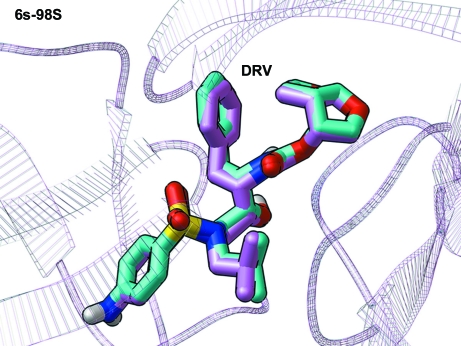
Crystal structures of the 6s-98S FIV protease chimera with darunavir and lopinavir bound have been determined at 1.7 and 1.8 Å resolution, respectively.
Keywords: chimeric FIV/HIV protease, feline immunodeficiency virus, darunavir, lopinavir
Abstract
A chimeric feline immunodeficiency virus (FIV) protease (PR) has been engineered that supports infectivity but confers sensitivity to the human immunodeficiency virus (HIV) PR inhibitors darunavir (DRV) and lopinavir (LPV). The 6s-98S PR has five replacements mimicking homologous residues in HIV PR and a sixth which mutated from Pro to Ser during selection. Crystal structures of the 6s-98S FIV PR chimera with DRV and LPV bound have been determined at 1.7 and 1.8 Å resolution, respectively. The structures reveal the role of a flexible 90s loop and residue 98 in supporting Gag processing and infectivity and the roles of residue 37 in the active site and residues 55, 57 and 59 in the flap in conferring the ability to specifically recognize HIV PR drugs. Specifically, Ile37Val preserves tertiary structure but prevents steric clashes with DRV and LPV. Asn55Met and Val59Ile induce a distinct kink in the flap and a new hydrogen bond to DRV. Ile98Pro→Ser and Pro100Asn increase 90s loop flexibility, Gln99Val contributes hydrophobic contacts to DRV and LPV, and Pro100Asn forms compensatory hydrogen bonds. The chimeric PR exhibits a comparable number of hydrogen bonds, electrostatic interactions and hydrophobic contacts with DRV and LPV as in the corresponding HIV PR complexes, consistent with IC50 values in the nanomolar range.
1. Introduction
The evolution of drug resistance is a fundamental challenge in the development of therapeutics to treat HIV/AIDS. At present there are over 25 FDA-approved drugs to treat AIDS, including nine which target the viral protease (PR; Anderson et al., 2009 ▶; Pokorná et al., 2009 ▶) and 14 which target reverse transcriptase (RT). Highly active anti-retroviral therapy (HAART) employs combinations of these drugs and has been very effective (Wensing et al., 2010 ▶), yet resistance continues to evolve. In HIV PR over 70 mutations at 38 positions of the 99-residue polypeptide have been observed (Bennett et al., 2009 ▶; Johnson et al., 2008 ▶, 2010 ▶). A strategy to understand mechanisms of inhibitor-resistance development is to employ FIV, a lentivirus with structural homology and pathogenetic similarity to HIV, as a model system (Elder et al., 2008 ▶, 2010 ▶). In particular, FIV PR is a 116-residue proteinase with 27 identical residues and 48% homology to HIV PR. Replacement of residues in FIV PR with homologous residues from HIV PR to create chimeric enzymes provides a system in which inhibitor specificity and pathways of drug-resistance development can be probed. Furthermore, it provides a format in which to develop broad-based inhibitors that are less sensitive to the evolution of resistance, e.g. TL-3, which has high affinity for both HIV and FIV PRs (K i values of 1.5 and 41 nM, respectively; Lee et al., 1998 ▶, 1999 ▶).
Recently, a chimeric FIV in which HIV residues were introduced at the equivalent positions in FIV PR has been constructed that is infectious in tissue culture while retaining sensitivity to the HIV PR-specific inhibitors darunavir (DRV) and lopinavir (LPV) (Lin et al., 2010 ▶). 12 replacements of eight residues in the active site were individually assessed for infectivity and for their ability to process Gag cleavage sites in a cell-based FIV Gag-Pol expression system. Combinations of these replacements were assessed for infectivity by transfection into glial cells. A mutant carrying six HIV PR residues gained significant activity, which continued to increase upon infection of fresh glial cells with the progeny virus. Analysis of the PR sequences showed that one of the six mutated residues, Ile98Pro, was further mutated to Ile98His or Ile98Ser. These chimeras, termed 6s-98H and 6s-98S, displayed acute sensitivity to DRV and LPV in the FIV Gag processing and infectivity assays, whereas wild-type FIV was not affected by these inhibitors. Thus, a chimeric FIV has been selected that is both infectious and sensitive to HIV PR inhibitors. These chimeric viruses can be used to monitor the evolution of drug resistance in FIV versus HIV, to better understand the molecular basis of PR-inhibitor specificity and to develop broad-based inhibitors. The chimeric FIV supports the development of less expensive and hazardous animal models for evaluating new inhibitors of drug-resistant HIV and the evolution of viral drug resistance in situ.
The originally designed 6s chimera contained six FIV→HIV replacements (FIV PR residue numbers), Ile37Val, Asn55Met, Val59Ile, Ile98Pro, Gln99Val and Pro100Asn, while the new, more infectious 6s-98S FIV PR contains the additional mutation Ile98Pro→Ile98Ser that arose during tissue-culture passage in the absence of selective drug pressure (Lin et al., 2010 ▶). In this paper, we describe the crystal structures of 6s-98S FIV PR bound to DRV and LPV and analyze the role of the HIV residues in conferring sensitivity to DRV and LPV without abolishing the Gag processing activity required for viral infectivity. The DRV-bound structure is also analyzed in terms of five mutants that do not support infectivity (Lin et al., 2010 ▶). Four additional mutants present in a non-infectious 12s FIV PR (Heaslet et al., 2007 ▶) are analyzed. The role of the mutants in affecting interactions with DRV and LPV is correlated with binding constants and the structures are compared with the corresponding wild-type HIV PR complexes (Tie et al., 2004 ▶; Stoll et al., 2002 ▶).
