Abstract
DNA damage is thought to play a critical role in the development of colorectal adenoma. Variation in DNA repair genes may alter their capacity to correct endogenous and exogenous DNA damage. We explored the association between common single-nucleotide polymorphisms (SNPs) in DNA repair genes and adenoma risk with a case–control study nested in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. A total of 1338 left sided, advanced colorectal adenoma cases and 1503 matched controls free of left-sided polyps were included in the study. Using DNA extracted from blood, 3144 tag SNPs in 149 DNA repair genes were successfully genotyped. Among Caucasians, 30 SNPs were associated with adenoma risk at P < 0.01, with four SNPs remaining significant after gene-based adjustment for multiple testing. The most significant finding was for a non-synonymous SNP (rs9350) in Exonuclease-1 (EXO1) [odds ratio (OR) = 1.30, 95% confidence interval (CI) = 1.11–1.51, P = 0.001)], which was predicted to be damaging using bioinformatics methods. However, the association was limited to smokers with a strong risk for current smokers (OR = 2.15, 95% CI = 1.27–3.65) and an intermediate risk for former smokers (OR = 1.45, 95% CI = 1.14–1.82) and no association for never smokers (OR = 0.98, 95% CI = 0.76–1.25) (Pinteraction = 0.002). Among the top findings, an SNP (rs17503908) in ataxia telangiectasia mutated (ATM) was inversely related to adenoma risk (OR = 0.75, 95% CI = 0.63–0.91). The association was restricted to never smokers (OR = 0.55, 95% CI = 0.40–0.76) with no increased risk observed among smokers (OR = 0.89, 95% CI = 0.70–1.13) (Pinteraction = 0.006). This large comprehensive study, which evaluated all presently known DNA repair genes, suggests that polymorphisms in EXO1 and ATM may be associated with risk for advanced colorectal adenoma with the associations modified by tobacco-smoking status.
Introduction
Colorectal cancer is the third most common cancer in the USA for both men and women (1). Epidemiological studies have shown that non-steroidal anti-inflammatory drugs, exogenous hormones and select dietary factors are risk factors of colorectal neoplasia (2,3). Genetic factors also contribute to risk with the heritability of colorectal cancer estimated to be 35% from a large twin study (4). Genome-wide association studies have identified at least 14 loci associated with the risk of colorectal cancer (5,6); however, additional loci are predicted to exist (7). Although there have been no genome-wide association studies exclusively of colorectal adenoma, a known precursor to colorectal cancer, studying genetic susceptibility to colorectal adenoma may give insight into the etiology of colorectal cancer.
Smoking has been consistently associated with an increased risk of colorectal adenoma (8,9). A recent meta-analysis found the risk estimate of adenoma for ever smokers to be 1.82 [95% confidence interval (CI) = 1.65–2.00] (8). Generally, the risk was stronger for current [odds ratio (OR) = 2.14, 95% CI = 1.86–2.46] as opposed to former smokers (OR = 1.47, 95% CI = 1.29–1.67) (8). Carcinogens generated from tobacco smoking interact with DNA to form DNA adducts, which can interfere with cell replication and if not repaired correctly, can cause somatic mutations leading eventually to cancer. Emerging evidence has also shown that tobacco smoking may interact with genetic factors, predisposing certain individuals to greater polyp susceptibility (10). However, the studies focused on limited number of candidate genes to date have only provided limited evidence for gene–environmental interactions (10).
Damage caused by smoking and other environmental exposures activates several different DNA repair pathways, including base excision repair, mismatch repair (MMR), nucleotide excision repair and double-strand break repair pathways (11). Rare germ line mutations in MMR have been shown to lead to hereditary non-polyposis colorectal cancer (HNPCC) (12) and mutations in the base excision repair gene, MUTYH, have been linked to a familial polyposis syndrome (13). Some significant associations have also been reported between common DNA repair gene polymorphisms and colorectal neoplasia risk (14–17). In particular, a common variant in the MLH1 gene region has been linked to the risk of colorectal cancers with microsatellite instability (18–21). However, most studies examined only small sets of single-nucleotide polymorphisms (SNPs) (14–17), and the associations remain to be confirmed. As a complex disease, multiple genetic variants with minor to moderate effects probably contribute to colorectal adenoma development, which makes scanning a large number of genes simultaneously rather than a small set of individual SNPs attractive.
Studies of genetic susceptibility to colorectal adenoma may give insight into the etiology of colorectal carcinogenesis. To examine the relationship between polymorphisms in DNA repair genes and colorectal adenoma risk as well as potential effect modification by tobacco smoking, we conducted a nested case–control study within the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. As most DNA repair genes have not been well characterized with regard to disease susceptibility, we decided to undertake a comprehensive approach including all DNA repair genes known to our knowledge to date (11). For each gene, we selected tag SNPs to thoroughly capture the common genetic variation in the region.
Methods
The PLCO Cancer Screening Trial
The PLCO is a clinical trial, designed to assess the efficacy of screening tests to reduce death from cancers of prostate, lung, colon and rectum and ovary. As described previously (22,23), 154 938 cancer-free men and women aged 55–74 were recruited from 10 sites in the USA between 1993 and 2001. Participants were randomly assigned to the control group or the intervention group (screening arm), where they underwent a 60 cm flexible sigmoidoscopy examination at study entry (T0) and year 3 (T3) or year 5 (T5) of the study. Those found to have a suspicious lesion were referred to their personal physician for subsequent diagnostic follow-up. Among the 64 658 men and women who underwent sigmoidoscopy at T0 in the PLCO Cancer Screening Trial, 8.8% were found to have adenoma (24). Cases of colorectal adenoma were pathologically verified according to medical records. Information on demographics, personal and family medical history and lifestyle factors (e.g. smoking and dietary intake) were collected by standard questionnaire at baseline. This trial was approved by the institutional review boards of the 10 screening centers and the National Cancer Institute in Bethesda, MD, USA, and all participants provided informed consent.
