thus, even though random breakage or fragile sites could explain genetic alterations after a massive genotoxic insult, they probably have a minor role in the genesis of CIN under normal circumstances.
The popular model: telomere erosion
After the identification of specific sequences that protect chromosome ends from erosion, telomeres have been suggested as major players in CIN (45). The initial description of the BFB cycle (27) included a role for the chromosome ends, which were first proposed to have a specialized structure—the telomere—around the same time (46,47). Whereas the original experiments introduced an interstitial break in a single chromosome (27), current models of telomere dysfunction are based on gradual erosion, also termed attrition (48). The theory that links telomere attrition to CIN gained popularity after the identification of telomerase activity and specific telomeric sequences (45), but the vast amount of data generated during the last decades has failed to provide a mechanism that adequately explains the role of telomeres in early carcinogenesis. Although induced telomere attrition in mouse mutants results in chromosome end-to-end fusions (49), this neither reproduces the CIN phenotype nor increases carcinogenesis if not aided by a tumor-inducing treatment (32,50). Long-term cultures of murine fibroblasts showed CIN in both wild-type and telomerase null cells and a large proportion of telomere-positive fusions (51); also human cell lines with chromosome fusions but a normal telomere length have been described (52), indicating no correlation between telomere length and CIN. In some models, telomere attrition has been associated with suppression of spontaneous tumorigenesis (53) and tumor regression (54).
To understand the effect of telomere attrition, it is important to recognize how telomeres protect chromosomes under physiological conditions. Since telomeres contain free DNA ends, they are capped by proteins from the non-homologous end joining (NHEJ) pathway (55). Normally, NHEJ catalyzes the fusion of free DNA ends, but the shelterin complex suppresses this reaction in telomeres (56). DNA repair pathways have dual functions, break repair and signalling for cell cycle arrest or apoptosis (31). When telomeres get shorter, the shelterin complex allows the activation of the damage signalling through ataxia telangiectasia mutated (ATM) but sustains the inhibition of repair (57). Through the combination of senescence signalling and repair inhibition, telomeres not only prevent unwanted cell proliferation but also chromosome fusion.
Detection of chromosomal breaks by the repair machinery leads to the local phosphorylation of histone H2A.X, usually described as γH2A.X (38,58). γH2A.X is rapidly dephosphorylated during break repair, with the goal to constrain damage signalling and to enable cell cycle progression. Telomere shortening also results in γH2A.X formation (59). In contrast to breaks, however, suppression of the DNA ligation step by shelterin (56) prevents telomere fusion while sustaining the γH2A.X signal. Persistent telomeric γH2A.X thus indicates that the protection against telomere fusion is functional.
Early carcinomas frequently show accumulation of γH2A.X in combination with an activated ATM pathway, culminating in p53-mediated cell cycle arrest (60,61). Inactivation of p53 allows cancer cells to proliferate in the presence of an activated ATM pathway (62), which explains the frequent mutation of p53 in advanced carcinomas. γH2A.X can also be found at telomeres of cancer cell lines, indicating ATM activation provides a long-term senescence signal (63). No fusion of γH2A.X-positive telomeres in cell lines has been reported, however, and even the short telomeres that prevail in senescent cells or cancer cells are able to recruit the ligation-suppressing shelterin complex (64,65). Taken together, the current data suggest that telomere attrition is an effective tumor suppression mechanism, capable of inducing senescence long before a critical telomere length is reached.
Cancer cells can attenuate an activated ATM pathway by telomere elongation. Like break repair, telomerase activity is augmented in cancer cells (66), but telomerase overexpression induces neither CIN nor cell transformation in model systems (67). The observation that telomerase reactivation occurs after cells have escaped from growth crisis (68) suggests that telomerase reactivation is selected for during tumor progression and thus is a consequence—not a cause—of CIN. The recent discovery of a role for telomerase in break repair indicates that telomerase upregulation in carcinomas might be a response to chromosome breakage in general, not just to telomere attrition (69,70). In conclusion, even though telomerase upregulation might give a growth advantage in the later stages of tumor development, the role of telomere attrition in the initiation of CIN is dubious. Examination of the genetic landscape in tumor samples might help to evaluate the contribution of telomeres in human cancer.
Centromere fission and spindle defects
Recently, a third pathway for breakage has been proposed in the form of centromeric breaks (11). Centromere fission was described—again using a plant as model system—even before the discovery of BFB (71). Unlike telomere attrition, no association between centromeric or pericentromeric breaks and CIN was suggested until recently. The latest data show that DNA damage can be generated under conditions that compromise the mitotic spindle (11,72), suggesting that a single mechanism causes aneuploidy and chromosome breakage (12). One spindle defect in particular, merotelic kinetochore attachment, seems important for centromere fission, because it is not efficiently corrected by the classic mitotic checkpoints (73) but can generate enough force to physically shear the kinetochore (11). A small percentage of merotelic attachments might even go undetected in normal cells; the associated centromere fission can be detected in ∼0.1% of all lymphocytes from healthy donors (74).