2. Materials and methods
2.1. Cloning, expression and purification
The 6s-98S chimeric PR gene (I37V/N55M/V59I/I98S/Q99V/P100N) was amplified by PCR from the viral genome and cloned into pET21a expression vector (EMD Chemicals Inc.) for expression using NdeI and HindIII cloning sites. Recombinant expression plasmid was transformed into the Rosetta 2(DE3)pLysS (Novagen Inc.) strain of Escherichia coli and PR expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at an OD600 of 0.8 for 4 h at 310 K. The inclusion bodies containing PR were isolated from the cell lysate by centrifugation at 8000 rev min−1 for 1 h and further purified by several washes with water. The purified inclusion bodies were solubilized in 7.5 M urea containing 20 mM Tris, 5 mM EDTA pH 8, and insoluble material was removed by centrifugation at 8000 rev min−1 followed by filtration through a 0.45 µm membrane. The clarified solution was subsequently purified by ion-exchange chromatography as described previously (Heaslet et al., 2007 ▶). Briefly, the solution was mixed with Whatman DE52 anion exchanger, incubated for 30 min and filtered. The filtered solution containing PR was then applied onto an RQ anion-exchange column equilibrated with 7.5 M urea containing 20 mM Tris, 5 mM EDTA pH 8. The column flowthrough was dialyzed against 25 mM phosphate buffer pH 7.2 containing 25 mM NaCl and 0.1% 2-mercaptoethanol overnight, followed by dialysis against 10 mM sodium acetate buffer pH 5.2, 0.1% 2-mercaptoethanol for 2 h. The refolded PR was clarified by centrifugation and filtration. The sample was then concentrated to 2–5 mg ml−1 for crystallization using an Amicon Ultracel-10K centrifugal device. The purified PR was separated using SDS–PAGE and verified by Western blotting.
2.2. Crystallization and structure determination
Lopinavir was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; darunavir was obtained from Tibotec Inc. 6s-98S PR (3.5 mg ml−1) was treated with saturating concentrations of either LPV or DRV by addition of 1/10th volume of the drug (final concentration of 3.0 mg ml−1) in DMSO (final concentration of 10% DMSO). After 15 min on ice, the mixture was centrifuged to remove the precipitated drug and the PR–drug complexes were then mixed in a 1:1 ratio with 0.5 M KSCN, 0.1 M MES buffer pH 6.1 in seated drops at 277 K. Long trigonal rod-shaped crystals formed after 24–72 h at 277 K. Crystals were frozen at 100 K using reservoir solution supplemented with 30% glycerol as a cryoprotectant. Data were collected on Stanford Synchrotron Radiation Lightsource beamline 11-1 (Soltis et al., 2008 ▶), integrated with MOSFLM (Leslie, 1999 ▶) and scaled with SCALA (Winn et al., 2011 ▶). The structures were solved by molecular replacement using MOLREP (Winn et al., 2011 ▶) with the 12s FIV PR structure (Heaslet et al., 2007 ▶) as a search model. Models were fitted to electron-density maps with MIFit (McRee, 1999 ▶) and were refined with REFMAC (Murshudov et al., 2011 ▶). Data-collection and refinement statistics are summarized in Table S11 and unbiased electron density for DRV and LPV is shown in Figs. S1 and S21. DRV and LPV are disordered about the local twofold axis of the PR dimer, with 0.50 occupancy for each orientation. Coordinates and structure factors have been deposited in the PDB with accession codes 3ogp for the DRV complex and 3ogq for the LPV complex. For the analysis in Tables S2 and S31, computed H-atom positions were added to the PR crystal structures using WHAT_IF (Vriend, 1990 ▶) and the protonation states of DRV and LPV were determined using the open-source software Avogadro v.1 (http://avogadro.openmolecules.net/). Distance and angle measurements were made with PMV (Sanner, 1999 ▶). Most figures were rendered with PyMOL (DeLano Scientific LLC, San Carlos, California, USA), but Figs. 3(b), 3(c) and 8 were created with PMV 1.5.4 (Sanner, 1999 ▶).
2.3. Docking calculations
The docking calculations presented in Table 2 were performed with the new program AutoDock Vina (Trott & Olson, 2010 ▶). The computed H-atom positions were added to the protease targets using WHAT_IF (Vriend, 1990 ▶) and the protonation states of DRV and LPV were determined using the open-source software Avogadro v.1 (http://avogadro.openmolecules.net/). Nonpolar H atoms were merged onto their respective heavy atoms for both the ligands and the targets using AutoDockTools (Sanner, 1999 ▶; Morris et al., 2009 ▶) prior to performing these docking calculations. All protease targets were superimposed with PyMOL onto the same reference structure (using their Cα atoms) prior to generating their pdbqt docking input files. The grid box was centered between the backbone amino groups of Ile50 and Ile50′ (for the HIV protease targets) and the size of the grid was 31.125 × 25.875 × 31.125 Å. Default parameters were used for the AutoDock Vina calculations and the best energy mode was selected from each docking job against a particular target for analysis.
3. Results and discussion
The crystal structures of 6s-98S FIV PR with DRV and LPV bound were determined to 1.7 and 1.8 Å resolution, respectively (Table S1, Figs. S1 and S21). To evaluate the effects of the 6s-98S replacements and their impact on Gag processing and DRV and LPV binding, the crystal structures were superposed and docking calculations were performed. Structural analysis included wild-type FIV PR with TL-3 bound (Wlodawer et al., 1995 ▶; Li et al., 2000 ▶; PDB entry 1b11), 6s-98S FIV PR with DRV bound (this paper; PDB entry 3ogp), 12s FIV PR with TL-3 bound (Heaslet et al., 2007 ▶; PDB entry 2hah) and wild-type HIV PR with DRV bound (Tie et al., 2004 ▶; PDB entry 2ien) (Table 1 ▶). Superposition based on secondary-structure alignment (Winn et al., 2011 ▶) over 174 Cα positions of the dimeric PRs resulted in r.m.s. deviations of 1.07 Å for 1b11 onto 2ien, 1.24 Å for 3ogp onto 2ien and 1.07 Å for 2hah onto 2ien. Analysis also included superposition of 6s-98S FIV PR with LPV bound (this paper; PDB entry 3ogq) versus wild-type HIV PR with LPV bound (Stoll et al., 2002 ▶; PDB entry 1mui; r.m.s. deviation of 1.25 Å).
Table 1. Amino-acid replacements in chimeric FIV PR versus HIV-1 PR.
| FIV segment | Residue | FIV wild type | FIV 6s-98S† | FIV 12s‡ | HIV-1 wild type§ |
|---|---|---|---|---|---|
| Active core | 37 | Ile | Val | Val | Val32 |
| Flap | 55 | Asn | Met | Met | Met46 |
| Flap | 56 | Met | Met¶ | Ile | Ile47 |
| Flap | 57 | Ile | Ile | Gly | Gly48 |
| Flap | 59 | Val | Ile | Ile | Ile50 |
| Flap | 62 | Gly | Gly | Phe | Phe53 |
| Flap | 63 | Lys | Lys | Ile | Ile54 |
| 90s loop | 97 | Leu | Leu¶ | Thr | Thr80 |
| 90s loop | 98 | Ile | Pro→Ser, His†† | Pro | Pro81 |
| 90s loop | 99 | Gln | Val | Val | Val82 |
| 90s loop | 100 | Pro | Asn | Asn | Asn83 |
| 90s loop | 101 | Leu | Leu | Ile | Ile84 |
| Structure‡‡ | 1b11 | 3ogp | 2hah | 2ien | |
| Inhibitor | TL-3 | DRV | TL-3 | DRV |
Supports the rate and specificity of Gag cleavage for infectious virus.