Study population
A nested case–control study was conducted among the PLCO participants randomized to the screening arm, who consented to participate in etiologic studies of cancer and related diseases, completed a risk factor questionnaire, provided a blood specimen and had no previous history of inflammatory bowel disease, colorectal polyps, Gardner’s syndrome, familial polyposis or cancer other than basal or squamous cell skin cancer. For this study, cases were participants found to have advanced colorectal adenoma (≥1 cm in size, containing villous/tubulovillous characteristics, or had severe dysplasia) of the distal colon or rectum at the T0 exam. Carcinoma in situ was classified as severe dysplasia. Controls were participants, who had successful sigmoidoscopy at T0 and were negative for polyps in the distal colon and rectum. Controls were frequency matched to cases on self-reported ethnicity (non-Hispanic Caucasian, non-Hispanic Black, Asian, Hispanic, Pacific Islander, American Indian or Alaskan Native or Unknown), gender and for a subset on age (55–59, 60–64, 65–69 and 70–74 years). Over 90% of the subjects were non-Hispanic Caucasians.
Genotyping
A total of 3338 tag SNPs from 149 genes (supplementary Table 1 is available at Carcinogenesis Online) involved in DNA repair pathways (11) were selected to comprehensively interrogate genetic variations across the candidate genes. Tag SNPs were selected for each gene, including the region 20 kb upstream and 10 kb downstream of the gene, using the CEU, JPT, CHB and YRI HapMap populations and the Carlson method (25) as implemented in Tagzilla with a r2 threshold of 0.8 and minor allele frequency ≥5%. SNPs with known or putative functional significance (i.e. non-synonymous, promoter, intron–exon splice sites) were also included whenever possible. The SNPs were genotyped on a custom iSelect panel utilizing Illumina’s GoldenGate platform.
Whole blood or buffy coat DNA was extracted with QIAamp DNA Blood Midi or Maxi Kits. For this study, sufficient DNA was available from 1342 cases and 1507 controls for genotyping. For quality control purposes, replicate samples from 195 individuals (∼7% of the population) were interspersed randomly within the plates. Genotyping was conducted at the National Cancer Institute Core Genotyping Facility, NIH. A total of 1338 cases and 1503 controls were successfully genotyped with over 90% of the genotypes for each subject having valid calls, and the overall concordance rate was >99% for replicated samples. After excluding the SNPs with call rate <90%, minor allele frequency <1%, or Hardy–Weinberg Equilibrium P-value <1 × 10−6 among Caucasian controls, 3144 SNPs of 3401 selected (92%) remained for analysis. For each gene, the percentage of tagSNPs passing our quality control criteria varied from 67 to 100% with an average of 93%.
Statistical analysis
The initial analyses were conducted using Plink, a whole genome association analysis toolset (26). Logistic regression was used to estimate the OR and 95% CI for the association between each SNP and colorectal adenoma risk assuming a log-additive model for the genotype, adjusting for age (55–59, 60–64, 65–69, 70–74 years), gender and ethnicity (non-Hispanic Caucasian, non-Hispanic Black and other). Another set of analysis was conducted among non-Hispanic Caucasians only.
For SNPs with main association P < 0.01, we tested if the SNP associations differed by smoking status (ever versus never) using the Breslow–Day test of homogeneity. The SNPs with evidence of heterogeneity (P < 0.05 between ever versus never smoker) among Caucasians were further tested for interaction with smoking status (never, former and current smoking) using a likelihood ratio test. The P-value for trend was generated by treating the smoking status (never = 0, former = 1 and current = 2) as a continuous variable in the interaction model, and the P-value for interaction was generated by treating the smoking status as categorical variable in the interaction model. Similarly, we also conducted analyses to examine the effect modification by smoking duration (<24 years, ≥24 years) and years since quitting smoking (<20 years, ≥20 years). These additional analyses were conducted with SAS 9.1.
Pairwise linkage disequilibrium measures (D′ and r2) were inferred from the Caucasian controls using the program Haploview (27). Haplotypes among Caucasians were estimated using an expectation–maximization algorithm for SNPs within the gene Exonuclease-1 (EXO1), which carried the SNP with lowest P-value in the current study, and risks for individual haplotypes were calculated assuming a log-additive model and using the generalized linear model implemented in R Haplostats package, adjusted for age and gender (28). For consistency with the SNP results, we used the haplotype containing the T allele at rs9350 as the reference haplotype.
To evaluate the potential for false-positive findings due to multiple testing, we adjusted the P values using a Bonferroni correction for the total number of (a) tag SNPs for each individual gene (gene based) as well as (b) all the SNPs tested in the current analysis, using the R multtest package.
Results
A total of 1338 colorectal adenoma cases and 1503 frequency-matched controls were included in the current analyses (Table I). Over 90% of the study subjects were Caucasian and <10% were African-American or other ethnicity. Compared with controls, cases were more probably to be current smokers, have a family history of colorectal cancer and be less educated.
Table I.