Apart from spindle control, other mitotic processes might contribute to centromere fission. In mammals, chromosome arms are liberated in prophase through the non-proteolytic removal of cohesin, but centromeres remain joined until the metaphase–anaphase transition (75). At least in theory, residual centromeric cohesin could resist the pulling force exerted by the spindle and thus contribute to centromere fission. In addition, mammalian centromeres comprise repetitive DNA, a feature of inducible fragile sites (41). No centromere fission, however, has been observed under the conditions that induce known fragile sites, suggesting that stalled replication unlikely causes centromeric DNA damage. The description of errors in replication initiation of a common fragile site but not in its replication progression (76), shows that sequence repetition not necessarily creates fragile DNA.
Given the connection between the mitotic spindle and breakage, DNA damage would be expected in mutants that undergo aneuploidy. Although studies that address spindle defects usually test only for aneuploidy, cells treated with spindle poisons and mutants in the spindle checkpoint gene Mad2 were shown to accumulate γH2A.X (14,77). In addition, haploinsufficiency or biallelic point mutation of spindle checkpoint genes such as Mad1 and BubR1 induces tumor development (18,78,79); knowing that aneuploidy alone cannot account for tumor formation, carcinogenesis in these mutants probably involves chromosome breakage. Even mild spindle defects, which lead to low levels of chromosome breakage but are compatible with development, suffice to produce anaphase bridges and translocations in mouse chromosomes (11,78). Interestingly, chromosome segregation defects have recently been implicated in pathways of early aging and senescence (80–82). Spindle defects thus are on a par with telomere attrition when concerning a possible link between aging and cancer.
Since breakage and aneuploidy are normally found together (6,7), a shared mechanism provides an attractive solution. In support of this hypothesis, centromere-driven instability has been proposed for CIN tumors such as liposarcoma (83), squamous cell carcinoma (84,85) or glioblastoma (86). Still, much of the data on centromere fission has been generated in experimental models and would therefore benefit from the same verification as random breakage and telomere erosion.
The three breakage pathways yield different products
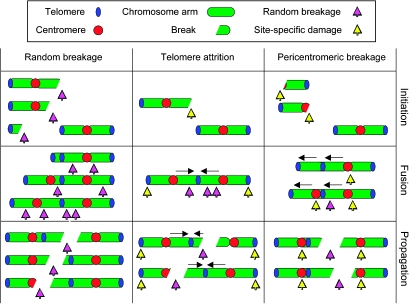
Based on the origins of DNA damage, three models can be proposed (Figure 2). The most obvious difference between the three pathways concern the place where breaks first occur and what kind of products are initially formed. Theories that depend on external break induction invariably assume random breakage, as it is hard to imagine how chromosomes can be specifically oriented on background radiation or reactive oxygen species. On the contrary, telomere fusion and centromere fission affect a specific chromosomal structure and might thus impose specificity on break sites. Even though telomere fusion and centromere fission originate at a specific chromosome site, the order of events in these two cases is very different; whereas breakage is a consequence of telomere–telomere fusion in the former model, centromeric breakage occurs before the actual fusion in the latter model. As a result, breaks are expected to appear in a random pattern in the telomere attrition model but specifically clustered around the centromere in the centromere fission hypothesis (Figure 2). Furthermore, the site of the first defect means that the initial fusion products have an antiparallel orientation in the telomere erosion model (two telomeres fuse) but adopt a tandem orientation after centromere fission (a broken centromere is fused to a telomere). In all three models, telomeres may capture broken ends (87) and incorporated into fusions as interstitial telomeric sequences (ITS). Exclusively in the centromere fission model, ITS are flanked by an intact chromosome and a tandem whole chromosome arm.
Fig. 2.
Comparison of chromosome breakage pathways Three models, random breakage (left), telomere fusion (middle), and centromere fission (right) are represented. For each model, initial defects are shown on the upper row, primary fusions on the second row and later products on the third row. Random breaks are depicted with purple arrows and site specificity is indicated with yellow arrows. Whereas random breaks are the main products in the random breakage and telomere fusion models, arm-level breaks are generated first in the centromere fission model and random breaks form only after secondary fusion has taken place. Fusions in the telomere erosion model are telomere to telomere (blue) and thus antiparallel, whereas fusions in the centromere fission model are centromere (red) to telomere and thus tandem (black arrows, oriented from the centromere to the telomere).