Does not support the rate and specificity of Gag cleavage for infectious virus.
Autolysis-resistant construct: Gln7Lys, Leu33Ile, Leu63Ile, Cys67Ala and Cys95Ala (Tie et al., 2004 ▶).
Single replacements Met56(Ile,Ala,Leu,Val) and Leu97Thr not tolerated for infectious virus.
The Ile98Pro replacement further mutated to Ser or His during selection ex vivo (Lin et al., 2010 ▶).
PDB code.
3.1. FIV PR replacements
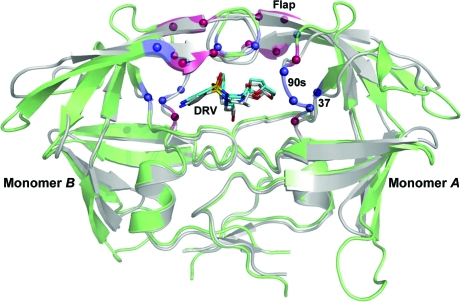
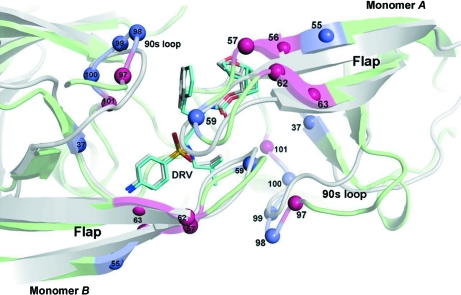
Figs. 1 ▶ and 2 ▶ show the distribution of the 6s-98S replacements in FIV PR with respect to the HIV PR fold. The 12 replacements per dimer are concentrated around the active site and are distributed within three regions: the catalytic core (Ile37Val), the flaps (Asn55Met and Val59Ile) and the 90s loop (Ile98Ser, Gln99Val and Pro100Asn) (Lin et al., 2010 ▶). Analysis of the crystal structures at these residues addresses their roles in Gag polyprotein cleavage and HIV–inhibitor interactions.
Figure 1.
The structure of the 6s-98S FIV PR dimer (green) with darunavir (DRV) bound (C atoms in cyan) superposed onto the structure of HIV PR with DRV bound (PDB code 2ien; gray). Cα positions of residues mutated in the 6s-98S chimera are indicated in blue; six additional residues mutated in the 12s chimera of FIV PR are indicated in red (Table 1 ▶). Mutations are in three regions of each monomer: the catalytic core (residue 37), the flaps and the 90s loop. Monomers A and B are labeled.
Figure 2.
Close-up view from the top of the superposition in Fig. 1 ▶ showing residue numbers for all mutated FIV PR residues in the 6s-98S and 12s chimeras. Residue identities are given in Table 1 ▶. The same color legend as in Fig. 1 ▶ applies to this image.
3.2. Ile37Val
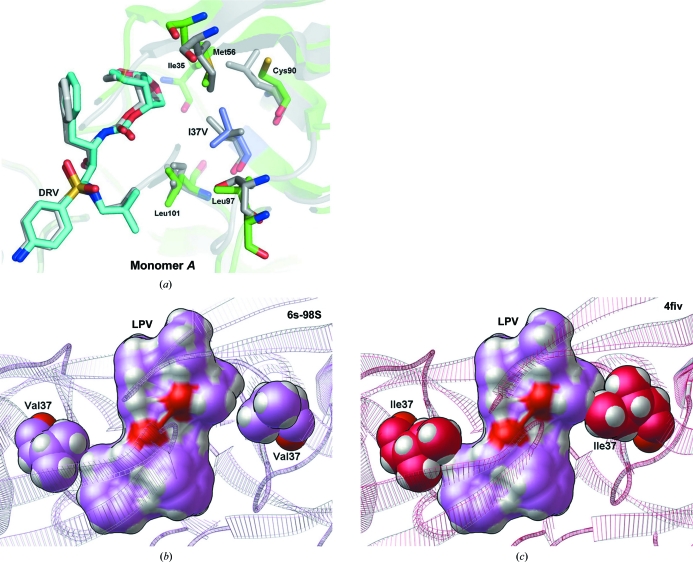
Ile37 and Val37 in wild-type and 6s-98S FIV PRs superpose so that Ile37Val results in loss of only a methyl group. The Val side chain can accommodate the P2-site residues of the MA–CA and NC–p2 cleavage sites (QAY-PIQ and NQM-QQA, respectively), accounting for the infectivity of the individual Ile37Val and Ile37Ala mutants (Lin et al., 2010 ▶). Contacts of Ile37 with Ile35, Met56, Cys90, Leu97 and Leu101 also occur with Val37 (Fig. 3 ▶ a), so that tertiary-structure packing is retained. With respect to HIV PR, loss of the terminal methyl group from Ile/Val permits favorable contacts with DRV (Fig. 3 ▶ a) and with LPV (Fig. 3 ▶ b). Although this Ile→Val substitution is required to prevent a steric clash with LPV (Fig. 3 ▶ c), the wild-type Ile did not display a significant steric clash with DRV when the FIV 6s-98S PR crystal structure was superimposed onto PDB entries 1b11 (Li et al., 2000 ▶), 2fiv (Laco et al., 1997 ▶), 3fiv (Laco et al., 1997 ▶) or 4fiv (Kervinen et al., 1998 ▶) by their Cα atoms. Hence, the structure at Ile37Val is consistent with its Gag cleavage activity, it permits inhibition by DRV and it is required for inhibition by LPV.
Figure 3.
The environment of the Ile37Val replacement. (a) The Val37 side chain preserves packing with five surrounding hydrophobic residues and permits favorable contacts with DRV. Only one orientation of the twofold-disordered inhibitor is shown. The superposition shows the structures of 6s-98S FIV PR (green) with DRV bound (C atoms in cyan) and HIV PR (gray) with DRV bound (C atoms in gray). Residues mutated in the 6s-98S chimera are shown in blue; only FIV 6s-98S residues are labeled. (b) Val37 in FIV 6s-98S does not display a steric clash with the crystallographic conformation of lopinavir. The top view of the ribbon model for FIV 6s-8S is shown as semi-transparent lines and Val37 is shown in CPK (spheres with light magenta C atoms). The crystallographic conformation of LPV is shown as a solvent-excluded surface with light magenta C atoms with the CPK representation for LPV displayed beneath it. (c) Ile37 in wild-type FIV crystal structures causes a steric clash with the superposed conformation of LPV. The top view of the ribbon model for wild-type FIV from PDB entry 4fiv (Kervinen et al., 1998 ▶) is shown as semi-transparent lines and Ile37 is shown in CPK with hot pink C atoms. The conformation of LPV from the presented FIV 6s-98S crystal structure was superposed onto 4fiv using the Cα atoms of PR and is shown as a solvent-excluded surface with light magenta C atoms (with the CPK representation for LPV displayed beneath it). Both Ile37 residues in the dimer clash with this conformation of LPV. The conformations of Ile37 in the other three crystal structures of wild-type FIV PR examined also cause a steric clash with LPV. Only one orientation of LPV is shown, but this trend applies to both orientations.
