Abstract
We have constructed a set of fragmentation vectors for the truncation of either the centromeric or the noncentromeric end of YACs containing a human DNA insert. These vectors carry ADE2 or HIS5 as the selectable marker, enabling direct use in AB1380, the host strain of most publicly available YAC libraries. Centromeric fragmentation vectors for AB1380 have not been reported previously; the noncentromeric vectors show high frequencies of fragmentation.
Yeast artificial chromosomes (YACs) consist of a large (50–2500 kb) DNA insert, flanked on both sides by a yeast selectable marker and a telomere, and on one side by a yeast centromere. An important application of YACs lies in the mapping of the human genome and the isolation of human disease genes. YAC contigs now cover nearly the entire human genome (Chumakov et al. 1995), but the depth of these contigs is often not sufficient to allow high-resolution ordering of markers. Because many candidate regions for disease genes have been allocated to YAC contigs still comprising several megabases, improvement of the resolution of these contigs is highly desirable. More recent, complementary resources in contig generation are bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) libraries. Although valuable in the ultimate generation of sequence-ready genomic regions, they preclude the delineation of the available YAC contig information in the megabase range, as the assembly of BAC/PAC contigs is a de novo endeavor. YAC fragmentation, that is, the creation of YACs with deletions from one end (Pavan et al. 1990), is an attractive method for obtaining more refined mapping while optimally using existing knowledge. Compared to the common route of constructing cosmid contigs, YAC fragmentation is a simple and quick alternative. Furthermore, YAC fragmentation provides the possibility of discarding the noninteresting parts of a YAC, which is useful for subsequent studies like the production of well-defined restriction maps, subcloning, gene searches, and gene characterizations, and as an attractive tool to focus YAC-derived transgene studies.
YAC fragmentation is based on the presence of sequences in the YAC insert homologous to a part of the fragmentation vector, which further contains a selectable marker, a telomere, and a centromere if the centromeric arm is to be targeted. Common repeats like the long interspersed repetitive element (LINE) and, in particular, Alu are attractive candidates for this purpose, as they occur frequently in YACs with a human insert. The series of fragmentation vectors described by Pavan et al. (1991) enable the construction of YAC fragmentation panels in a his3− background. However, all publicly available YAC libraries, like the Centre d’Etude du Polymorphisme Humain (CEPH), Imperial Chemical Industries PLC (ICI), and Imperial Cancer Research Fund (ICRF) libraries (Albertsen et al. 1990; Anand et al. 1990; Larin et al. 1991), have been constructed in yeast strain AB1380, which is not his3−. Therefore, YACs must first be transferred to a his3− background, through meiosis or kar1 transfer (Spencer et al. 1994), before these vectors can be used. To bypass this problem, we have adapted existing fragmentation vectors with the ADE2 gene and/or CEN6, thereby creating a set of vectors that can be used directly in AB1380 to fragment YACs from both ends.
RESULTS AND DISCUSSION
For fragmentation of YACs from libraries in AB1380 a vector with a suitable selectable marker is required. AB1380 has the genotype MATa ura3 trp1 ade2, his5, lys2-1, can1-100, allowing the use of ADE2, HIS5, or LYS2, as the URA3 and TRP1 genes are in use for selection of the YAC and the ile and thr mutations are not characterized. We chose the ADE2 gene and inserted it into the fragmentation vectors pBP108 and pBP109, containing the Alu target sequence in opposite orientations (Pavan et al. 1991). The resulting plasmids pBP108/ADE and pBP109/ADE (Fig. 1A) were tested for their ability to create fragmentation panels on YACs from different parts of the human genome (see Table 1 and below). As a result of the fragmentation, YACs lose the URA3 gene while acquiring the ADE2 gene, which is selected for on media lacking adenine. Replica plating to media lacking uracil showed that the majority of the transformants (80%–97%) had the correct Ade+Ura− phenotype, indicating a high recombination rate at the Alu repeats of the YAC. The remaining colonies had not become uracil dependent, which might be explained by integration of the pBP108(109)/ADE vectors elsewhere in the genome, by maintenance as a circular plasmid, or by the presence in some of the cells of two YACs, one original and one fragmented. Pulsed-field gel electrophoresis (PFGE) analysis of a fragmentation panel derived from YAC 939_h_7, showed that all Ade+Ura− colonies contained correctly retrofitted YACs, ranging in size from ∼150 to 2300 kb. Figure 2 shows a selection of fragmented YACs, ordered by size. The panel was analyzed with several rare-cutting restriction endonucleases and no inconsistencies were found (Van de Vosse et al. 1997).