Demographic characteristics of cases and controls, PLCO Cancer Screening Trial baseline exam 1993–2001
| Controls (%) N = 1503 | Cases (%) N = 1338 | |
| Race | ||
| White | 1370 (91) | 1241 (93) |
| Black | 66 (4) | 50 (4) |
| Other | 67 (5) | 47 (3) |
| Gender | ||
| Male | 950 (63) | 853 (64) |
| Female | 553 (37) | 485 (36) |
| Age | ||
| 55–59 | 498 (33) | 390 (29) |
| 60–64 | 460 (31) | 419 (31) |
| 65–69 | 344 (23) | 333 (25) |
| 70–75 | 201 (13) | 196 (15) |
| Smoking status | ||
| Never | 650 (43) | 451 (34) |
| Former cigarette smoker | 679 (45) | 627 (47) |
| Current cigarette smoker | 105 (7) | 197 (15) |
| Cigar or pipe smoker | 69 (5) | 62 (5) |
| Family history of colorectal cancer | ||
| Yes | 141 (9) | 162 (12) |
| No | 1362 (91) | 1176 (88) |
| Body mass index (BMI) | ||
| <25 | 451 (30) | 369 (28) |
| 25–29.9 | 680 (46) | 594 (45) |
| ≥30 | 354 (24) | 364 (27) |
| Education | ||
| <12 years | 439 (29) | 470 (35) |
| Post high school | 489 (33) | 479 (36) |
| College graduate or postgraduate | 574 (38) | 389 (29) |
| Regular non-steroidalanti-inflammatory drug use | ||
| Yes | 935 (62) | 776 (58) |
| No | 565 (38) | 559 (42) |
Of the 3144 SNPs analyzed, 129 SNPs (supplementary Table 2 is available at Carcinogenesis Online) were associated with adenoma risk among all subjects and 127 SNPs with risk among Caucasians only at P < 0.05. The SNPs associated with colorectal adenoma risk at P ≤ 0.01 level among Caucasians and are shown in Table II. After adjusting for multiple testing for all the SNPs tested in the analysis, none of these 30 SNPs remained statistically significant. However, six SNPs remained associated with adenoma risk among Caucasians at P ≤ 0.05 level after a gene-based multiple testing correction: EXO1 rs9350, FANCC rs400727, ERCC1 rs10412761, DCLRE1A rs2301180, RAD54B rs3762053 and POLE rs11614717. Further adjustment for smoking status did not substantially alter the results in Table II (results not shown). The most statistically significant SNP associated with risk was EXO1 rs9350 with heterozygotes displaying a 1.95-fold risk (95% CI = 1.05–3.62) and CC homozygotes displaying a 2.39-fold risk (95% CI = 1.30–4.38) comparing with TT homozygotes.
Table II.
OR of colorectal adenoma for polymorphisms in DNA repair genes with P < 0.01, PLCO Cancer Screening Trial baseline exam 1993–2001
| Caucasian subjectsa |
All subjectsb |
||||||||
| SNP | Gene | Risk allele | RAFc | OR | 95% CI | P | OR | 95% CI | P |
| rs9350 T→C | EXO1 | C | 0.82 | 1.30 | 1.11–1.51 | 0.001 | 1.24 | 1.07–1.44 | 0.003 |
| rs400727 T→G | FANCC | G | 0.91 | 1.44 | 1.15–1.80 | 0.002 | 1.40 | 1.14–1.72 | 0.001 |
| rs3762053 G→T | RAD54B | G | 0.16 | 1.25 | 1.09–1.44 | 0.002 | 1.23 | 1.07–1.41 | 0.004 |
| rs11614717 T→G | POLE | T | 0.14 | 1.27 | 1.09–1.49 | 0.002 | 1.24 | 1.07–1.44 | 0.005 |
| rs4703564 T→C | XRCC4 | C | 0.89 | 1.42 | 1.13–1.78 | 0.002 | 1.31 | 1.09–1.59 | 0.005 |
| rs242448 A→T | FANCC | T | 0.92 | 1.42 | 1.13–1.78 | 0.002 | 1.41 | 1.14–1.75 | 0.002 |
| rs4658535 A→G | EXO1 | G | 0.79 | 1.26 | 1.08–1.45 | 0.002 | 1.25 | 1.09–1.42 | 0.002 |
| rs1699499 C→T | FANCC | T | 0.97 | 1.70 | 1.20–2.40 | 0.003 | 1.67 | 1.19–2.35 | 0.003 |
| rs17503908 G→T | ATM | G | 0.08 | 1.34 | 1.11–1.62 | 0.003 | 1.33 | 1.10–1.60 | 0.003 |
| rs10412761 G→A | ERCC1 | A | 0.57 | 1.19 | 1.06–1.33 | 0.003 | 1.19 | 1.07–1.33 | 0.001 |
| rs3885676 C→T | XRCC4 | T | 0.87 | 1.36 | 1.11–1.66 | 0.003 | 1.31 | 1.10–1.56 | 0.003 |
| rs4465523 A→G | APEX1 | A | 0.33 | 1.18 | 1.05–1.32 | 0.006 | 1.20 | 1.08–1.34 | 0.001 |
| rs17277375 T→C | APEX1 | T | 0.32 | 1.18 | 1.05–1.33 | 0.006 | 1.19 | 1.06–1.33 | 0.003 |