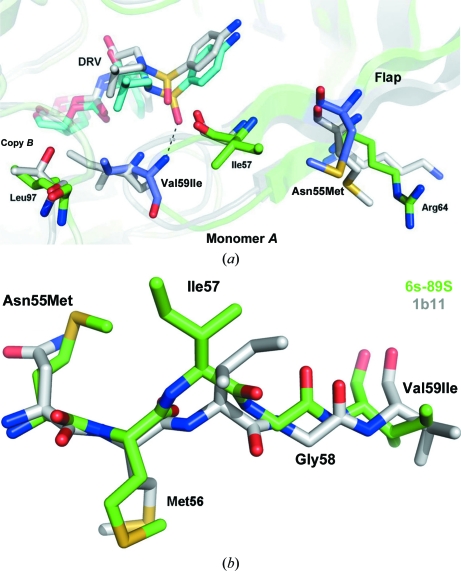
3.3. Asn55Met and Val59Ile
In FIV PR the side chains of residues 55, 57 and 64 project outward from the surface of the flap (Fig. 4 ▶ a); in wild-type PR an interaction between Asn55 and Arg64 stabilizes the conformation of the antiparallel β-strands. In the Asn55Met mutant the methionine side chain retains contacts with aliphatic atoms of Arg64 but acquires hydrophobic contacts with Ile57; the latter interaction flips the Ile57 side chain (Fig. 4 ▶ b) and introduces a distinct kink in the flap, which is particularly noticeable in monomer A (Fig. 2 ▶). As a consequence, Ile59 at the tip of the flap in monomer A is retracted 1.4 Å, and 1.7 Å relative to HIV PR, allowing the Ile59 amide to hydrogen bond directly to DRV (Fig. 4 ▶ a and Table S21). In contrast, Ile50 in HIV PR interacts with the sulfonamide of DRV only via the bound ‘flap’ H2O molecule. The Ile59 side chain also makes hydrophobic contacts with DRV in both monomers A and B (Table S41). Hence, the Asn55Met replacement works in synergy with other flap residues to promote specific recognition of DRV.
Figure 4.
(a) Top view of the flap of monomer A showing the Asn55Met and Val59Ile replacements and the adjacent FIV PR residues Ile57 and Arg64. Interaction of these residues stabilizes a kinked conformation of the flap, enabling a direct hydrogen bond (dashed line) between Ile59 and the sulfonamide of DRV. Only one orientation of the twofold-disordered inhibitor is shown. The superposition shows the structures of 6s-98S FIV PR (green) with DRV bound (C atoms in cyan) and HIV PR (gray) with DRV bound (C atoms in gray). Residues mutated in the 6s-98S chimera are shown in blue; only FIV residues are labeled. (b) Residues 55–59 in the flap of FIV PR showing the shift in Ile57 in the 6s-98S chimera (green) correlated with the Asn55Met replacement. The wild-type FIV PR structure (PDB entry 1b11) is shown in gray. Hydrophobic interaction between Met55 and Ile57 shifts the Cα atom of Ile57 by 1.7 Å and also inverts the side chain (180° rotation about the χ1 torsion). Consequently, the flap in 6s-98S is kinked and Ile59 is retracted by 1.4 Å, allowing the amide of this residue to hydrogen bond directly to the sulfonamide of DRV (A).
Kinking of the flap, however, does not affect Gag processing. The Asn55Met replacement is on the enzyme surface and the single Val59Ile mutant is tolerated (Lin et al., 2010 ▶) as Ile59 retains a contact with Leu97 from the opposite subunit (Fig. 4 ▶ a). Relative to wild-type FIV PR the monomer B flap is kinked less and Ile59 is shifted 0.7 Å; hence, the flaps exhibit plasticity in positioning Ile59 in the P1 site.
3.4. Ile98Ser, Gln99Val and Pro100Asn
The 90s loop in wild-type FIV PR, Glu93-Asp94-Asn95-Ser96-Leu97-Ile98-Gln99, contains a three-residue insertion (shown in bold) relative to HIV PR based on alignment of the flanking β-strands. The 90s loop bulges out at the site of this insertion and is also shifted away from the flap of the opposite subunit (Figs. 2 ▶ and 5 ▶). The expansion and displacement of the 90s loop relative to HIV PR may allow greater mobility of the flaps and play a role in accommodating the FIV Gag substrate. Salt bridges involving Glu93 and Asp94 in the insertion may also participate in flap opening (Wlodawer et al., 1995 ▶). Replacements at residues 98, 99 and 100 (Table 1 ▶) are therefore considered in terms of 90s loop flexibility.
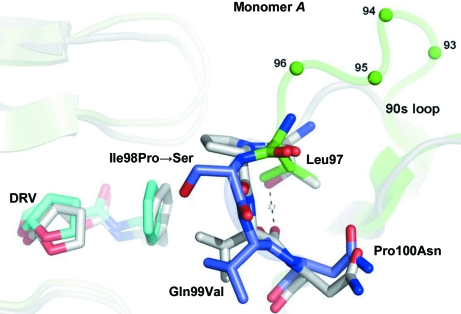
Figure 5.
Relative to HIV PR, the 90s loop of FIV PR contains an insertion at residues 93–95 followed by a reverse turn at residues 97–100. The replacement Ile98Pro further mutated to Ser during ex vivo selection, implicating this residue in interactions with Gag substrates. The replacements Gln99Val and Pro100Asn mimic HIV PR with contacts to DRV and stabilizing hydrogen bonds. The Leu97Thr replacement, which knocks out activity, would stabilize the reverse turn via a hydrogen bond, as in HIV PR (dashed line). Together, these mutants suggest a role for 90s loop flexibility in Gag recognition. The superposition shows the structures of 6s-98S FIV PR (green) with DRV bound (C atoms in cyan) and HIV PR (gray) with DRV bound (C atoms in gray). Residues mutated in the 6s-98S chimera are shown in blue; only FIV residues are labeled.