Figure 1.
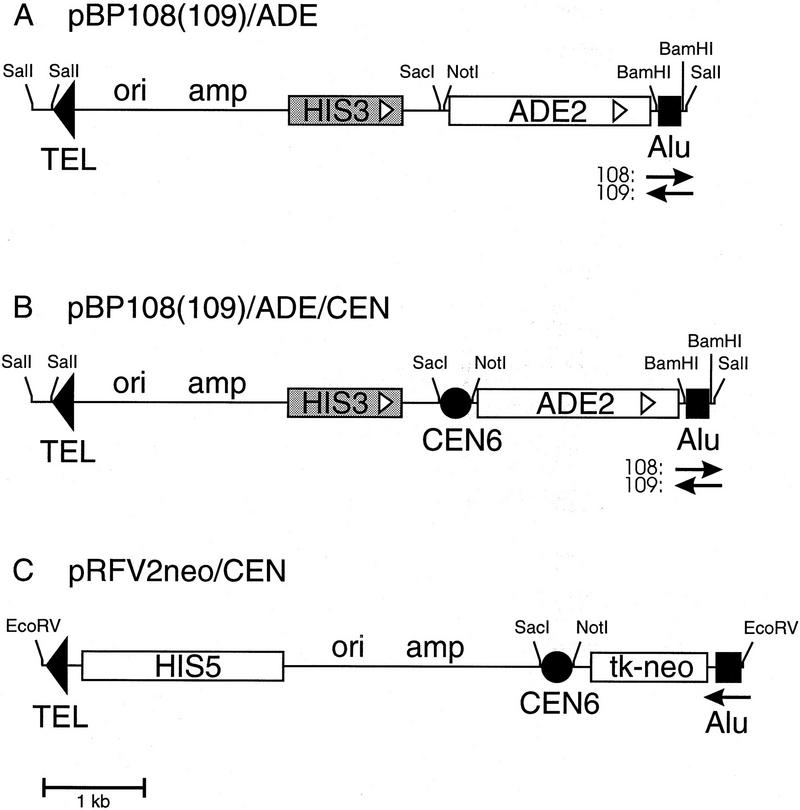
Schematic representation of the new fragmentation vectors, linearized between the Alu and telomere (TEL) fragments. (CEN6) Centromere from yeast chromosome VI; (tk–neo) thymidine kinase promoter coupled to the neomycin gene conferring G418 resistance to mammalian cells. Relevant restriction sites are indicated. The open arrowheads within HIS3 and ADE2 boxes indicate the transcriptional direction.
Table 1.