Telomeric fusions give rise to dicentric chromosomes, which are highly instable and liberate new reactive ends upon breakage (27,88). New breaks in dicentric chromosomes are situated between the two centromeres; all fragments therefore contain a centromere and fuse to form new dicentrics. Some of the secondary products in the telomere erosion model might be formed due to pericentromeric breaks, but these products occur after break randomization and would thus represent a minor proportion. In contrast to telomere erosion, a pericentromeric break can yield either a centromere-containing or centromere-less fragment. In this way, unstable as well as stable fusions can be formed, all of which adopt a tandem orientation. This means that stable tandem fusions, which contain a single centromere and do not enter BFB cycles, are formed exclusively by a pericentromeric break followed by fusion to a chromosome end. To determine which pathways govern the early steps of CIN in human carcinogenesis, a comparison of these models to chromosome alterations in tumor samples might yield important clues.
What tumor samples and cell lines show
To determine which of the three models most faithfully describes CIN in human cancer, we have to examine samples from actual neoplasms. The first clue comes from breakpoint localization; traditional marker analyses showed that nearly half of all breakpoints on the largest chromosome map to the centromeric and pericentromeric region and only a small proportion to the arms and telomeres (89). Recent advances in microarrays have enabled large-scale analysis of copy number changes in tumor samples (43), and massive sequencing has identified the pattern of breakpoints in a large panel of cancer cell lines (28). Chromosome breakage and the location of breakpoints can be inferred from the limits of regions that show copy number alteration; if a normally continuous DNA tract is found in two different amounts, this tract must have been broken and one of the fragments must have been amplified or lost to yield a different number of copies than the other fragment (Figure 1). By analyzing the copy number of a large number of chromosomal regions, breakpoints can be mapped with kilobase precision. Even though the latest studies focussed on the identification of small regions, the most evident switches in copy number localize to centromeres and pericentromeric regions (28,43,90). These data show that the most common alterations involve whole chromosome arms—arm-level alterations are more frequent than aneuploidy in some studies (43)—and that breaks frequently localize adjacent to centromeres (Figure 3). The high frequency of arm-level alterations corroborates the centromere fission model, because this is the only mechanism that specifically splits a chromosome into two arms.
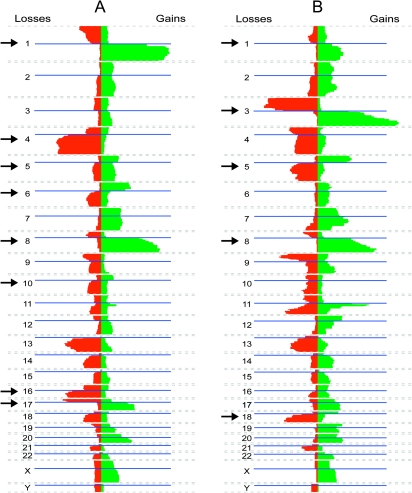
Fig. 3.
Chromosome breaks frequently liberate whole arms Copy number analysis of 844 liver carcinomas (A) and 1827 head and neck squamous cell carcinomas (B) were retrieved from the Progenetix database (90). For each type of carcinoma, gains are indicated in green (right) and losses in red (left). Blue lines indicate centromere positions. Chromosomes that bear evidence of centromere fission are indicated with an arrow. Analysis of 3131 profiles showed a similar preference for whole arm gains and losses in a wide range of carcinomas (43). The biphasic pattern of chromosome 11q probably is a result of whole arm amplification followed by loss of the distal segment.
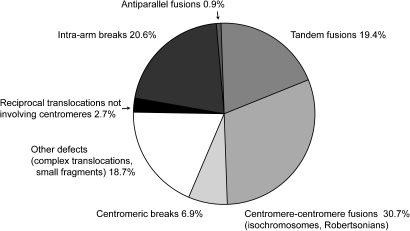
The analysis of copy number changes alone does not depict the topology of breakage and fusion. Microscopy based techniques, for example spectral karyotyping and fluorescent in situ hybridization, yield a much more visual image of chromosome fragments and their instability. Optical techniques showed that chromosome arms are normally not found alone but rapidly fuse to other fragments or intact chromosomes (83,91). Some of the copy number changes observed in the large-scale analyses are therefore brought about by chromosome arms that ‘hitch a ride’ on the ends of healthy chromosomes (Figure 4). When viewed by microscopy, fused chromosomes appear to grow from a telomere; this observation might have contributed to the focus on telomere defects. Close inspection of the formed products, however, shows that the fusion point comprises a centromere and a telomere, and analysis by banding techniques shows that a fragment the size of a whole arm attaches to the end of a healthy chromosome in a tandem orientation (91). A classification of products described in literature (94) and spectral karyotyping databases (93) indicate that tandem fusions are much more common than antiparallel fusions (Figure 5). Also the other products of centromere fission—for example centromeric fusion of two arms and isochromosomes (84,95)—are common in tumor samples.
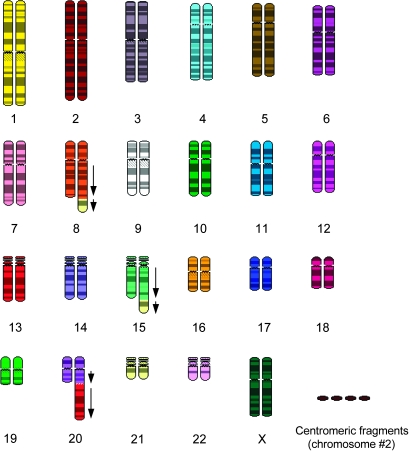
Fig. 4.