Individually, the replacements Ile98Pro, Gln99Val and Pro100Asn support cleavage of the MA–CA junction and viral infectivity (Lin et al., 2010 ▶). In the context of the 6s-98S chimera, infectivity greatly increased upon evolution of Pro98 into His or Ser (Lin et al., 2010 ▶); i.e. the more rigid Pro at this position is disfavored (Fig. 5 ▶). Ser at this position also arose in HIV PR in response to the novel inhibitor PL-100 (Dandache et al., 2008 ▶). The orientation of Ser98 (or His98) in the P1 site implies that this residue could interact with Gag substrate. With respect to residue 99, Val replaces Gln and projects into the P1 site. However, a Gln99–Glu15 hydrogen bond is lost, indicating that Gln99Val may also impart greater flexibility. At Pro100Asn, the main-chain atoms superpose and the side chains are oriented away from the active site. Substitution of Pro100 may be compensated by new hydrogen bonds involving Asn100, the amide of Asn39 and the side chain of Asp42. The 90s loop replacements in 6s-98S FIV PR, together with the three-residue insertion, support a requirement for greater flexibility in this loop relative to HIV PR and a role for it in interacting with Gag.
3.5. 12s FIV PR and loss-of-function mutants
A chimeric FIV PR with 12 substitutions per monomer (i.e. 24 per dimer) mimicking HIV PR can process Gag-Pol polyprotein ex vivo, but does not support the production of infectious virus owing to differences in the order and rate of the cleavage reactions (Lin et al., 2006 ▶, 2010 ▶). The 12s chimera contains the 6s-98S substitutions (except that it contains Ile98Pro) together with Met56Ile, Ile57Gly, Gly62Phe, Lys63Ile, Leu97Thr and Leu101Ile; i.e. four additional replacements in the flap and two in the 90s loop (Figs. 1 ▶ and 2 ▶). The two types of substitutions, those which support infectivity and those which do not, can be compared in the 6s-98S and 12s FIV PR structures (Table 1 ▶).
Met56 to Ile, Ala, Leu and Val as individual mutations do not produce infectious viruses (Lin et al., 2010 ▶). Met56 has multiple hydrophobic packing contacts in the core of the protein (Fig. 3 ▶ a) that stabilize the closed form of the flap when in contact with Gag cleavage sites. Any substitution with a shorter or branched side chain at this position would be destabilizing, consistent with the lack of infectivity for seven chimeras that contain the Met56 mutation (Lin et al., 2010 ▶). Ile57Gly and Gly62Phe were not assayed individually (Lin et al., 2010 ▶), but consideration of the flap structure (Fig. 4 ▶ a) and interactions within 12s PR (Heaslet et al., 2007 ▶) suggest that Ile57Gly and Gly62Phe might form compensatory hydrophobic interactions and be tolerated.
With respect to additional 90s loop mutants in 12s, Thr97 hydrogen bonds across the reverse turn to the carbonyl of Val99, as in HIV PR (Fig. 5 ▶), making the loop less flexible and presumably reducing affinity for FIV Gag. This interaction is consistent with a loss of infectivity in 9s and 12s chimeras containing Leu97Thr as well as in the Leu97Thr mutant itself (Lin et al., 2010 ▶). In the Leu101Ile mutant Ile101 places only one methyl group in the P1 site, potentially reducing the affinity for substrate and contributing to loss of infectivity of the 12s chimera, although this mutant was assayed only in combination with the deleterious Met56Ile and Leu97Thr mutants. Finally, Lys63Ile removes polar interactions with Asp94 and Ser96 in the 90s loop, but it gains a hydrophobic packing contact with Leu92, so that individually this replacement might be tolerated.
3.6. Inhibition by DRV and LPV
The 6s-98S mutations in chimeric FIV PR allow Gag processing to occur with a sufficiently correct order and rate to support viral infectivity while conferring sensitivity to the HIV PR inhibitors DRV and LPV. It is of interest to understand how the mutations increase affinity for HIV PR inhibitors in the context of the FIV protein. In terms of IC50 values, the 6s-98S mutations increase the affinity for DRV from 1927 nM for wild-type FIV PR to 36 nM for the chimera and the affinity for LPV from 3779 to 30 nM (Lin et al., 2010 ▶); i.e. an ∼50-fold to 125-fold increase in the affinity for the two drugs. In HIV PR, DRV and LPV have around tenfold smaller IC50 values of 3 nM (Koh et al., 2003 ▶) and 2 nM (Bulgheroni et al., 2004 ▶), respectively. In contrast, the broad-based inhibitor TL-3 (Lee et al., 1998 ▶, 1999 ▶) has IC50 values of 42, 28 and 6 nM against FIV PR, 6s-98S FIV PR and HIV PR, respectively (Lin et al., 2010 ▶; Bühler et al., 2001 ▶). HIV-like drug sensitivity of the 6s-98S chimera is further apparent in ex vivo infectivity assays, where DRV and LPV concentrations of up to 800 nM have no effect on wild-type FIV, whereas 50 nM DRV abolishes activity of the 6s-98S chimeric virus and 50 nM LPV reduces activity by ∼50% (Lin et al., 2010 ▶). The crystal structures of 6s-98S FIV PR with DRV and LPV bound are shown in Figs. 6 ▶ and 7 ▶, respectively (chemical structures of the inhibitors are shown in Figs. S3 and S41, respectively).
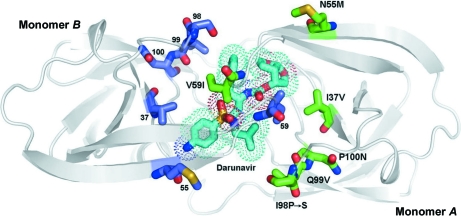
Figure 6.
The structure of 6s-98S FIV PR with DRV bound viewed along the local twofold axis of the dimer. All atoms of the mutated residues are shown (monomer A, C atoms in green; monomer B, C atoms in blue). DRV C atoms are shown in cyan, O atoms in red, N atoms in blue and S atoms in yellow. The dots represent the van der Waals surface for DRV. Monomers A and B are labeled.
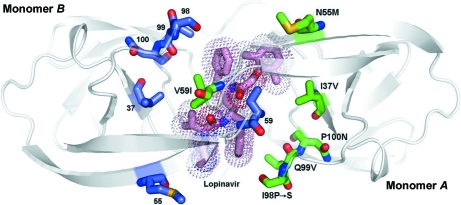
Figure 7.
The structure of 6s-98S FIV PR with LPV bound viewed along the local twofold axis of the dimer. All atoms of the mutated residues are shown (monomer A, C atoms in green; monomer B, C atoms in blue). LPV C atoms are shown in pink, O atoms in red, N atoms in blue and S atoms in yellow. The dots display the van der Waals surface for LPV. Monomers A and B are labeled.
