Abstract
The MHC-II locus encodes a cluster of highly polymorphic genes HLA-DR, -DQ, and -DP that are co-expressed in mature B lymphocytes. Two cell lines were established over 30 years ago from a patient diagnosed with acute lymphocytic leukemia. Laz221 represented the leukemic cells of the patient; whereas Laz388 represented the normal B cells of the patient. Whereas Laz388 expressed both HLA-DR and HLA-DQ surface and gene products, Laz221 expressed only HLA-DR genes. The discordant expression of HLA-DR and HLA-DQ genes was due to epigenetic silencing of the HLA-DQ region CTCF-binding insulators that separate the MHC-II subregions by DNA methylation. These epigenetic modifications resulted in the loss of binding of the insulator protein CTCF to the HLA-DQ flanking insulator regions and the MHC-II specific transcription factors to the HLA-DQ promoter regions. These events led to the inability of the HLA-DQ promoter regions to interact with flanking insulators that control HLA-DQ expression. Inhibition of DNA methylation by treatment with 5’deoxyazacytidine reversed each of these changes and restored expression of the HLA-DQ locus. These results highlight the consequence of disrupting an insulator within the MHC-II region and may be a normal developmental mechanism or one used by tumor cells to escape immune surveillance.
Keywords: MHC, gene regulation, transcription, DNA methylation, chromatin
Introduction
The major histocompatibility complex class II (MHC-II) genes encode cell surface α/β heterodimeric glycoproteins that present foreign antigens to antigen specific CD4+T lymphocytes in order to initiate, control, and/or maintain adaptive immune responses 1. There are three MHC-II isotypes HLA-DR, -DQ, and –DP encoded within the MHC-II locus. This process is aided by two other MHC-II associated molecules, HLA-DM and –DO, which are also α/β heterodimers with sequence and structural homology to MHC-II proteins 2, 3. Although the levels of mRNA and protein from each of the above genes differ, all are typically coexpressed in a tissue specific manner. MHC-II genes are expressed constitutively in B-lymphocytes, macrophages, dendritic cells, and thymic epithelia and can also be induced in most other cell types following exposure to interferon-γ (reviewed in 4).
The transcription of MHC-II genes is coordinated by a conserved group of transcriptional factors, RFX (regulatory factor X), CREB (cAMP response element binding protein), and NF-Y (nuclear factor-Y), which bind to highly conserved, promoter-proximal sequences termed the X1, X2, and Y boxes, respectively 4–6. CIITA (class II transactivator), a non-DNA binding transcription factor, is recruited to the DNA bound X and Y box transcription factors and mediates the interaction between co-factors, chromatin remodeling factors, and general transcription machinery 5, 7, 8. CIITA expression is limiting and is highly regulated 9–11. In addition to the promoter proximal elements, distal elements contribute to the expression of the genes in this locus 12–14. The MHC-II locus is punctuated by a series of CCCTC transcription factor (CTCF) binding sites 15. CTCF binding to its target DNA sequences is methylation sensitive 16, 17, and CTCF has been shown to demarcate and insulate regions of regulatory activity within the genome by functioning as an enhancer blocker 18 or by preventing the spread of heterochromatin into active genes 19–21. CTCF has been proposed to function in part by forming the nexus of long-range chromatin loops. Recently, CTCF was found to be required for the expression of all MHC-II genes 15, 22. The binding of CTCF to the insulator site between HLA-DR and HLA-DQ, termed XL9, results in the formation of long-range chromatin loops between XL9 and the promoter proximal regulatory X-Y box regions of the flanking genes, HLA-DRB1 and HLA-DQA1. This interaction was coincident with gene expression and dependent on CTCF and CIITA binding to their respective sites 22. The depletion of CTCF by RNAi, resulted in the loss of HLA-DRB1 and HLA-DQA1 gene expression. These studies predict that disruption of CTCF binding to one of the MHC-II insulator sites may have a significant consequence on MHC-II gene expression.