Broken arms fuse to the end of chromosomes. Example of a molecular cytogenetic analysis of a neuroectodermal tumor (92). The skygram was retrieved from the NCBI SKY/CGH database (93). The main structural alterations observed correspond to centromeric breaks (chromosomes 13 and 21) followed by fusion to telomeres (chromosomes 8, 15 and 20). Arrows indicate tandem orientation of the fusions. The fusion of chromosome 21 to chromosomes 8 and 15 classifies as a jumping translocation.
Fig. 5.
Classification of segmental defects in tumor samples. Spectral karyotyping analyses corresponding to 98 human carcinomas from the NCBI SKY/CGH database (93) were inspected for structural alterations. Samples without apparent structural alterations (seven cases) or bearing >25 alterations (four cases) were discarded. Alterations were classified according to breakpoint and fusion products. Multiple copies of the same alteration in a single sample were counted as a single event, as they correspond to aneuploid state of entire fusion products and do not involve de novo breakage. The majority of products involve centromeric fission, and only a minor proportion can be attributed to telomere–telomere fusion.
In all three models, ITS can be formed when telomeres capture broken ends (87). No comprehensive search for ITS in tumor samples has been conducted, but the studies carried out until now show ITS at antiparallel fusion sites formed after centromere fission (96,97). In some cases, a combination of probes has enabled direct detection of the fusion between a pericentromeric chromosomal region and telomere (97). In conclusion, the most frequent chromosomal defects do not seem to be the result of random breakage or telomere erosion, although genuine telomere–telomere fusions have occasionally been detected (98–100). Mitotic centromere fission followed by NHEJ (11,101) provides a simple model that faithfully describes the pattern of common chromosome alterations in CIN tumors (Figure 6).
Fig. 6.
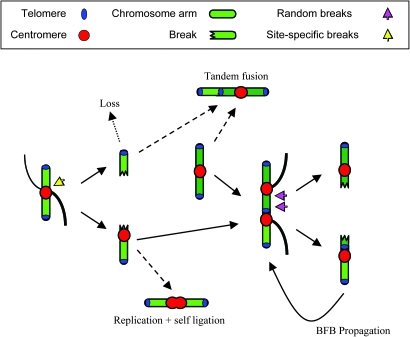
Early events leading to CIN. An uncorrected merotelic attachment causes chromosome breakage at the centromeric region (yellow arrow), resulting in two reactive chromosome arms. The arm without a centromere is easily lost in subsequent cell divisions or can be rescued by a translocation. The centromere-containing arm can replicate and segregate. The centromere-containing arm thus forms a reactive species that persists during subsequent cell divisions. The arm bearing the first break can self-ligate, forming a pseudodicentric chromosome, or fuse to a healthy chromosome (dark green), forming a dicentric chromosome bearing an interstitial telomere. Capture of a dicentric chromosome by two spindle poles leads to secondary breaks (purple arrows), propagating BFB cycles. Dashed lines indicate events that stabilize chromosome arms with breaks. Figure adapted from (12).
From single breaks to ongoing instability
A question that arises is how events such as centromere fission can lead to ongoing instability. The analysis of tumor samples provides us with a large number of individual ‘snapshots’, that hint at centromere fission as an important early step. Long-time cultures of cancer cell lines (102) and serial sampling of patients (103), however, indicate that tumor karyotypes evolve over time.
The high levels of CIN in advanced tumors suggest that these have acquired adaptations to gene dosage. Cancer cells seem to cope with gene dosage by increasing proteasomal protein degradation, used as a target for chemotherapy (104). Also in yeast, mutations that increase activity of the ubiquitin–proteasome pathway generate tolerance for aneuploidy (26). Additional screenings for compounds that preferentially inhibit growth of aneuploid cells identified protein folding and autophagy as potential targets (105). Although many of these adaptations promote tumor survival individually (106), they probably are induced together as a result of gene dosage. As a consequence, these pathways might cause a general ‘spillover’ effect on key cell cycle regulators (107) and promote further genetic instability.
Initial steps in this gene dosage cycle probably are quite small, as whole arm gains add a considerable amount of DNA to the genome and are associated with reduced viability. Since small chromosomes can be present in extra copies without compromising viability, the arms from small chromosomes might target larger chromosomes for instability (Figure 4). In addition, commonly amplified arms on large chromosomes contain ‘strong’ oncogenes such as c-Myc (8q), ErbB2 (17q) or Pik3CA (3q). Still, these changes add or remove a considerable amount of genetic material and are probably to cause a spillover effect to important regulatory pathways. In conclusion, gene dosage is a mechanism that can inhibit cell transformation in its first steps but might promote ongoing CIN and malignancy when overcome by adaptation.