3.7. Darunavir complexes
Regarding the types of contacts with DRV, the number of hydrogen bonds, electrostatic interactions and hydrophobic contacts are comparable in HIV and FIV 6s-98S PRs (Tables S2 and S41, respectively). DRV adopts very similar conformations in the HIV and 6s-98S FIV PR complexes, with an average difference between corresponding atoms of 0.38 Å (Fig. 3 ▶ a). Hydrogen bonds involving main-chain amides and carbonyls, a key determinant of the high affinity of DRV for multi-drug-resistant strains of HIV (Tie et al., 2004 ▶; Koh et al., 2003 ▶), are retained in the 6s-98S complex. As noted, an additional main-chain hydrogen bond from Ile59 to DRV is gained owing to the kinking of the flap in monomer A (Fig. 4 ▶ a). This interaction contrasts with the ‘flap’ H2O-mediated interaction of Ile50 with DRV in HIV PR. A direct hydrogen bond is also observed between Ile50 of HIV PR and the sulfonamide of the novel inhibitor PL-100, which displays a high genetic barrier to resistance (Dandache et al., 2008 ▶). Retraction of one flap is associated with a difference between monomers and five fewer hydrophobic contacts in monomer B versus monomer A of 6s-98S (Table S41). Asymmetry in the dimer may enhance affinity for the non-C 2-symmetric inhibitor DRV (Heaslet et al., 2006 ▶).
In terms of the six chimeric residues, the Ile37Val replacement allows DRV to form favorable hydrophobic interactions with the active site (Fig. 3 ▶ a), but Ile does not display an obvious steric clash with DRV in the four wild-type FIV PR crystal structures examined. Asn55Met and Pro100Asn are not in the active site but contribute to kinking of the flap and positioning of the 90s loop, respectively. Val59Ile retains favorable hydrophobic contacts with DRV and Gln99Val mimics those in HIV (Table S41). A loss of hydrophobic contacts owing to Ile98Ser is compensated by new contacts with DRV involving Met56 and Leu101 (Table S41). Overall, the structure of the DRV complex of 6s-98S is consistent with a 50-fold reduction in IC50 compared with wild-type FIV PR.
Similarly, when DRV was docked against three different wild-type HIV PR crystal structures, the two presented crystal structures of FIV 6s-98S PR and two crystal structures of wild-type FIV PR, the expected trend in affinity values against these three proteases was reproduced by the AutoDock Vina results (Table 2 ▶). When judging the docking results by either (i) the average of the predicted binding energies against the different structures for a particular protease or (ii) the best energy produced against any particular structure from each of the three proteases, DRV docked with the highest affinity against wild-type HIV PR (average energy = −9.28 kcal mol−1; best energy = −9.7 kcal mol−1) followed by FIV 6s-98S (average energy = −8.8 kcal mol−1; best energy = −9.2 kcal mol−1), with wild-type FIV PR displaying the worst affinity (average energy = −8.45 kcal mol−1; best energy = −8.5 kcal mol−1). Although the differences in these binding energy values were smaller than the standard error in the AutoDock Vina scoring function, the fact that the positive-control redocking experiment with DRV produced a perfect reproduction of its crystallographic binding mode (Fig. 8 ▶) supports the observed trend in the docking results.
Table 2. AutoDock Vina results for docking DRV and LPV against wild-type HIV PR, 6s-98S FIV PR and wild-type FIV PR.
Crystal structures of the targets are labeled by their PDB accession codes. 1mui is a lopinavir-bound crystal structure of wild-type HIV protease (Stoll et al., 2002 ▶). 2ien is a darunavir-bound crystal structure of wild-type HIV protease (Tie et al., 2004 ▶). 3kfn is a complex of wild-type HIV protease with TL-3 and allosteric fragment 4D9 (Perryman et al., 2010 ▶). 1b11 is a TL-3-bound crystal structure of wild-type FIV protease (Li et al., 2000 ▶). 4fiv is an Lp-130-bound crystal structure of wild-type FIV protease (Kervinen et al., 1998 ▶). All energies are in kcal mol−1 (1 kcal mol−1 = 4.184 kJ mol−1).
| Target structure | DRV results | LPV results |
|---|---|---|
| Wild-type HIV 1mui | −9.7 | −10.9 |
| Wild-type HIV 2ien | −9.0 | −10.3 |
| Wild-type HIV 3kfn | −9.1 | −10.3 |
| Average of three wild-type HIV crystal structures | −9.28 | −10.5 |
| FIV 6s-98S–DRV crystal | −9.2 | −10.5 |
| FIV 6s-98S–LPV crystal | −8.4 | −10.3 |
| Average of two FIV 6s-98S crystal structures | −8.8 | −10.4 |
| Wild-type FIV 1b11 | −8.4 | −10.3 |
| Wild-type FIV 4fiv | −8.5 | −10.3 |
| Average of two wild-type FIV crystal structures | −8.45 | −10.3 |
Figure 8.
AutoDock Vina calculations produced a docked mode for DRV that superposed perfectly on the crystallographic conformation of FIV 6s-98S PR. The ribbon representation for FIV 6s-98S PR is shown as semi-transparent lines and the crystallographic conformation of DRV is shown as sticks with light magenta C atoms. The docked conformation of DRV is displayed as sticks with cyan C atoms. For both modes, O atoms are red, S atoms are yellow and N atoms are blue. For the docked mode of DRV, the H atoms are shown as white sticks.
3.8. Lopinavir complexes
There are 56 favorable interactions of 6s-98S PR with DRV and 60 with LPV (Tables S3 and S41, respectively), consistent with the similar IC50 values for the two compounds (36 and 30 nM, respectively). The hydrophobic packing of Ile37Val and Val59Ile in 6s-98S is similar to HIV PR and the Ile37Val substitution removes steric clashes with both sides of the drug (Figs. 3 ▶ b and 3 ▶ c). Main-chain hydrogen bonds to LPV in HIV PR become electrostatic interactions in 6s-98S PR. The loss of these hydrogen bonds is compensated by a gain of interactions involving Gly32 for both orientations of LPV. Unlike the DRV complex, a ‘flap’ H2O is bound between LPV carbonyl groups and the amides of Ile59 (Fig. S21). A significant difference is that many fewer hydrophobic contacts are formed owing to the Ile98Ser replacement (Table S41 and Fig. 7 ▶), but these are compensated by additional contacts involving Leu28, Ala33 and Met56 in 6s-98S. The 6s-98S PR–LPV structure displays greater asymmetry between monomers than for HIV PR (r.m.s. of 0.61 Å versus 0.32 Å for monomer A onto monomer B), perhaps accommodating the non-C 2-symmetric drug. The conformation of LPV itself is similar in HIV and 6s-98S PR, but the positions and torsion angles of the phenyl rings in the P1 and P1′ sites differ significantly (average difference of 0.82 Å for all atoms of LPV in the two complexes). Torsional freedom may contribute to the 2.5-fold greater improvement in IC50 for LPV with the chimera compared with DRV.