In 1978, Lazirus et al. established the Laz221 cell line from a patient with acute lymphoblastic leukemia 23. Laz221 cells were similar in phenotype to the ALL cells of the patient and were considered to represent null cells at the time. Later Laz221 cells were determined to be most similar to a pre-B lymphocyte and were thought to arise from these cells 24, 25. The cell line Laz388 was established from the same patient by transforming with Epstein-Barr-virus 23, 26 and represented a normal peripheral B cell. One curious difference between the cells was that whereas Laz388 expressed both HLA-DR and HLA-DQ proteins, Laz221 only expressed the HLA-DR proteins 24, 25. Analysis of the cells showed that the HLA-DR genes were expressed in Laz221 but neither HLA-DQA or HLA-DQB was expressed 24. Examination of the cis-regulatory elements by transfection of DNA reporter constructs into each of the cell lines and the use of EMSAs suggested that the HLA-DQB1 gene was regulated differently and that the Laz221 cell line was lacking a critical component 24, 27. Because Laz221 expressed HLA-DR molecules at normal levels, this meant that each of the MHC-II specific transcription factors RFX5, RFX-B, RFXAP, and CIITA were present and fully active and that some other aspect of the system was defective. Upon discovery of XL9 and its role in HLA-DRB1 and HLA-DQA1 transcription, we hypothesized that the differential expression of the HLA-DQ locus in Laz221 might involve the use of XL9. We found that the region between and including the two insulators XL9 and C2 was hypermethylated in Laz221 but not Laz388 cells. DNA methylation resulted in the loss of CTCF binding and long distance interactions between the XL9 and MHC-II gene promoters. Critically, treatment with 5’-deoxyazacytadine (5-azaC) reverted each of the molecular defects in Laz221 cells, resulting in the reexpression of the HLA-DQ genes. Thus, these data point to DNA methylation of an MHC-II subregion and insulator as a potential mechanism by which cells may attempt to avoid detection by the immune system through the loss of an MHC-II isotype.
Results
CTCF does not bind to the MHC insulators that surround the HLA-DQ subregion in Laz221 cells
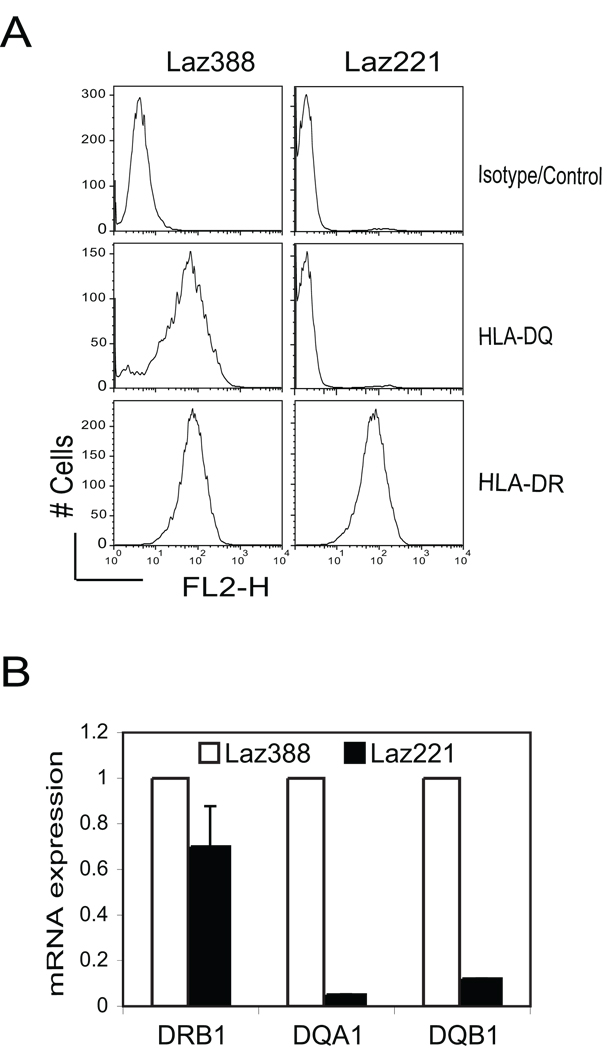
Previous analyses demonstrated differential expression of HLA-DR and HLA-DQ surface protein levels between the Laz221 and Laz388 cell lines 24, 25. This data is recapitulated in Figure 1A, which shows two distinct expression profiles for these cells. Previous work also showed that HLA-DQB mRNA was not expressed in Laz221 24. Confirmation of that result showed that compared to Laz388, Laz221 cells had extremely low levels of HLA-DQB1 mRNA. Additionally in Laz221 cells HLA-DQA1 mRNA levels were very low, suggesting that the cause of expression may be related to the HLA-DQ locus. Similar levels of HLA-DRB1 transcripts were observed between the cell lines (Figure 1B). Southern hybridization experiments showed that HLA-DRB1 and HLA-DQB1 were similar in both cell lines, indicating no major deletion in the genes 24. Since the previous analyses of these cell lines, we now know that the RFX complex (RFX5, RFX-B, and RFXAP) and CIITA are specific to and required for the expression of all MHC-II genes 4. The finding that HLA-DR is expressed on the surface and that HLA-DRB1 mRNA is present in Laz221 cells argues that the RFX subunits, CIITA, and other necessary transcription factors for MHC-II transcription are fully functional in Laz221 cells.
Figure 1. Laz221 cells express HLA-DR but not HLA-DQ gene products.