| rs2301180 C→T | DCLRE1A | T | 0.71 | 1.18 | 1.05–1.34 | 0.006 | 1.16 | 1.03–1.30 | 0.01 |
| rs6864054 G→A | XRCC4 | A | 0.94 | 1.44 | 1.11–1.87 | 0.006 | 1.33 | 1.05–1.70 | 0.02 |
| rs5744990 A→G | POLE | A | 0.13 | 1.24 | 1.06–1.45 | 0.006 | 1.23 | 1.06–1.43 | 0.007 |
| rs6573333 T→G | MNAT1 | G | 0.98 | 2.36 | 1.26–4.40 | 0.007 | 1.18 | 0.80–1.74 | 0.39 |
| rs3102854 G→A | RAD54B | G | 0.24 | 1.19 | 1.05–1.35 | 0.007 | 1.18 | 1.04–1.33 | 0.009 |
| rs7829886 A→T | RAD54B | T | 0.73 | 1.19 | 1.05–1.35 | 0.008 | 1.16 | 1.03–1.31 | 0.02 |
| rs12050102 T→G | APEX1 | T | 0.33 | 1.17 | 1.04–1.32 | 0.008 | 1.19 | 1.06–1.33 | 0.002 |
| rs4150628 G→A | GTF2H1 | G | 0.44 | 1.16 | 1.04–1.23 | 0.008 | 1.14 | 1.03–1.27 | 0.02 |
| rs228606 T→G | ATM | T | 0.39 | 1.16 | 1.04–1.30 | 0.008 | 1.15 | 1.04–1.28 | 0.009 |
| rs3212986 A→C | ERCC1 | C | 0.74 | 1.19 | 1.04–1.35 | 0.008 | 1.19 | 1.05–1.34 | 0.006 |
| rs10838192 C→T | ALKBH3 | C | 0.21 | 1.19 | 1.05–1.36 | 0.009 | 1.19 | 1.04–1.35 | 0.008 |
| rs828918 G→C | XRCC5 | G | 0.13 | 1.24 | 1.06–1.45 | 0.009 | 1.19 | 1.02–1.38 | 0.03 |
| rs3131382 T→C | MSH5 | C | 0.93 | 1.36 | 1.08–1.71 | 0.009 | 1.37 | 1.09–1.72 | 0.007 |
| rs1150793 G→A | MSH5 | G | 0.06 | 1.34 | 1.08–1.67 | 0.009 | 1.26 | 1.02–1.55 | 0.03 |
| rs707915 A→T | MSH5 | A | 0.06 | 1.34 | 1.08–1.67 | 0.009 | 1.26 | 1.02–1.55 | 0.03 |
| rs11615 G→A | ERCC1 | A | 0.58 | 1.16 | 1.04–1.31 | 0.009 | 1.15 | 1.02–1.28 | 0.02 |
| rs5744903 T→C | POLE | T | 0.06 | 1.32 | 1.07–1.62 | 0.01 | 1.28 | 1.04–1.57 | 0.02 |
OR per risk allele assuming a log-additive model, adjusted for age and sex.
OR per risk allele assuming a log-additive model, adjusted for age, sex and race.
RAF, risk allele frequency among Caucasian controls.
Given the importance of smoking as a risk factor for adenoma, we examined the heterogeneity in risk by smoking status (ever versus never). Of the SNPs with a P < 0.01 for their main association, three SNPs displayed significant heterogeneity in risk by smoking status (P < 0.05) and were selected for further analyses stratified by cigarette smoking status (never, former and current) (Table III). The risk of colorectal adenoma at EXO1 rs9350 was significantly modified by smoking status (Pinteraction = 0.006) with a 2-fold increased risk among current smokers (OR = 2.15; 95% CI: 1.27–3.65), a modest increased risk among former smokers (OR = 1.45; 95% CI: 1.14–1.82), and no association among never smokers (OR = 0.98; 95% CI: 0.76–1.25) (Ptrend = 0.002). A stronger increased risk was also observed among individuals with longer smoking duration (≥24 years: OR = 1.59; 95% CI: 1.20–2.10) compared with shorter duration (<24 years: OR = 1.48; 95% CI: 1.09–1.99) (Pinteraction = 0.02), as well as individuals who quit more recently (<20 years: OR = 1.64; 95% CI: 1.24–2.18) (Pinteraction = 0.02) compared with those who quit a longer time ago (≥20 years: OR = 1.42; 95% CI: 1.0–1.99). Similarly, another SNP in EXO1, rs4658535, also showed a monotonically increasing pattern of risk from never, former, to current smokers (Ptrend = 0.002). These two SNPs in EXO1 were strongly correlated (r2 = 0.82), making it difficult to differentiate the associations of one from the other statistically. When both SNPs were put in the same model, neither of them remained significantly associated with adenoma risk (P > 0.05 for both) due to the high correlation. Combining former and current smokers, the C allele at rs9350 and G allele at rs4658535 were associated with increased risk among ever smokers with ORs of 1.54 (95% CI = 1.26–1.89) and 1.46 (95% CI = 1.21–1.77) for the two SNPs, respectively.
Table III.