Concluding remarks
Large-scale genetic analyses of tumor samples and cancer cell lines show that the vast majority of copy number changes correspond to intact chromosomes or entire chromosome arms (28,43), which points at the centromeric region as a breakage hotspot. One has to admit that also the other pathways might occasionally generate a pericentromeric break. The vast numbers of arm level translocations in tumor samples, however, show that pericentromeric breaks must be formed preferentially. In advanced tumors, chromosome segments are more probably to undergo several rounds of breakage; the biphasic pattern of the 11q arm (Figures 1 and 3) probably has its origin in a centromeric break causing amplification of the whole arm, followed by loss of the distal part. In this way, cells gain extra copies of the proximal CycD1 oncogene but repress the distal tumor suppressor Chk1. Note that the reverse pattern is very rare because the centromeric break anchors the proximal part to acceptor chromosomes. Thus, even though oncogenes and tumor suppressor genes govern the proliferation of cancer cells, the genetic signature of carcinomas points at centromere fission as the mechanism that confers genetic plasticity.
Among the most striking structures resulting from the fusion between whole arms and intact chromosomes are ITS. Although ITS can be powerful markers, reliable detection of ITS seems challenging and even the largest ancestral ITS in our genome (on chromosome 2q13) is distinguished in only 10% of all metaphases (98). The absence of detectable ITS from a fusion site might therefore illustrate a technical instead of a biological problem. Although cancer ITS are rarely examined in detail, the studies done until now show ITS in tandem fusions between single arms and intact chromosomes (96). Most ITS are tandem repeats—not the inverted repeats generated in telomere–telomere fusions—and probably formed through the capture of a non-telomeric break on a chromosome end (87).
Telomeres might be the preferred substrate for the chromosome arms that are liberated in mitosis, because telomeres come preloaded with components of the NHEJ pathway (55,108) and recombination has little activity after mitotic exit (31). Although telomere fusion is repressed by the shelterin complex, the fusion of arms to intact chromosomes shows that this protection is not waterproof. The fully activated NHEJ machinery on centromeric breaks might circumvent the shelterin inhibition of the telomeres. Nonetheless, the considerable percentage of isochromosomes (Figure 5) illustrates the effectiveness of shelterin, because isochromosomes are formed by centromeric breaks that persist alongside telomeres until the arm has been replicated and repaired. Recently described phenomena such as chromothripsis (109), found in a few percent of carcinomas, might be ascribed to shelterin function; efficient telomere protection helps the confinement of BFB propagation (Figure 6) to a small pool of chromosomes, which results in reutilization of broken chromosome arms and limits copy number states. The high frequency of breakage in chromothripsis, together with the low percentage of samples that show this phenomenon, hints at unstable dicentric chromosomes.
ITS have been analyzed extensively in congenital syndromes (110,111) and evolutionary biology (112,113). Although the origin of ITS in evolutionary biology is a matter of debate, it is accepted that congenital ITS are formed in meiosis. Like mitosis, the final meiotic divisions can suffer from chromosome segregation defects (114); also in meiosis, spindle defects seem to have a critical role when it comes to genetic instability. Practically, all congenital ITS result from the translocation of a single donor segment onto different—random—chromosome recipients, termed jumping translocations (97,115). Repair by NHEJ agrees with the random nature of jumping translocations (97,111,115) because this repair pathway non-selectively fuses unprotected chromosome ends (31). Analysis of jumping translocations in congenital defects again shows whole arm fusion to intact chromosomes, frequently in combination with isochromosome formation of the other arm. In conclusion, a single mechanism, centromere fission, might be responsible for genetic instability on the cellular, individual and evolutionary scale.
Funding
Fondo de Investigación en Salud (PS09/00572 and RD08/0075/0010) and Ministerio de Ciencia y Investigación (BFU 2009-08395-E). K.v.W. is supported by a JAE-doc fellowship from the Spanish National Research Council (CSIC). The Department of Immunology and Oncology was founded and is supported by the CSIC and by Pfizer.