A) Surface MHC-II expression patterns were assessed using flow cytometry with isotype control, anti-HLA-DQ, and anti-HLA-DR antibodies. This figure represents one of the two independent experiments. B) mRNA levels for HLA-DRB1, -DQA1, and -DQB1 genes were determined by the real-time RT-PCR using RNAs prepared from the Laz388 and Laz221 cells. These data represent an average derived from three biological replicates plotted with respect to the expression in Laz388. Standard error is shown for Laz221 cells.
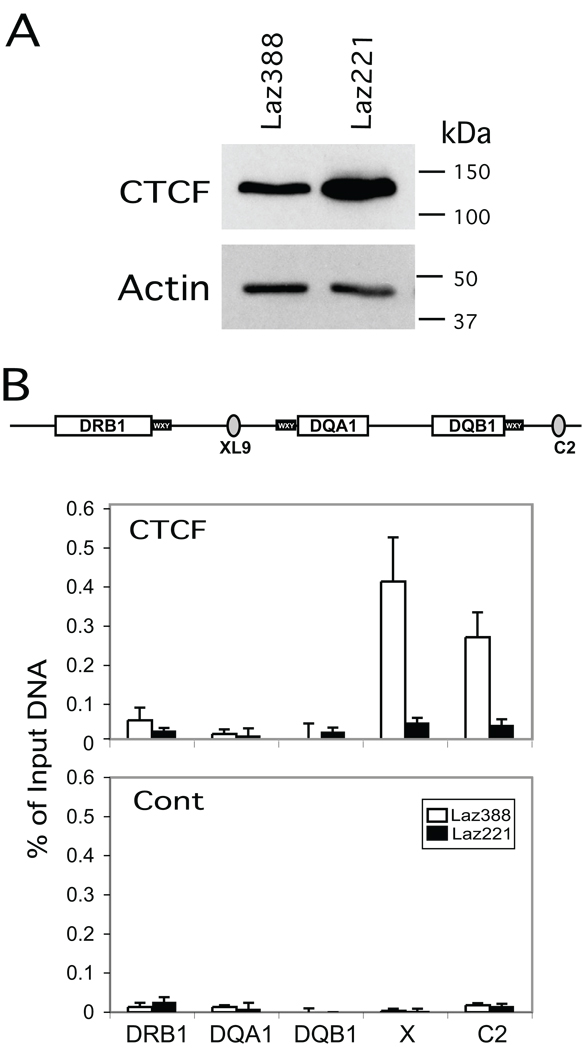
Our recent discovery that HLA-DQA1 utilized the XL9 insulator as a distal regulator of HLA-DQA1 expression 22, led us to examine if XL9 was mutated in Laz221 cells. However, DNA sequencing of genomic clones isolated from Laz221 and Laz388 showed identical sequences (data not shown). We also tested whether CTCF, the insulator factor at XL9 that is responsible for its MHC-II regulatory activity, was normally expressed Laz221 cells. Western blotting of both Laz388 and Laz221 revealed that the overall levels of CTCF are similar with Laz221 expressing slightly more CTCF than Laz388 cells (Figure 2A).
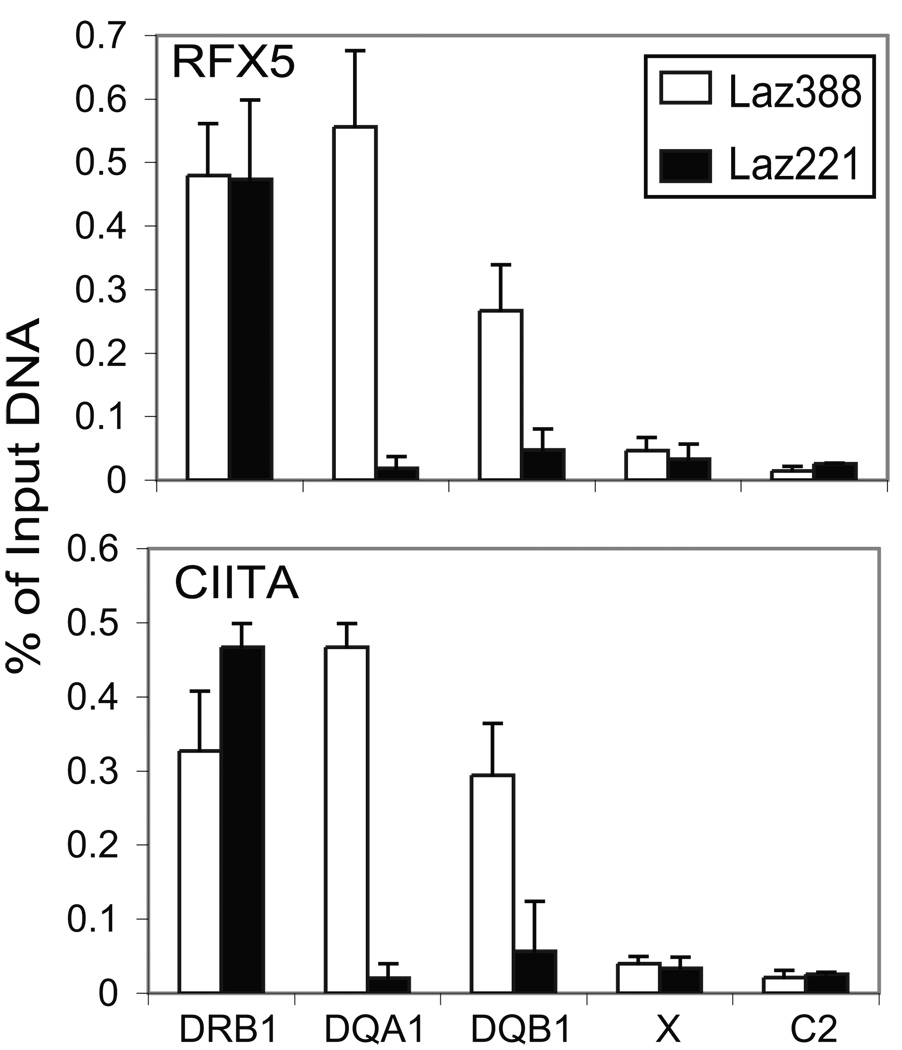
Figure 2. CTCF does not bind to HLA-DQ flanking insulator elements XL9 and C2.
A) Western blot of cell lysates from Laz388 and Laz221 stained with anti-CTCF and anti-β-actin antibodies demonstrated near equal levels of CTCF in both cell lines. B) Quantitative real-time ChIP assays were performed using chromatin