OR of colorectal adenoma for select DNA repair SNPs stratified by smoking status among Caucasiansa, PLCO Cancer Screening Trial baseline exam 1993–2001
| Never |
Former |
Current |
P trendb | P interactionc | ||||||
| Gene | Risk allele | OR | P | OR | P | OR | P | |||
| rs9350 | EXO1 | C | 0.98 (0.76–1.25) | 0.86 | 1.45 (1.14–1.82) | 0.002 | 2.15 (1.27–3.65) | 0.004 | 0.002 | 0.006 |
| rs4658535 | EXO1 | G | 0.98 (0.77–1.23) | 0.84 | 1.34 (1.08–1.68) | 0.008 | 2.13 (1.30–3.49) | 0.003 | 0.002 | 0.006 |
| rs17503908 | ATM | T | 0.55 (0.40–0.76) | 0.0003 | 0.84 (0.64–1.11) | 0.22 | 1.25 (0.71–2.22) | 0.44 | 0.008 | 0.02 |
SNPs selected here were those with significant main effect (adjusted for age, sex) (P < 0.01) and gene–smoking (ever versus never smoking) interaction (P < 0.05).
P-value for trend was calculated assuming smoking status (never = 0, former = 1 and current = 2) as continuous variable in the interaction model.
P-value for interaction was calculated assuming smoking status (never = 0, former = 1 and current = 2) as categorical variable in the interaction model.
The risk of adenoma at rs17503908 in ataxia telangiectasia mutated (ATM) was also modified by smoking status (Pinteraction = 0.02) with the T allele displaying a decreased risk for adenoma only among never smokers (OR = 0.55, 95% CI = 0.40–0.76) (Table III) but not among ever smokers (OR = 0.89, 95% CI = 0.70–1.13). Notably, rs17503908 showed an intermediate risk for adenoma also among former smokers (OR = 0.84, 95% CI = 0.64–1.11) compared with never smokers (OR = 0.55, 95% CI = 0.40–0.76) and current smokers (OR = 1.25, 95% CI = 0.71–2.22) (Ptrend = 0.008). Similar patterns were also observed when the results were stratified by smoking duration and time since quitting (data not shown).
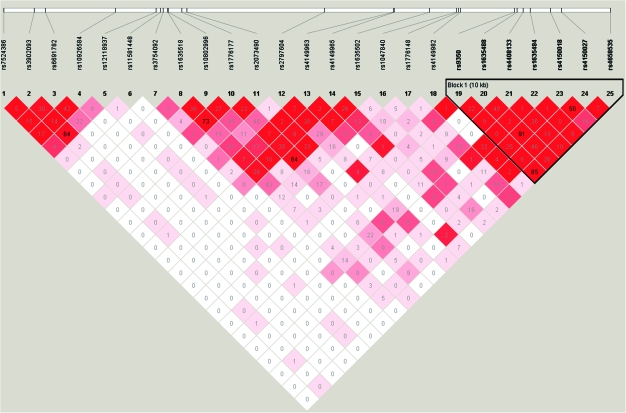
Given that the associations of two SNPs in EXO1 appeared to be modified by smoking status, we explored this region in greater detail. A total of 25 SNPs were genotyped in EXO1. Seven SNPs (rs9350, rs1635488, rs4408133, rs1635484, rs4150018, rs4150027 and rs4658535), including the two SNPs significantly associated with risk, were in strong linkage disequilibrium as measured by D′ (Figure 1) and mildly to strongly correlated (r2 range: 0.23–0.84). The five most frequent haplotypes comprised of these seven SNPs were analyzed in association with adenoma risk by smoking status (never and ever smoking, but not never, former and current smoking status due to limited power) (supplementary Table 3 is available at Carcinogenesis Online). Compared with the haplotype containing the T allele at rs9350 (rs9350-rs1635488-rs4408133-rs1635484-rs4150018-rs4150027-rs4658535: TCGCGTA), three haplotypes, each containing the risk alleles at rs9350 and rs4658535, were significantly associated with an increased risk of adenoma with risk estimates ranging from 1.23 to 1.30. The associations were stronger among smokers (40–60% increased risk) compared with never smokers for which none of the haplotypes were associated with risk. The test for the haplotype–smoking interaction was marginally significant (P = 0.06).
Fig. 1.
Linkage disequilibrium plot of EXO1 among Caucasian controls from the PLCO Cancer Screening Trial baseline exam 1993–2001. The colors represent the extent of pairwise linkage disequilibrium as measured by D′ with red indicating D′ = 1.0 and shades of pink to white indicating D′ <1.0. The numeric values are the pairwise r2 values.
Discussion
DNA repair has long been implicated in colorectal cancer with the discovery that germ line mutations in MMR genes lead to HNPCC (29) and mutations in the base excision repair gene, MUTYH, lead to a familial polyposis syndrome. However, the etiological role of common genetic variation in DNA repair genes in colorectal adenoma and cancer has not been comprehensively studied in the context of epidemiological studies. Although some common SNPs in DNA repair genes have been reported to be associated with colorectal cancer and/or adenoma (18–21,30,31), with the exception of the MHL1-93G > A variant with microsatellite instable tumors (18–21), most associations have not been replicated (30,31). Furthermore, data on effect modifications by important environmental factors are sparse.
In our study, >3000 SNPs from 153 DNA repair genes were evaluated simultaneously among 2841 study subjects, which is the largest and most comprehensive study for colorectal adenoma risk focusing on DNA repair genes to date. Among the SNPs associated with risk, we found that genetic polymorphisms in EXO1 and ATM significantly modified the effect of cigarette smoking on risk, predisposing smokers to greater adenoma susceptibility. When stratified by genotype, smoking was only significantly associated with increased adenoma risk among individuals homozygous for the risk allele at EXO1 rs9350 (OR = 1.80, 95% CI = 1.47–2.19 for ever smokers with the CC genotype) and ATM rs17503908 (OR = 1.69, 95% CI = 1.40–2.03 for ever smokers with the GG genotype).