YAC Fragmentation Frequencies of pBP108 (109)/ADE, pBP108 (109)/ADE/CEN, and pRFV2neo/CEN
| Vector | YAC | No. of coloniesa | No. of correct phenotype/no. testedb | No. truncated YACs/ no. tested by PFGE |
|---|---|---|---|---|
| A pBP108/ADE | 939_h_7 | 200–300 | 29/30 (97%) | 29/29 (100%) |
| pBP109/ADE | 939_h_7 | 200–300 | 31/37 (84%) | 31/31 (100%) |
| pBP108/ADE | 960_a_5 | 21 | 20/21 (95%) | 20/20 (100%) |
| pBP109/ADE | 960_a_5 | 43 | 21/23 (91%) | 21/21 (100%) |
| B pBP108/ADE/CEN | 766_a_12 | 408c | 47/100 (47%) | 44/44 (100%) |
| pBP109/ADE/CEN | 766_a_12 | 481c | 38/100 (38%) | 32/32 (100%) |
| pBP108/ADE/CEN | 932_e_9 | 831c | 26/100 (26%) | 23/24 (96%) |
| pBP109/ADE/CEN | 932_e_9 | 940c | 22/100 (22%) | 08/09 (89%) |
| C pRFV2neo/CEN | 766_a_12 | 600–700 | 53/149 (36%) | 06/06 (100%) |
Total number of colonies found on different spreads of the same transformation mixture per 5 μg of vector.
Number of correct phenotype: Ade+ Ura− (A); Ade+ Trp− (B); and His+ Trp− (C). Number tested: Number of colonies transferred manually or by replica plating to a plate selecting for the targeted YAC arm; upon loss of the original YAC arm, colonies become Ura− (A) or Trp− (B and C).
16%–38% of the colonies were red; these were not analyzed further.
Figure 2.
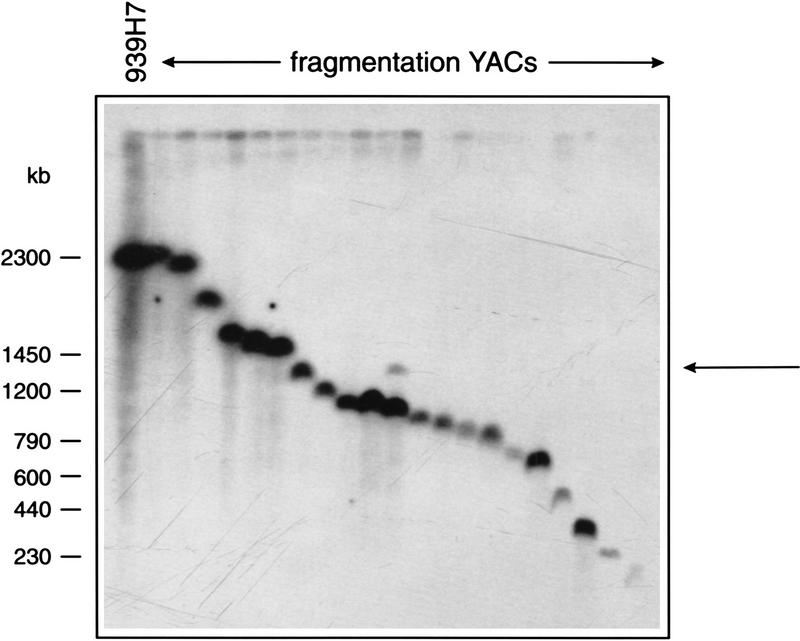
YAC fragmentation panel of YAC 939_h_7 made using pBP108/ADE2 and pBP109/ADE2. Fragmentation YACs were ordered by size (deduced from an initial gel) and separated on a PFGE gel, which was blotted and probed with a 270-bp Alu fragment (isolated from pBP108 by BamHI digestion). The conditions used were 0.6% SeaKem Gold agarose, 1× PFG buffer (0.045 m Tris, 0.045 m boric acid, 0.2 mm EDTA at pH 8.3), for 44 hr, at 100 V with 20–500 sec pulse time. The extra band indicated by the arrow may be the result of an impure YAC clone.