prepared from the Laz388 and Laz221 cells with anti-CTCF and non-specific control antibodies to determine the in vivo occupancy of CTCF at XL9, C2, and promoter regions of HLA-DRB1, -DQA1, and –DQB1 genes. The schematic shows the relative positions of the amplicons used to assess CTCF binding across this region. Strong CTCF binding was detected at the XL9 and C2 elements in Laz388 cells while no significant binding was detected at these sites in Laz221 cells. These results are represented of the compiled data from three biological replicates and are presented with standard error as the percentage of input chromatin used for the ChIP assay.
In addition to XL9, another CTCF binding insulator termed C2, was recently identified that lies between HLA-DQB1 and the pseudogene HLA-DQA2 (Figure 2B, Majumder and Boss, unpublished results). C2 interacts with XL9, and like XL9 forms long-distance interactions with the proximal promoters of HLA-DQA1 and HLA-DQB1 (Majumder and Boss, unpublished results). Thus, to determine if CTCF binding to Laz221 was similar to that in Laz388, chromatin immunoprecipitation assays were performed at XL9 and C2 (Figure 2B). Whereas CTCF bound to XL9 and C2 in Laz388 cells, it did not bind to these sites in Laz221 cells. The binding of CTCF was specific as shown by the non-specific DNA and antibody controls presented (Figure 2B). This suggested that a potential cause for the lack of HLA-DQ expression could be do to the inability to bind CTCF at XL9 and C2.