When LPV was docked against three different wild-type HIV PR crystal structures, the two presented crystal structures of FIV 6s-98S PR and two crystal structures of wild-type FIV PR, the average of the predicted binding energies against the different PR targets also followed the expected trend in relative affinity, but over a narrower range (Table 2 ▶). However, the positive-control redocking experiment for LPV against FIV 6s-98S was not as successful at reproducing its crystallographic binding mode as for DRV (Fig. 8 ▶). For LPV only half of the backbone and half of the side chains superposed onto the crystallographic mode, while the other half of LPV displayed slight rotations away from its crystallographic mode owing to the greater number of ‘active torsions’ (16 in LPV versus 14 in DRV) and the fact that the side chains of LPV are more similar to each other than those of DRV.
3.9. Changes in mobility
Temperature factors were analyzed for the DRV and LPV complexes of the chimera and were compared with those of the TL-3 complex of wild-type FIV PR (Li et al., 2000 ▶) to identify regions of above-average mobility. The FIV PR structures have similarly flexible regions at the N-terminus (residues 5–9) and within a solvent-exposed loop (residues 75–84). However, the other mobile region encompasses residues 53–64 in 6s-98S (i.e. the tip of the flap), whereas in the wild type it includes residues 44–54 (i.e. the preceding loop and β-strand). This suggests that the 6s-98S mutations Asn55Met and Val59Ile impart greater flexibility to the flaps, allowing adaptation to the HIV inhibitors and contributing to the binding affinity for Gag substrate sequences.
4. Conclusions
Crystal structures of 6s-98S FIV PR in complex with DRV and LPV reveal how the six substitutions contribute to retention of wild-type FIV PR activity while conferring on the chimera specific recognition of the HIV inhibitors. Ile37Val retains tertiary structure, while steric clashes with the crystallographic conformations of Ile37 in the four wild-type FIV PR structures examined explains the necessity of this substitution for LPV affinity of the chimeric PR. At the same time, the docking results in Table 2 ▶ reproduce the trend in relative affinity of DRV for wild-type HIV PR versus FIV 6s-98S PR versus wild-type FIV PR. These molecular-modeling results help demonstrate why the six substitutions in FIV 6s-98S are needed to enable inhibition by DRV. The Asn55Met mutant induces new hydrophobic contacts with Ile57, introducing a distinct kink in the flap of monomer A of the dimer. Consequently, the Val59Ile amide interacts directly with the sulfonamide of DRV, in contrast to a ‘flap’ H2O-mediated interaction in HIV PR. The Ile98Ser, Gln99Val and Pro100Asn replacements increase 90s loop flexibility via substitution of one Pro with Asn and mutation of another to Ser; the latter greatly increases infectivity, suggesting a role for residue 98 in interaction with Gag. At the same time, Val99 provides hydrophobic contacts to DRV and LPV, while Asn100 compensates for hydrogen bonds lost from Gln99. In contrast, additional substitutions in the 12s chimeric FIV PR destabilize the closed form of the flaps or confer greater rigidity to the 90s loop, accounting for the loss of infectivity when these additional mutations are present.
Overall, 6s-98S FIV PR acquires a comparable number of hydrogen bonds, electrostatic interactions and hydrophobic contacts to DRV and LPV as in the corresponding HIV PR complexes, consistent with the dramatically increased sensitivity to these inhibitors and IC50 values in the nanomolar range. Accordingly, when LPV was docked to crystal structures of HIV PR and FIV 6s-98S (Table 2 ▶), the average docking energies were comparable. Similarly, main-chain hydrogen bonds to DRV are retained in the 6s-98S chimera in both the crystal structure and docking results. The loss of hydrophobic contacts arising from the Ile98Ser substitution is compensated by contacts involving Met56 and Leu101 with DRV and Leu28, Ala33 and Met56 with LPV. Increased asymmetry in the dimer and greater mobility in the flaps contribute to the capacity of 6s-98S PR to specifically recognize both DRV and LPV while accommodating Gag substrate sequences.
Both DRV and LPV are new-generation FDA-approved drugs for HIV-1 PR with broad-spectrum efficacy against wild type and drug-resistant mutants. Since the substitutions in FIV 6s-98S gave this chimeric PR affinities for DRV and LPV similar to those of wild-type HIV PR, studying the common interactions that are displayed when binding to either PR system provides a guide that can assist future drug discovery and design research. To develop new anti-HIV drugs that display a broad spectrum of activity and higher genetic barriers to the evolution of drug resistance (such as DRV displays), when analyzing the results of virtual screens and when performing hit-to-lead development or lead optimization studies extra weight should be given to the potential significance of candidate compounds that display these common interactions. Specifically, when discovering new hits or when optimizing the affinity of current HIV PR inhibitors, the presented results support the potential utility of compounds that can form hydrogen bonds to the backbone carbonyl O atom of Gly27, the backbone amide of Asp29, the backbone amide of Asp30 and/or the backbone carbonyl O atom of Asp30 (using HIV residue numbers). Of course, the candidate compounds should also form a hydrogen bond to the side chain of the catalytic Asp25. In addition, direct hydrogen bonding from flap residues to the inhibitor with displacement of the ‘flap’ H2O increases affinity. Finally, forming electrostatic interactions or hydrogen bonds with the side chain of Arg8 is useful for affinity and may increase specificity against these viral PRs (Sherman & Tidor, 2008 ▶). The chimeric FIV PR system is being used to screen for new broad-spectrum inhibitors, to select mutants resistant to DRV/LPV and to study the evolution pathway of resistance.
Supplementary Material
PDB reference: 6s-98S FIV protease with darunavir bound, 3ogp
PDB reference: with lopinavir bound, 3ogq
Supplementary material file. DOI: 10.1107/S0907444911011681/hv5180sup1.pdf
Acknowledgments
This research was supported by NIH grants P01 GM083658 (AJO, BET, JHE and CDS) and R01 AI081585 (JHE). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences. Lopinavir was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. This research was carried out to support and extend the FightAIDS@Home project (a part of IBM’s ‘World Community Grid’).
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: HV5180). Services for accessing this material are described at the back of the journal.