Fragmentation vectors for centromeric fragmentation of YACs have been described (Pavan et al. 1991; Larionov et al. 1996). However, as these vectors carry the HIS3 marker, they do not allow direct fragmentation of AB1380 YACs. For the construction of more convenient CEN fragmentation vectors, we cloned CEN6 into pBP108/ADE and pBP109/ADE (see above) as well as into pRFV2neo (Emanuel et al. 1995). The resulting CEN plasmids (Fig. 1B,C) were used to fragment two overlapping YACs, 766_a_12 (1050 kb) and 932_e_9 (1400 kb). The results indicate that both pBP108/ADE/CEN and pBP109/ADE/CEN truncate YACs efficiently: ∼25%–40% of the colonies had lost the TRP1 gene from the centromeric YAC arm and had become shorter, judged by PFGE and Southern blotting (Table 1). pRFV2neo/CEN was also used successfully: All six fragmentation YACs tested had become shorter and hybridized to a CEN6 probe as well as to an Alu probe (data not shown). If the ultimate aim of fragmentation is transferring the YAC into mammalian cells, this vector directly supplies a marker (neoR) for selection.
Compared to the HIS5 and LYS2 fragmentation vectors for the noncentromeric arm of YACs, pBP108/ADE2 and pBP109/ADE2 give a high percentage of correctly fragmented YACs [80%–97% vs. 21%–46% (Emanuel et al. 1995) and 27%–49% (Lewis et al. 1992)]. However, no direct comparisons have been performed. Both vectors have been used successfully to obtain a detailed rare-cutter restriction map of a 2300-kb region on the X chromosome (Van de Vosse et al. 1997). Two other groups have also obtained truncated YACs using these plasmids (G. Williams and M. Cruts, unpubl.).
We believe that these vectors are a valuable addition to the existing sets of fragmentation vectors, allowing direct and efficient truncation from both ends of YACs directly in AB1380, thus alleviating the need of an additional time-consuming step, that is, swapping the YACs through meiosis or kar1 transfer to another genetic background. All vectors are available upon a mailed request to J. den Dunnen.
METHODS
Vector Constructions
Using partially filled-in fragments, a 2.0-kb BglII fragment from pASZ11 (Stotz and Lindner 1990), containing the ADE2 gene, was cloned into the XbaI site of pBP108 and pBP109 (Pavan et al. 1991). Both ADE2 transcriptional orientations were obtained. The initial fragmentation experiments showed that the vectors in which ADE2 transcription is in the direction of the Alu fragment gave slightly better results and were therefore used in the fragmentation experiments and for the subsequent addition of CEN6. For the construction of pBP108(109)/ADE/CEN, CEN6 was amplified by PCR from pLA433 (Panzeri and Philippsen 1982), using the following primers: 5′-AAGGAAAAAAGCGGCCGCGTTATGGAACCTGTCG-3′ and 5′-GGAAGGAAGAGCTCTTTCGTGTCGGTCGTCC-3′ (NotI and SacI sites are underlined), and cloned between the NotI and SacI sites of pBP108(109)/ADE and pRFV2neo (Emanuel et al. 1995).
Yeast Transformation and Analysis of Fragmented YACs
The Alkali Cation Kit of BIO101 was used for all fragmentation experiments. Before transformation, vectors were linearized with SalI (the pBP derivatives) or EcoRV (pRFV2neo/CEN). YACs were grown in SD+ medium [synthetic dextrose media containing 8 grams/liter of Bacto yeast nitrogen base (YNB), 20 grams/liter of dextrose, 55 mg/liter of adenine, 55 mg/liter of tyrosine, 14 grams/liter of Bacto casaminoacids]. After transformation, cells were plated on SD− media [SD without casaminoacids, but supplemented with arginine (20 mg/liter), methionine (20 mg/liter), isoleucine (30 mg/liter), phenylalanine (50 mg/liter), leucine (60 mg/liter), valine (150 mg/liter), histidine (20 mg/liter), tryptophan (20 mg/liter), uracil (20 mg/liter), lysine (30 mg/liter)]. For the pBP vectors, cells were spread on SD− plates lacking adenine and tryptophan (when truncating from the noncentromeric end of the YAC) or adenine and uracil (truncating from the centromeric end), whereas for pRFV2neo/CEN SD− plates lacking histidine and uracil were used. After 3–4 days at 30°C, colonies were either manually transferred or replica plated to SD− plates lacking uracil or tryptophan (for fragmentations of the URA3 and TRP1 arm, respectively) to screen out the false-positive colonies resulting from integration in the yeast genome or circular maintenance of the fragmentation plasmid. Agarose plugs were made from colonies with the correct genotype and subsequently analysed by pulsed field gel electrophoresis.