XL9 and the HLA-DQ region are hypermethylated in Laz221 cells
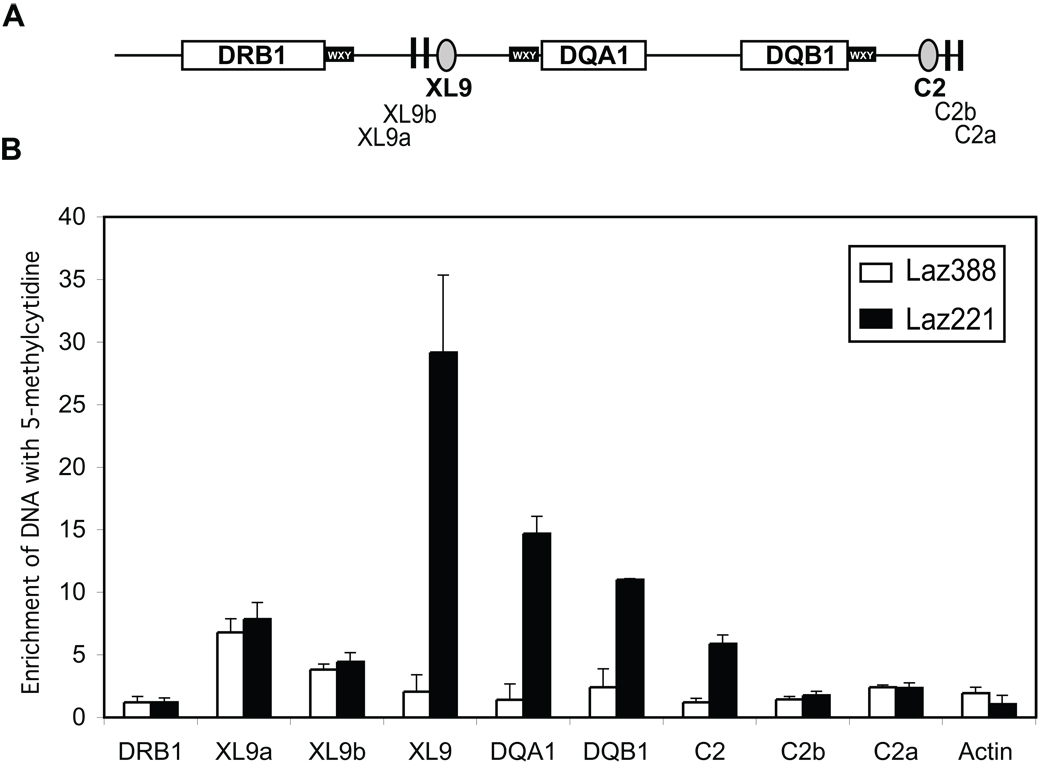
DNA methylation is a heritable mark that can result in reducing the accessibility of DNA to transcription factors by controlling the local chromatin structure. DNA methylation is also known to prevent CTCF binding to its target sequences 16, 17, 31; suggesting a mechanism by which CTCF binding may be abrogated in Laz221 cells. To determine if DNA methylation may play a role in the differential expression of the HLA-DQ genes, methyl-DNA immune precipitation assays (MeDIP) were conducted across the HLA-DQ subregion 29. In MeDIP, DNA is fragmented by sonication to produce fragments of 600–1,000 bp in length. A methylcytosine specific antibody is used to immunoprecipitate the DNA prior to analysis by real-time PCR. The MeDIP technique circumvents the sequence bias that is present in the methylation sensitive restriction digestion technologies and can be rapidly applied across a region. A series of amplicons from just upstream of XL9 to downstream of C2 was designed and used to assess the locus. The results showed that the sequences surrounding XL9 were highly enriched for cytosine methylation in Laz221 cells but showed no methylation in Laz388 (Figure 3). Additionally, each of the promoter regions surrounding the HLA-DQA1 and HLA-DQB1 genes were also methylated in Laz221 cells but not in Laz388. Lastly, the C2 insulator was also hypermethylated in Laz221 cells compared to Laz388. In contrast, the HLA-DRB1 proximal promoter region and that of a CpG island in the β-actin gene showed no enrichment for methylcytosine containing DNA. Intriguingly, the sequences represented by amplicons C2a and C2b, which lie outside of the XL9-C2 defined HLA-DQ region showed no cytosine DNA methylation, suggesting that C2 was the boundary of this epigenetic mark. Sequences XL9a and XL9b, which represent random regions also outside of the XL9-C2 domain, exhibited DNA methylation irrespective of the cell line, suggesting that methylation of these sequences is normally associated with this intergenic region. While the number of methylated cytosines correlates with signal strength in the MeDIP assay, the data collected were not simply due to the number of potential CpGs within a region as XL9 has fewer CpG sites within the 500 bp surrounding DNA than HLA-DQA1 or HLA-DQB1 (Table 1).
Figure 3. The HLA-DQ subregion is hypermethylated in Laz221 cells but not in Laz388.
A) Schematic diagram shows the HLA-DRB1- C2 region of MHC-II locus and position of primers used for the real time PCR following immunoprecipitation. B) Denatured sonicated genomic DNAs from Laz388 and Laz221 were incubated with and without anti-5-methyl-cytosine (α-5mC) antiserum, and the methylated DNAs were collected by immunoprecipitation. The relative enrichment in the bound DNA over input fractions was determined by quantitative real-time PCR for the regions shown in A. These data represent the average with standard error derived from three independent cell cultures.
Table 1.