Acknowledgments
The authors thank Catherine Mark for editorial assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ATM
ataxia telangiectasia mutated
- BFB
breakage-fusion-bridge
- CIN
chromosomal instability
- ITS
interstitial telomeric sequence
- MIN
microsatellite instability
- NHEJ
non-homologous end joining
References
- 1.Draviam VM, et al. Chromosome segregation and genomic stability. Curr. Opin. Genet. Dev. 2004;14:120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Pihan G, et al. Mutations and aneuploidy: co-conspirators in cancer? Cancer Cell. 2003;4:89–94. doi: 10.1016/s1535-6108(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 3.Baranovskaya S, et al. Functional significance of concomitant inactivation of hMLH1 and hMSH6 in tumor cells of the microsatellite mutator phenotype. Proc. Natl Acad. Sci. USA. 2001;98:15107–15112. doi: 10.1073/pnas.251234498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wind N, et al. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 5.Royrvik EC, et al. Slip slidin' away: a duodecennial review of targeted genes in mismatch repair deficient colorectal cancer. Crit. Rev. Oncog. 2007;13:229–257. doi: 10.1615/critrevoncog.v13.i3.20. [DOI] [PubMed] [Google Scholar]
- 6.Duesberg P, et al. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl Acad. Sci. USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabarius A, et al. Instability of chromosome structure in cancer cells increases exponentially with degrees of aneuploidy. Cancer Genet. Cytogenet. 2003;143:59–72. doi: 10.1016/s0165-4608(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 8.Boveri T. Zur Frage der Entstehung maligner Tumoren. Jena, Germany: Fischer Verlag; 1914. [Google Scholar]
- 9.Miller DA, et al. The quinacrine fluorescent and Giemsa banding karyotype of the rat, Rattus norvegicus, and banded chromosome analysis of transformed and malignant rat liver cell lines. Cancer Res. 1972;32:2375–2382. [PubMed] [Google Scholar]
- 10.Geigl JB, et al. Defining 'chromosomal instability'. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Alonso Guerrero A, et al. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc. Natl Acad. Sci. USA. 2010;107:4159–4164. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-A C, et al Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle. 2010;9:2275–2280. doi: 10.4161/cc.9.12.11865. [DOI] [PubMed] [Google Scholar]
- 13.Jallepalli PV, et al. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 14.Burds AA, et al. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc. Natl Acad. Sci. USA. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schliekelman M, et al. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalitsis P, et al. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver BA, et al. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 19.Weaver BA, et al. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson SL, et al. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bersu ET, et al. Growth characteristics of the murine trisomy 19 thymus. Teratology. 1982;26:85–94. doi: 10.1002/tera.1420260112. [DOI] [PubMed] [Google Scholar]
- 23.Carr DH. The chromosome abnormality in mongolism. Can. Med. Assoc. J. 1962;87:490–495. [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver BA, et al. The role of aneuploidy in promoting and suppressing tumors. J. Cell Biol. 2009;185:935–937. doi: 10.1083/jcb.200905098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker DJ, et al. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson SL, et al. Mechanisms of chromosomal instability. Curr. Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg G, et al. Alternative lengthening of telomeres-an enhanced chromosomal instability in aggressive non-MYCN amplified and telomere elongated neuroblastomas. Genes Chromosomes Cancer. 2011;50:250–262. doi: 10.1002/gcc.20850. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein C, et al. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat. Res. 2002;511:145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 32.Blasco MA, et al. Evolving views of telomerase and cancer. Trends Cell. Biol. 2003;13:289–294. doi: 10.1016/s0962-8924(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 33.Lengauer C, et al. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 34.Taccioli GE, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Y, et al. Efficient repair of DNA double-strand breaks in malignant cells with structural instability. Mutat. Res. 2010;683:115–122. doi: 10.1016/j.mrfmmm.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosoi Y, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int. J. Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- 37.Pucci S, et al. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene. 2001;20:739–747. doi: 10.1038/sj.onc.1204148. [DOI] [PubMed] [Google Scholar]
- 38.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki K, et al. Qualitative and quantitative analysis of phosphorylated ATM foci induced by low-dose ionizing radiation. Radiat. Res. 2006;165:499–504. doi: 10.1667/RR3542.1. [DOI] [PubMed] [Google Scholar]
- 40.Endt H, et al. Detailed analysis of DNA repair and senescence marker kinetics over the life span of a human fibroblast cell line. J. Gerontol. A. Biol. Sci. Med. Sci. 66:367–375. doi: 10.1093/gerona/glq197. [DOI] [PubMed] [Google Scholar]