Acknowledgments
We thank Dr. S. Pestka for kindly providing pRFV2neo and Dr. P. Philippsen for pLA433.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ddunnen@ruly46.medfac.leidenuniv.nl; FAX +31(0)71-527 60 75.
REFERENCES
- Albertsen HM, Abderrahim H, Cann HM, Dausset J, Le Paslier D, Cohen D. Construction and characterization of a yeast artificial chromosome library containing seven haploid human genome equivalents. Proc Natl Acad Sci. 1990;87:4256–4260. doi: 10.1073/pnas.87.11.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Villasente A, Tyler-Smith C. Construction of yeast artificial chromosome libraries with large inserts using fractionation by pulsed-field gel electrophoresis. Nucleic Acids Res. 1990;17:3425–3433. doi: 10.1093/nar/17.9.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov IM, Rigault P, Le Gall I, Bellanné-Chantelot C, Billault A, Guillou S, Soularue P, Guasconi G, Poullier E, Gros I, Belova M, Lander ES, Weissenbach J, Le Paslier D. A YAC contig map of the human genome. Nature. 1995;377:175–298. doi: 10.1038/377175a0. [DOI] [PubMed] [Google Scholar]
- Emanuel SL, Cook JR, O’Rear J, Rothstein R, Pestka S. New vectors for manipulation and selection of functional yeast artificial chromosomes (YACs) containing human DNA inserts. Gene. 1995;55:167–174. doi: 10.1016/0378-1119(94)00852-j. [DOI] [PubMed] [Google Scholar]
- Larin Z, Monaco AP, Lehrach H. Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc Natl Acad Sci. 1991;88:4123–4127. doi: 10.1073/pnas.88.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BC, Shah NP, Braun BS, Denny CT. Creation of a yeast artificial chromosome fragmentation vector based on lysine-2. Genet Anal Tech Appl. 1992;9:86–90. doi: 10.1016/1050-3862(92)90003-n. [DOI] [PubMed] [Google Scholar]
- Panzeri L, Philippsen P. Centromeric DNA from chromosome VI in Saccharomyces cerevisiae strains. EMBO J. 1982;1:1605–1611. doi: 10.1002/j.1460-2075.1982.tb01362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan WJ, Hieter P, Reeves RH. Generation of deletion derivatives by targeted transformation of human-derived yeast artificial chromosomes. Proc Natl Acad Sci. 1990;87:1300–1304. doi: 10.1073/pnas.87.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan WJ, Hieter P, Sears D, Burkhoff A, Reeves RH. High-efficiency yeast artificial chromosome fragmentation vectors. Gene. 1991;106:125–127. doi: 10.1016/0378-1119(91)90576-w. [DOI] [PubMed] [Google Scholar]
- Spencer F, Hugerat Y, Simchen G, Hurko O, Connelly C, Hieter P. Yeast kar1 mutants provide an effective method for YAC transfer to new hosts. Genomics. 1994;22:118–126. doi: 10.1006/geno.1994.1352. [DOI] [PubMed] [Google Scholar]
- Stotz A, Lindner P. The ADE2 gene from Saccharomyces cerevisiae: Sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- Van de Vosse, E., P. van der Bent, J.J. Heus, G.J.B. van Ommen, and J.T. den Dunnen. 1997. High resolution mapping by YAC fragmentation of a 2.5 Mb Xp22 region containing the human RS, KFSD, and CLS disease genes. Mamm. Genome (in press). [DOI] [PubMed]