CpG dinucleotides per locus
| Locus | CpGs* |
|---|---|
| HLA-DRB1 proximal promoter region | 6 |
| XL9a (4000 bp from the core XL9)** | 14 |
| XL9b (2000 bp from the core XL9) | 4 |
| XL9 | 3 |
| HLA-DQA1 proximal promoter region | 14 |
| HLA-DQB1 proximal promoter region | 70 |
| C2 | 8 |
| C2b (2000 bp from the core XL9) | 4 |
| C2a (4000 bp from the core XL9) | 9 |
| b-Actin promoter region | 110 |
CpGs in the 500 bp that encompasses the locus
See Schematic for relative location
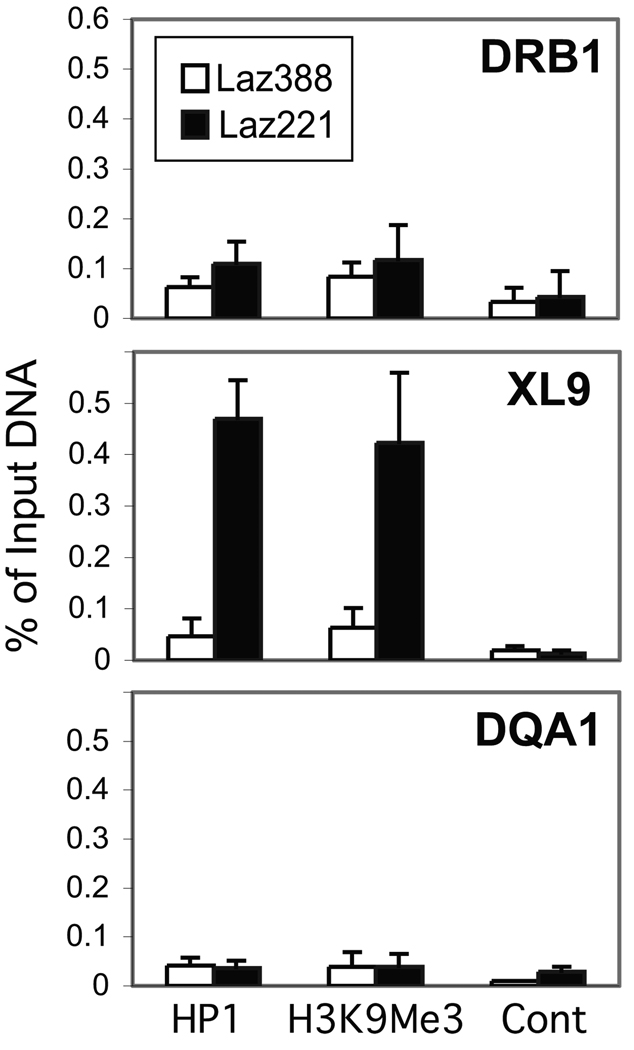
In addition to DNA methylation chromatin accessibility is modulated through a variety of histone modifications that serve to recruit heterochromatin-modulating factors. One such factor is heterochromatin protein-1 (HP-1), which recognizes chromatin that contains histone H3K9me3 posttranslational modifications 32. To determine if the regions bearing DNA methylation carried heterochromatin modifications, ChIP assays were carried out for HP-1 and H3K9me3 (Figure 4). The results showed that the repressive histone modification and HP-1 were present at XL9 chromatin but not at the proximal promoters in Laz221 cells. Neither the mark nor HP-1 was found at these sites in Laz388 cells. Together, these data demonstrate that the HLA-DQ subregion is hypermethylated in Laz221 cells and that the XL9 insulator region is packaged in a repressive chromatin structure that would be predicted to silence its function.
Figure 4. XL9 chromatin is marked by histone H3K9me3 modifications and bound by HP-1.
Quantitative real-time ChIP assays were conducted using control, anti-histone H3K9me3, and anti-HP-1 antibodies on chromatin prepared from Laz388 and Laz221 cells with amplicons representing XL9 and the proximal promoter regions of HLA-DRB1 and HLA-DQA1 genes. These data represent an average with standard error derived from three independent chromatin preparations. The data are plotted with respect to the percent of input chromatin for that region.
The HLA-DQ proximal promoter regions do not bind RFX and CIITA
The DNA methylation patterns over the proximal promoter regions of the HLA-DQA1 and HLA-DQB1 genes suggest that RFX and CIITA may not be able to bind to their respective sequences. To test this hypothesis, the in vivo occupancies of RFX5 and CIITA to the HLA-DRB1 and HLA-DQA1 in Laz221 and Laz388 cells were determined by ChIP. As expected RFX5 and CIITA bound to the promoter proximal regions of HLA-DRB1 in both cell lines and to HLA-DQA1 and HLA-DRB1 in Laz388. However, RFX5 and CIITA did not bind to the promoter proximal region of HLA-DQA1 or HLA-DQB1 (Figure 5). This suggests that DNA methylation may prevent the assembly of the active proximal promoter regulatory regions and actions of XL9 and C2.
Figure 5. RFX and CIITA do not bind to the HLA-DQA1 and HLA-DQB1 promoter proximal WXY box regions.