- 41.Lukusa T, et al. Human chromosome fragility. Biochim. Biophys. Acta. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Arlt MF, et al. Common fragile sites. Cytogenet. Genome Res. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- 43.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue H, et al. Sequence of the FRA3B common fragile region: implications for the mechanism of FHIT deletion. Proc. Natl Acad. Sci. USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greider CW, et al. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 46.Muller HJ. The remaking of chromosomes. Collecting Net. 1938;13:81–88. [Google Scholar]
- 47.Rapoport JA. Mutations restituting the telomere. Comptes Rendus De L Academie Des Sciences De Ĺ URSS. 1941;31:266–269. [Google Scholar]
- 48.Lundblad V. Genome instability: McClintock revisited. Curr. Biol. 2001;11:R957–R960. doi: 10.1016/s0960-9822(01)00573-5. [DOI] [PubMed] [Google Scholar]
- 49.Hande MP, et al. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang S, et al. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao LY, et al. Genomic instability in both wild-type and telomerase null MEFs. Chromosoma. 2004;113:62–68. doi: 10.1007/s00412-004-0291-7. [DOI] [PubMed] [Google Scholar]
- 52.Saltman D, et al. Telomeric structure in cells with chromosome end associations. Chromosoma. 1993;102:121–128. doi: 10.1007/BF00356029. [DOI] [PubMed] [Google Scholar]
- 53.Cosme-Blanco W, et al. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007;8:497–503. doi: 10.1038/sj.embor.7400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pathak S, et al. Spontaneous regression of cutaneous melanoma in sinclair swine is associated with defective telomerase activity and extensive telomere erosion. Int. J. Oncol. 2000;17:1219–1224. doi: 10.3892/ijo.17.6.1219. [DOI] [PubMed] [Google Scholar]
- 55.Bailey SM, et al. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bombarde O, et al. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29:1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Misri S, et al. Detecting ATM-dependent chromatin modification in DNA damage and heat shock response. Methods Mol. Biol. 2009;523:395–410. doi: 10.1007/978-1-59745-190-1_26. [DOI] [PubMed] [Google Scholar]
- 58.Rogakou EP, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 59.d'Adda di Fagagna F. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 60.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 61.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 62.Davoli T, et al. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakamura AJ, et al. Telomere-dependent and telomere-independent origins of endogenous DNA damage in tumor cells. Aging (Albany NY) 2009;1:212–218. doi: 10.18632/aging.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlseder J, et al. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 65.Xu L, et al. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lantuejoul S, et al. Telomerase expression in lung preneoplasia and neoplasia. Int. J. Cancer. 2007;120:1835–1841. doi: 10.1002/ijc.22473. [DOI] [PubMed] [Google Scholar]
- 67.Venkatesan RN, et al. Telomerase expression in chickens: constitutive activity in somatic tissues and down-regulation in culture. Proc. Natl Acad. Sci. USA. 1998;95:14763–14768. doi: 10.1073/pnas.95.25.14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Degerman S, et al. Telomerase upregulation is a postcrisis event during senescence bypass and immortalization of two Nijmegen breakage syndrome T cell cultures. Aging Cell. 2010;9:220–235. doi: 10.1111/j.1474-9726.2010.00550.x. [DOI] [PubMed] [Google Scholar]
- 69.Cong Y, et al. Actions of human telomerase beyond telomeres. Cell Res. 2008;18:725–732. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- 70.Masutomi K, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl Acad. Sci. USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Darlington CD. Misdivision and the genetics of the centromere. J. Genet. 1939;37:341–U19. [Google Scholar]
- 72.Dalton WB, et al. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 2007;67:11487–11492. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rupa DS, et al. Detection of chromosomal breakage in the 1cen-1q12 region of interphase human lymphocytes using multicolor fluorescence in situ hybridization with tandem DNA probes. Cancer Res. 1995;55:640–645. [PubMed] [Google Scholar]
- 75.Hauf S, et al. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 76.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 77.Quignon F, et al. Sustained mitotic block elicits DNA breaks: one-step alteration of ploidy and chromosome integrity in mammalian cells. Oncogene. 2007;26:165–172. doi: 10.1038/sj.onc.1209787. [DOI] [PubMed] [Google Scholar]
- 78.Iwanaga Y, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 79.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 80.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 81.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maehara K, et al. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sirvent N, et al. Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer. 2000;29:117–129. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1014>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 84.Hermsen M, et al. Centromeric chromosomal translocations show tissue-specific differences between squamous cell carcinomas and adenocarcinomas. Oncogene. 2005;24:1571–1579. doi: 10.1038/sj.onc.1208294. [DOI] [PubMed] [Google Scholar]