EXO1, located at chromosome 1q42–q43, encodes a protein with 5′→ 3′ and 3′→ 5′ double-stranded DNA exonuclease activity. It also exhibits some endonuclease activity correcting 5′-overhanging flap structures. EXO1 is involved in DNA MMR, recombination, replication and telomere stabilization (32). EXO1-mutant cells showed increased microsatellite instability and incomplete MMR capability (33). Mice with EXO1 knockout were found to have lower survival rates and higher mutation rates as well as higher susceptibility to lymphomas (33). EXO1 has been implicated in hereditary HNPCC due to its role in DNA MMR; however, studies investigating rare germ line variants in EXO1 have not shown consistent findings as reviewed by Liberti et.al. (34).
We observed an increased colorectal adenoma risk among individuals carrying a C allele at rs9350 and a G allele at rs4658535 in EXO1. The SNPs were highly correlated (r2 = 0.82), making it impossible to differentiate the associations of each statistically. Using the PolyPhen database, we found that rs9350 was predicted to be ‘probably damaging’ (position-specific independent counts score difference = 2.17) with the C to T substitution causing a non-synonymous amino acid change by replacing proline with leucine (35), suggesting that rs9350 may be a causal variant. The substitution was also predicted to be ‘deleterious’ using the SIFT database. Several studies have examined the association between common polymorphisms in EXO1 and the risk of lung, oral, brain and colorectal cancer (36–42); however, data are limited and inconclusive due to the small sample size and differences in the SNPs genotyped in the studies (36–42). No studies have examined the association between this polymorphism and adenoma. Consistent with our findings, two case–control studies of colorectal cancer found a decreased cancer risk for individuals carrying T allele at EXO1 rs9350 compared with C allele (40,43). No association was observed with rs9350 in two studies of lung cancer (37,38), one study of oral cancer (39) and one study of breast cancer (44), suggesting that the association of C allele at rs9350 may be organ/tissue specific. In addition, the previous studies in other cancers (37–39) were relatively small (N ≤ 680 cases each) and conducted in Asian populations, where differences in environmental exposures may modify the association of rs9350 and cancer risk compared with Caucasians.
Smoking is an important risk factor for adenoma with current smokers having an 1.8-fold increased risk (95% CI = 1.5–2.1) in the full PLCO cohort (45). We observed a stronger association between adenoma risk and rs9350 in EXO1 among smokers compared with non-smokers and hypothesize that the C allele at EXO1 rs9350 may increase risk among smokers by reducing the protein’s capacity or efficiency to repair the damage caused by smoking exposure. Tsai et al. also reported an increased risk of oral cancer among smokers who carried the A allele at rs1047840 (r2 = 0.18 with rs9350 in our study) in EXO1 but not among non-smokers (39). The haplotype results further confirmed the strong association between the EXO1 region encompassing rs9350 and adenoma risk.
We observed a significant decreased risk among never smokers for the T allele at rs17503908 in ATM. ATM, located at chromosome 11q22.3, encodes a cell cycle checkpoint kinase which regulates many downstream proteins, including the tumor suppressor proteins p53 and BRCA1, the checkpoint kinase CHK2, checkpoint proteins RAD17 and RAD9 and the DNA repair protein NBS1. ATM is thought to be a master controller of the cell cycle checkpoint-signaling pathways and functions to repair DNA damage and maintain genome stability. Persons with ataxia telangiectasia, an autosomal recessive diseased caused by rare missense or truncating mutations in ATM, have an increased sensitivity to ionizing radiation and an increased risk of cancer (46). Heterozygous carriers of these rare ATM mutations have an increased risk of several cancers including colorectal cancer (47).
Several lines of evidence have suggested the etiological role for ATM in colorectal carcinogenesis (47,48). Polymorphisms at rs1800056 and rs1800057 in ATM have been associated with colorectal cancer risk (49) and although the results were not replicated in a follow-up study (50), rs1801516 has been associated with disease penetrance among HNPCC carriers (51). In our study, the T allele at ATM rs1801516 was also marginally associated with adenoma risk (P = 0.011 for Caucasians and P = 0.016 for all subjects), but no association was observed with rs1800056 (P = 0.22). To date, no study has reported an association between ATM rs17503908 and colorectal adenoma, which is located in an intronic region of ATM. We observed an inverse association for the T allele at rs17503908. Differences in sensitivity to DNA damage or higher expression levels of ATM could reduce risk for neoplastic transformation or subsequent proliferation by activating p53 (52). In stratified analyses, this inverse association was restricted to never smokers. It is possible that smokers do not benefit from carrying this allele due to an antagonistic effect between the SNP and smoking exposure. An in vitro study observed that smoking exposure activated ATM in human pulmonary adenocarcinoma cells through phosphorylation in a dose-dependent manner (53). Similarly, benzo[a]pyrene diol epoxide, a polycyclic aromatic hydrocarbon found in tobacco smoke, has been shown to bind to ATM (54) and induce ATM expression in esophageal cancer cell lines (55), suggesting that ATM plays an active role in responding to tobacco smoke exposure. We speculate that the kinase encoded by the ATM may be saturated by smoking exposure, which may prevent its protective effect.