Quantitative ChIP assays were performed as in Figures 2 and 4 with the indicated antibodies to assess the in vivo occupancy of the RFX5 and CIITA at the promoter proximal regions of HLA-DRB1 and HLA-DQ genes and the XL9 and C2 elements. No significant binding of RFX5 and CIITA was observed at the HLA-DQ promoter proximal region in Laz221 cells. In contrast to this, high levels of RFX5 and CIITA were detected at the same region in Laz388 cells. High levels of RFX5 and CIITA were observed on HLA-DRB1 promoter where no significant difference was found in both cell lines. XL9 and C2 do not bind RFX or CIITA. These data were derived from three independent chromatin preparations.
Inhibition of DNA methyltransferase reverses the differential expression of HLA-DQ genes in Laz221 cells
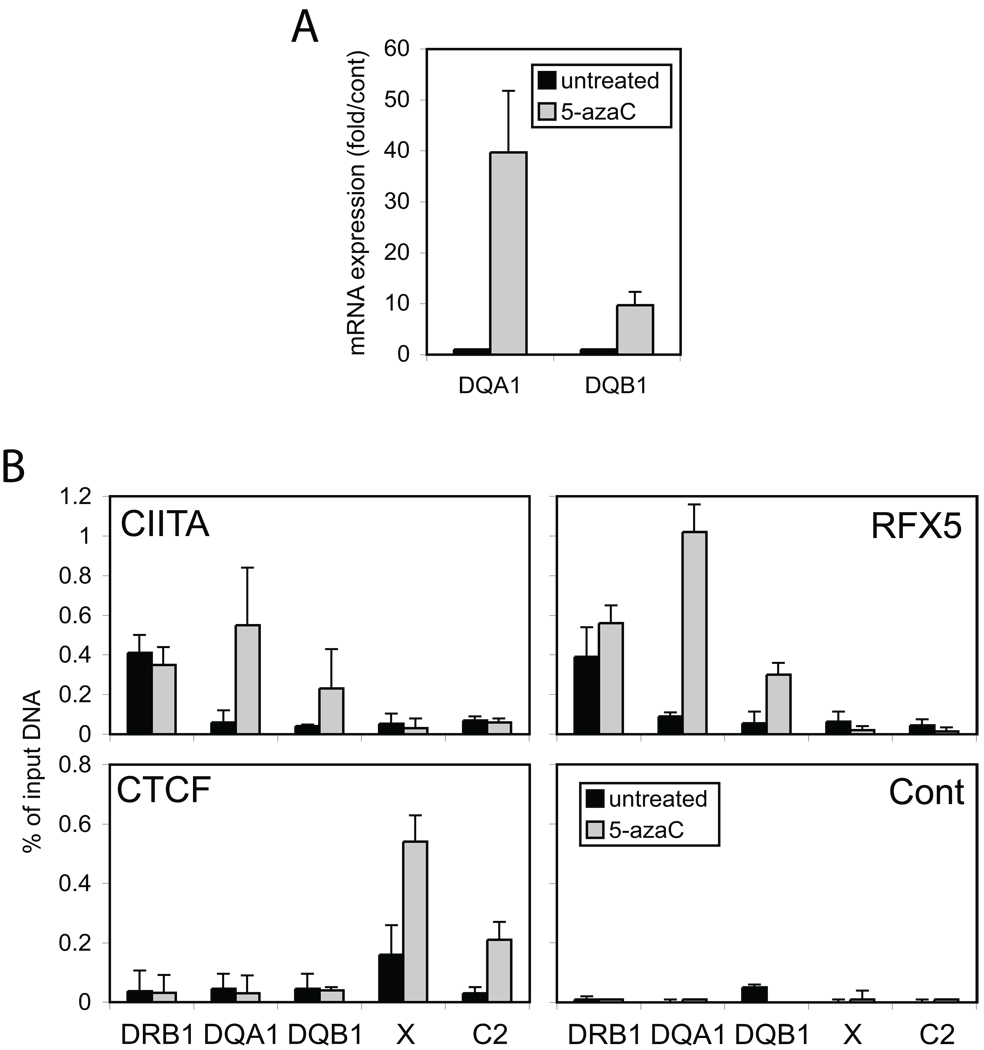
To determine if DNA methylation was responsible for the Laz221 phenotype, Laz221 cells were treated for two days with 5azaC, an inhibitor of the maintenance DNA methyltransferase DMNT1 33–35. Total RNA was isolated and analyzed for the expression of HLA-DRB1, HLA-DQA1, and HLA-DQB1 mRNA levels (Figure 6A). While 5azaC treatment increased the levels of HLA-DRB1 by 3-4-fold (data not shown); the levels of HLA-DQA1 were increased by 40-fold and levels of HLA-DQB1 were increased by 7.5 fold over the untreated cultures. Moreover, 5azaC treatment also resulted in the ability of RFX5, CIITA, and CTCF to bind their target sites in Laz221 cells (Figure 6B).