References
- Anderson, J., Schiffer, C., Lee, S.-K. & Swanstrom, R. (2009). Handb. Exp. Pharmacol. 189, 85–110. [DOI] [PMC free article] [PubMed]
- Bennett, D. E. et al. (2009). PLoS One, 4, e4724. [DOI] [PMC free article] [PubMed]
- Bühler, B., Lin, Y.-C., Morris, G., Olson, A. J., Wong, C.-H., Richman, D. D., Elder, J. H. & Torbett, B. E. (2001). J. Virol. 75, 9502–9508. [DOI] [PMC free article] [PubMed]
- Bulgheroni, E., Citterio, P., Croce, F., Lo Cicero, M., Viganò, O., Soster, F., Chou, T.-C., Galli, M. & Rusconi, S. (2004). J. Antimicrob. Chemother. 53, 464–468. [DOI] [PubMed]
- Dandache, S., Coburn, C. A., Oliveira, M., Allison, T. J., Holloway, M. K., Wu, J. J., Stranix, B. R., Panchal, C., Wainberg, M. A. & Vacca, J. P. (2008). J. Med. Virol. 80, 2053–2063. [DOI] [PubMed]
- Elder, J. H., Lin, Y.-C., Fink, E. & Grant, C. K. (2010). Curr. HIV Res. 8, 73–80. [DOI] [PMC free article] [PubMed]
- Elder, J. H., Sundstrom, M., de Rozieres, S., de Parseval, A., Grant, C. K. & Lin, Y.-C. (2008). Vet. Immunol. Immunopathol. 123, 3–13. [DOI] [PMC free article] [PubMed]
- Heaslet, H., Kutilek, V., Morris, G. M., Lin, Y.-C., Elder, J. H., Torbett, B. E. & Stout, C. D. (2006). J. Mol. Biol. 356, 967–981. [DOI] [PubMed]
- Heaslet, H., Lin, Y.-C., Tam, K., Torbett, B. E., Elder, J. H. & Stout, C. D. (2007). Retrovirology, 4, 1. [DOI] [PMC free article] [PubMed]
- Johnson, V. A., Brun-Vézinet, F., Clotet, B., Günthard, H. F., Kuritzkes, D. R., Pillay, D., Schapiro, J. M. & Richman, D. D. (2008). Top. HIV Med. 16, 138–145. [PubMed]
- Johnson, V. A., Brun-Vézinet, F., Clotet, B., Günthard, H. F., Kuritzkes, D. R., Pillay, D., Schapiro, J. M. & Richman, D. D. (2010). Top. HIV Med. 18, 156–163. [PubMed]
- Kervinen, J., Lubkowski, J., Zdanov, A., Bhatt, D., Dunn, B. M., Hui, K. Y., Powell, D. J., Kay, J., Wlodawer, A. & Gustchina, A. (1998). Protein Sci. 7, 2314–2323. [DOI] [PMC free article] [PubMed]
- Koh, Y. et al. (2003). Antimicrob. Agents Chemother. 47, 3123–3129. [DOI] [PMC free article] [PubMed]
- Laco, G. S., Schalk-Hihi, C., Lubkowski, J., Morris, G., Zdanov, A., Olson, A. J., Elder, J. H., Wlodawer, A. & Gustchina, A. (1997). Biochemistry, 36, 10696–10708. [DOI] [PubMed]
- Lee, T., Laco, G. S., Torbett, B. E., Fox, H. S., Lerner, D. L., Elder, J. H. & Wong, C.-H. (1998). Proc. Natl Acad. Sci. USA, 95, 939–944. [DOI] [PMC free article] [PubMed]
- Lee, T., Le, V.-D., Lim, D., Lin, Y.-C., Morris, G. M., Wong, A. L., Olson, A. J., Elder, J. H. & Wong, C.-H. (1999). J. Am. Chem. Soc. 121, 1145–1155.
- Leslie, A. G. W. (1999). Acta Cryst. D55, 1696–1702. [DOI] [PubMed]
- Li, M., Morris, G. M., Lee, T., Laco, G. S., Wong, C.-H., Olson, A. J., Elder, J. H., Wlodawer, A. & Gustchina, A. (2000). Proteins, 38, 29–40. [DOI] [PubMed]
- Lin, Y.-C., Brik, A., de Parseval, A., Tam, K., Torbett, B. E., Wong, C.-H. & Elder, J. H. (2006). J. Virol. 80, 7832–7843. [DOI] [PMC free article] [PubMed]
- Lin, Y.-C., Torbett, B. E. & Elder, J. H. (2010). J. Virol. 84, 6799–6809. [DOI] [PMC free article] [PubMed]
- McRee, D. E. (1999). J. Struct. Biol. 125, 156–165. [DOI] [PubMed]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S. & Olson, A. J. (2009). J. Comput. Chem. 30, 2785–2791. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Perryman, A. L., Zhang, Q., Soutter, H. H., Rosenfeld, R., McRee, D. E., Olson, A. J., Elder, J. E. & Stout, C. D. (2010). Chem. Biol. Drug Des. 75, 257–268. [DOI] [PMC free article] [PubMed]
- Pokorná, J., Machala, L., Řezáčová, P. & Konvalinka, J. (2009). Viruses, 1, 1209–1239. [DOI] [PMC free article] [PubMed]
- Sanner, M. F. (1999). J. Mol. Graph. 17, 57–61. [PubMed]
- Sherman, W. & Tidor, B. (2008). Chem. Biol. Drug Des. 71, 387–407. [DOI] [PMC free article] [PubMed]
- Soltis, S. M. et al. (2008). Acta Cryst. D64, 1210–1221. [DOI] [PMC free article] [PubMed]
- Stoll, V., Qin, W., Stewart, K. D., Jakob, C., Park, C., Walter, K., Simmer, R. L., Helfrich, R., Bussiere, D., Kao, J., Kempf, D., Sham, H. L. & Norbeck, D. W. (2002). Bioorg. Med. Chem. 10, 2803–2806. [DOI] [PubMed]
- Tie, Y., Boross, P. I., Wang, Y.-F., Gaddis, L., Hussain, A. K., Leshchenko, S., Ghosh, A. K., Louis, J. M., Harrison, R. W. & Weber, I. T. (2004). J. Mol. Biol. 338, 341–352. [DOI] [PubMed]
- Trott, O. & Olson, A. J. (2010). J. Comput. Chem. 31, 455–461. [DOI] [PMC free article] [PubMed]
- Vriend, G. (1990). J. Mol. Graph. 8, 52–56. [DOI] [PubMed]
- Wensing, A. M., van Maarseveen, N. M. & Nijhuis, M. (2010). Antiviral Res. 85, 59–74. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wlodawer, A., Gustchina, A., Reshetnikova, L., Lubkowski, J., Zdanov, A., Hui, K. Y., Angleton, E. L., Farmerie, W. G., Goodenow, M. M., Bhatt, D., Zhang, L. & Dunn, B. M. (1995). Nature Struct. Biol. 2, 480–488. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: 6s-98S FIV protease with darunavir bound, 3ogp
PDB reference: with lopinavir bound, 3ogq
Supplementary material file. DOI: 10.1107/S0907444911011681/hv5180sup1.pdf