- 85.Uchida K, et al. Molecular cytogenetic analysis of oral squamous cell carcinomas by comparative genomic hybridization, spectral karyotyping, and fluorescence in situ hybridization. Cancer Genet. Cytogenet. 2006;167:109–116. doi: 10.1016/j.cancergencyto.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 86.Squire JA, et al. Molecular cytogenetic analysis of glial tumors using spectral karyotyping and comparative genomic hybridization. Mol. Diagn. 2001;6:93–108. doi: 10.1054/modi.2001.22745. [DOI] [PubMed] [Google Scholar]
- 87.Bailey SM, et al. Dysfunctional mammalian telomeres join with DNA double-strand breaks. DNA Repair (Amst) 2004;3:349–357. doi: 10.1016/j.dnarep.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 88.Jager D, et al. Stabilization of dicentric chromosomes in Saccharomyces cerevisiae by telomere addition to broken ends or by centromere deletion. EMBO J. 1989;8:247–254. doi: 10.1002/j.1460-2075.1989.tb03370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brito-Babapulle V, et al. Break points in chromosome #1 abnormalities of 218 human neoplasms. Cancer Genet. Cytogenet. 1981;4:215–225. doi: 10.1016/0165-4608(81)90015-7. [DOI] [PubMed] [Google Scholar]
- 90.Baudis M, et al. Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17:1228–1229. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 91.Raimondi SC, et al. Multiple telomeric associations of a trisomic whole q arm of chromosome 1 in a child with acute lymphoblastic leukemia. Cancer Genet. Cytogenet. 1987;24:87–93. doi: 10.1016/0165-4608(87)90085-9. [DOI] [PubMed] [Google Scholar]
- 92.Bayani J, et al. Molecular cytogenetic analysis of medulloblastomas and supratentorial primitive neuroectodermal tumors by using conventional banding, comparative genomic hybridization, and spectral karyotyping. J. Neurosurg. 2000;93:437–448. doi: 10.3171/jns.2000.93.3.0437. [DOI] [PubMed] [Google Scholar]
- 93.Knutsen T, et al. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Padilla-Nash HM, et al. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. [DOI] [PubMed] [Google Scholar]
- 95.Sawyer JR, et al. Translocation (1;19) (q21;q13.3) is a recurrent reciprocal translocation in meningioma. Cancer Genet. Cytogenet. 2002;134:88–90. doi: 10.1016/s0165-4608(01)00610-0. [DOI] [PubMed] [Google Scholar]
- 96.Busson Le Coniat M, et al. Interstitial telomere repeats in translocations of hematopoietic disorders. Leukemia. 2000;14:1630–1633. doi: 10.1038/sj.leu.2401876. [DOI] [PubMed] [Google Scholar]
- 97.Reddy KS, et al. Fusion of 9 beta-satellite and telomere (TTAGGG)n sequences results in a jumping translocation. Hum. Genet. 2000;107:268–275. doi: 10.1007/s004390000360. [DOI] [PubMed] [Google Scholar]
- 98.Azzalin CM, et al. Fluorescence in situ hybridization with a synthetic (T2AG3)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet. Cell Genet. 1997;78:112–115. doi: 10.1159/000134640. [DOI] [PubMed] [Google Scholar]
- 99.Sawyer JR, et al. Evidence for telomeric fusions as a mechanism for recurring structural aberrations of chromosome 11 in giant cell tumor of bone. Cancer Genet. Cytogenet. 2005;159:32–36. doi: 10.1016/j.cancergencyto.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 100.Sawyer JR, et al. A role for telomeric and centromeric instability in the progression of chromosome aberrations in meningioma patients. Cancer. 2000;88:440–453. [PubMed] [Google Scholar]
- 101.Alonso Guerrero A, et al. Merotelic attachments and non-homologous end joining are the basis of chromosomal instability. Cell Div. 2010;5:13. doi: 10.1186/1747-1028-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whang-Peng J, et al. Cytogenetic studies of human breast cancer lines: MCF-7 and derived variant sublines. J. Natl Cancer Inst. 1983;71:687–695. [PubMed] [Google Scholar]
- 103.van Tilborg AA, et al. The random development of LOH on chromosome 9q in superficial bladder cancers. J. Pathol. 2002;198:352–358. doi: 10.1002/path.1215. [DOI] [PubMed] [Google Scholar]
- 104.Chen D, et al. The ubiquitin-proteasome system as a prospective molecular target for cancer treatment and prevention. Curr. Protein Pept. Sci. 2010;11:459–470. doi: 10.2174/138920310791824057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang YC, et al. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Loda M, et al. Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nat. Med. 1997;3:231–234. doi: 10.1038/nm0297-231. [DOI] [PubMed] [Google Scholar]
- 108.Samper E, et al. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boutouil M, et al. Fragile site and interstitial telomere repeat sequences at the fusion point of a de novo (Y;13) translocation. Hum. Genet. 1996;98:323–327. doi: 10.1007/s004390050216. [DOI] [PubMed] [Google Scholar]
- 111.Rivera H, et al. Centromere-telomere (12;8p) fusion, telomeric 12q translocation, and i (12p) trisomy. Clin. Genet. 1999;55:122–126. doi: 10.1034/j.1399-0004.1999.550209.x. [DOI] [PubMed] [Google Scholar]
- 112.Lin KW, et al. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutat. Res. 2008;658:95–110. doi: 10.1016/j.mrrev.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Ropiquet A, et al. A paradox revealed: karyotype evolution in the four-horned antelope occurs by tandem fusion (Mammalia, Bovidae, Tetracerus quadricornis) Chromosome Res. 2010;18:277–286. doi: 10.1007/s10577-010-9115-1. [DOI] [PubMed] [Google Scholar]
- 114.Sugawara S, et al. An experimental approach to the analysis of mechanisms of meiotic nondisjunction and anaphase lagging in primary oocytes. Cytogenet. Cell Genet. 1980;28:251–264. doi: 10.1159/000131538. [DOI] [PubMed] [Google Scholar]
- 115.Berger R, et al. Jumping translocations. Genes Chromosomes Cancer. 2007;46:717–723. doi: 10.1002/gcc.20456. [DOI] [PubMed] [Google Scholar]