Our study has several advantages and limitations. First, it is the largest study to date to investigate a broad range of DNA repair gene polymorphisms for colorectal adenoma risk. Although still somewhat underpowered to examine gene–environment interactions for the SNPs of moderate association, our findings provide promising leads for replication in future pooled analyses. Future analyses exploring these interactions with regard to colorectal cancer may lead to additional insight into colorectal neoplasm progression. Our study included only advanced adenoma cases and so the results may not be generalizable to non-advanced adenomatous polyps; however, advanced adenomas are more to progress to colorectal cancer and therefore clinically more relevant. Moreover, our study only included left-sided adenomas in the distal colon and rectum. Thus, the results of our study may not be generalizable to adenomas observed in the proximal colon, which may be more probably to occur as the result deficiencies in MMR. In addition, our study population came from a cancer prevention screening trial in which participants were generally more probably to be Caucasian, more educated, less probably to smoke and more physically active than the general population (56). Thus, our results may not be broadly generalizable to the entire population or to other ethnicities. However, since our case–control study was nested within a randomized population-based colorectal cancer screening trial, this reduces the potential for selection bias often inherited in clinic-based case–control studies of adenoma, where persons may undergo endoscopy for reasons other than routine screening, such as gastrointestinal symptoms, blood in their stools, diagnostic follow-up or because they have a family history of colorectal cancer.
In summary, in this large comprehensive study of DNA repair gene polymorphisms and colorectal adenoma risk, we found that an SNP in EXO1 predicted to deleteriously alter function was associated with increased adenoma risk. The association was restricted to ever smokers and stronger in current smokers than former smokers. Although additional studies are needed to confirm our findings, this intriguing result suggests that genetic variation in EXO1 may modify susceptibility to colorectal adenoma, particularly among smokers.
Supplementary material
Supplementary Tables 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health (NIH).
Supplementary Material
Acknowledgments
The authors thank Drs Christine Berg and Philip Prorok, Division of Cancer Prevention, NCI, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr Thomas Riley and staff at Information Management Services and Ms Barbara O’Brien and staff at Westat for their contributions to the PLCO Cancer Screening Trial. Finally, we acknowledge the study participants for donating their time and making this study possible.
Author contributions: Y.G., S.B., R.H. and W.-Y.H. designed the study. Y.G. and S.B. also analyzed data and wrote the manuscript. L.B., M.Y. and S.C. were instrumental in the genotyping for this project. All authors read, gave comments and approved the final version of the manuscript. All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ATM
ataxia telangiectasia mutated
- CI
confidence interval
- EXO1
Exonuclease-1
- HNPCC
hereditary non-polyposis colorectal cancer
- MMR
mismatch repair
- OR
odds ratio
- PLCO
Prostate, Lung, Colorectal and Ovarian
- SNP
single-nucleotide polymorphism
References
- 1.Jemal A, et al. Cancer statistics, 2009. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Terry MB, et al. Risk factors for advanced colorectal adenomas: a pooled analysis. Cancer Epidemiol. Biomarkers Prev. 2002;11:622–629. [PubMed] [Google Scholar]
- 3.Huxley RR, et al. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int. J. Cancer. 2009;125:171–180. doi: 10.1002/ijc.24343. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Tenesa A, et al. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat. Rev. Genet. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 6.Houlston RS, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botteri E, et al. Cigarette smoking and adenomatous polyps: a meta-analysis. Gastroenterology. 2008;134:388–395. doi: 10.1053/j.gastro.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Giovannucci E, et al. Tobacco, colorectal cancer, and adenomas: a review of the evidence. J. Natl Cancer Inst. 1996;88:1717–1730. doi: 10.1093/jnci/88.23.1717. [DOI] [PubMed] [Google Scholar]
- 10.Raimondi S, et al. Gene-smoking interaction on colorectal adenoma and cancer risk: review and meta-analysis. Mutat. Res. 2009;670:6–14. doi: 10.1016/j.mrfmmm.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Wood RD, et al. Human DNA repair genes, 2005. Mutat. Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Kinzler KW, et al. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen M, et al. Analysis of MUTYH genotypes and colorectal phenotypes in patients With MUTYH-associated polyposis. Gastroenterology. 2009;136:471–476. doi: 10.1053/j.gastro.2008.10.056. [DOI] [PubMed] [Google Scholar]
- 14.Berndt SI, et al. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67:1395–1404. doi: 10.1158/0008-5472.CAN-06-1390. [DOI] [PubMed] [Google Scholar]
- 15.Stern MC, et al. XRCC1, XRCC3, and XPD polymorphisms as modifiers of the effect of smoking and alcohol on colorectal adenoma risk. Cancer Epidemiol. Biomarkers Prev. 2006;15:2384–2390. doi: 10.1158/1055-9965.EPI-06-0381. [DOI] [PubMed] [Google Scholar]
- 16.Skjelbred CF, et al. Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: a case control study. BMC Cancer. 2006;6:67. doi: 10.1186/1471-2407-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bigler J, et al. DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps. Cancer Epidemiol. Biomarkers Prev. 2005;14:2501–2508. doi: 10.1158/1055-9965.EPI-05-0270. [DOI] [PubMed] [Google Scholar]
- 18.Mrkonjic M, et al. Specific variants in the MLH1 gene region may drive DNA methylation, loss of protein expression, and MSI-H colorectal cancer. PLoS One. 2010;5:e13314. doi: 10.1371/journal.pone.0013314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raptis S, et al. MLH1 -93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J. Natl Cancer Inst. 2007;99:463–474. doi: 10.1093/jnci/djk095. [DOI] [PubMed] [Google Scholar]
- 20.Samowitz WS, et al. The MLH1 -93 G>A promoter polymorphism and genetic and epigenetic alterations in colon cancer. Genes Chromosomes Cancer. 2008;47:835–844. doi: 10.1002/gcc.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell PT, et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58:661–667. doi: 10.1136/gut.2007.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes RB, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat. Res. 2005;592:147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Prorok PC, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control. Clin. Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 24.Weissfeld JL, et al. Flexible sigmoidoscopy in the PLCO cancer screening trial: results from the baseline screening examination of a randomized trial. J. Natl Cancer Inst. 2005;97:989–997. doi: 10.1093/jnci/dji175. [DOI] [PubMed] [Google Scholar]
- 25.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Schaid DJ, et al. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch HT, et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat. Rev. Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 31.Foulkes WD. Inherited susceptibility to common cancers. N. Engl. J. Med. 2008;359:2143–2153. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 32.Tran PT, et al. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst.) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Wei K, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberti SE, et al. Is hEXO1 a cancer predisposing gene? Mol. Cancer Res. 2004;2:427–432. [PubMed] [Google Scholar]
- 35.Ramensky V, et al. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang JS, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol. Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu NY, et al. Lung cancer susceptibility and genetic polymorphisms of Exo1 gene in Taiwan. Anticancer Res. 2009;29:725–730. [PubMed] [Google Scholar]
- 38.Jin G, et al. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: a case-control analysis. Lung Cancer. 2008;60:340–346. doi: 10.1016/j.lungcan.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Tsai MH, et al. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral Oncol. 2009;45:e90–e94. doi: 10.1016/j.oraloncology.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto H, et al. Single nucleotide polymorphisms in the EXO1 gene and risk of colorectal cancer in a Japanese population. Carcinogenesis. 2005;26:411–416. doi: 10.1093/carcin/bgh335. [DOI] [PubMed] [Google Scholar]
- 41.Yoshiya G, et al. Influence of cancer-related gene polymorphisms on clinicopathological features in colorectal cancer. J. Gastroenterol. Hepatol. 2008;23:948–953. doi: 10.1111/j.1440-1746.2008.05307.x. [DOI] [PubMed] [Google Scholar]
- 42.Zienolddiny S, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 43.Haghighi MM, et al. Impact of EXO1 polymorphism in susceptibility to colorectal cancer. Genet. Test. Mol. Biomarkers. 2010;14:649–652. doi: 10.1089/gtmb.2010.0034. [DOI] [PubMed] [Google Scholar]
- 44.Wang HC, et al. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer Res. 2009;29:3897–3901. [PubMed] [Google Scholar]
- 45.Ji BT, et al. Tobacco smoking and colorectal hyperplastic and adenomatous polyps. Cancer Epidemiol. Biomarkers Prev. 2006;15:897–901. doi: 10.1158/1055-9965.EPI-05-0883. [DOI] [PubMed] [Google Scholar]
- 46.Morrell D, et al. Mortality and cancer incidence in 263 patients with ataxia-telangiectasia. J. Natl Cancer Inst. 1986;77:89–92. [PubMed] [Google Scholar]
- 47.Thompson D, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J. Natl Cancer Inst. 2005;97:813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 48.Kweekel DM, et al. Explorative study to identify novel candidate genes related to oxaliplatin efficacy and toxicity using a DNA repair array. Br. J. Cancer. 2009;101:357–362. doi: 10.1038/sj.bjc.6605134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb EL, et al. Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14 704 first-degree relatives. Hum. Mol. Genet. 2006;15:3263–3271. doi: 10.1093/hmg/ddl401. [DOI] [PubMed] [Google Scholar]
- 50.Jones JS, et al. ATM polymorphism and hereditary nonpolyposis colorectal cancer (HNPCC) age of onset (United States) Cancer Causes Control. 2005;16:749–753. doi: 10.1007/s10552-005-1540-7. [DOI] [PubMed] [Google Scholar]
- 51.Maillet P, et al. A polymorphism in the ATM gene modulates the penetrance of hereditary non-polyposis colorectal cancer. Int. J. Cancer. 2000;88:928–931. doi: 10.1002/1097-0215(20001215)88:6<928::aid-ijc14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 52.Concannon P, et al. Variants in the ATM gene associated with a reduced risk of contralateral breast cancer. Cancer Res. 2008;68:6486–6491. doi: 10.1158/0008-5472.CAN-08-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorgensen ED, et al. DNA damage response induced by exposure of human lung adenocarcinoma cells to smoke from tobacco- and nicotine-free cigarettes. Cell Cycle. 2010;9:2170–2176. doi: 10.4161/cc.9.11.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang Z, et al. Identification of benzo(a)pyrene diol epoxide-binding DNA fragments using DNA immunoprecipitation technique. Cancer Res. 2003;63:1470–1474. [PubMed] [Google Scholar]
- 55.Jiang Y, et al. Ataxia-telangiectasia mutated expression is associated with tobacco smoke exposure in esophageal cancer tissues and benzo[a]pyrene diol epoxide in cell lines. Int. J. Cancer. 2007;120:91–95. doi: 10.1002/ijc.22121. [DOI] [PubMed] [Google Scholar]
- 56.Pinsky PF, et al. Evidence of a healthy volunteer effect in the prostate, lung, colorectal, and ovarian cancer screening trial. Am. J. Epidemiol. 2007;165:874–881. doi: 10.1093/aje/kwk075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.