Figure 6. Inhibition of DNA methylation derepresses HLA-DQ gene expression.
Laz221 cells were treated with 5azaC for two days as described in the materials and methods. Total RNA and chromatin were purified from the cells after two days and compared to control, untreated Laz221 cells. A) mRNA levels for HLA-DQA1 and -DQB1 genes were determined by the real-time RT-PCR. Results are shown with respect to the levels of GAPDH mRNA, which did not change appreciably (data not shown). B) Quantitative real-time ChIP assays were performed as above with indicated antibodies to assess the in vivo occupancies of RFX, CIITA, and CTCF at HLA-DQA1, -DQB1, -DRB1, XL9 and C2. The average of three independent experiments is presented.
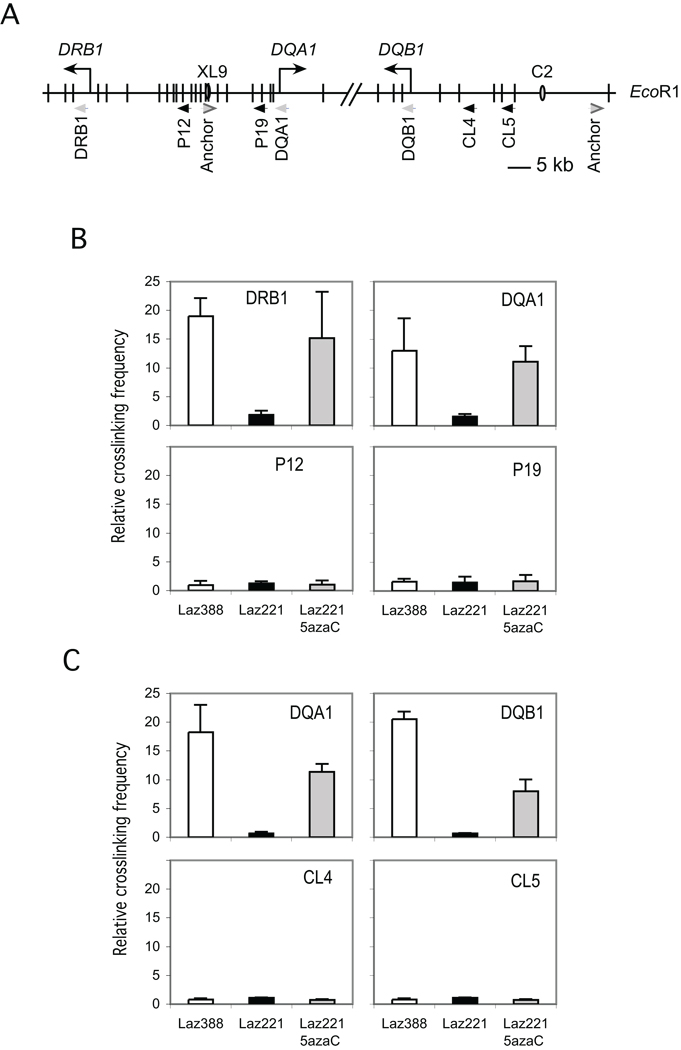
XL9 functions by interacting directly with the proximal promoter regions of the HLA-DRB1 and HLA-DQA1 genes 22. C2 was found to interact with HLA-DQA1 and HLA-DQB1 (Majumder and Boss unpublished). Disruption of these interactions by depleting CTCF results in a loss of expression from these promoters 22. Thus, it was of interest to determine if the DNA methylation played a role in these interactions. Interactions between distal elements can be observed using the chromatin conformation capture assay, termed 3C 22, 30. The formation of a 3C product provides a measure of the spatial proximity of the elements in question compared to a randomly selected element. Here, the ability of XL9 and C2 to interact with the promoter proximal restriction fragments of HLA-DRB1 and HLA-DQA1 was compared to random sequences located within the locus. Previous analyses have characterized these elements and the controls required to demonstrate that the system was operational 22 (Majumder and Boss, unpublished). In Laz388, 3C products were formed between XL9 and HLA-DRB1 and HLA-DQA1, but not with the control fragments P12 and P19 (Figure 7B). In Laz221, 3C products were not observed between XL9 and either promoter proximal restriction fragment. Similarly, C2 interacted with HLA-DQA1 and HLA-DQB1 in Laz388 but not in Laz221 (Figure 7C). These results indicate that the insulator regions were not active in modulating MHC-II activity in Laz221 cells. The inability to form 3C products in Laz221 was not due to restriction site accessibility as the conditions used still allowed enzyme access to the sites of the isolated chromatin (Supplemental Figure 1). Treatment of Laz221 cells with 5azaC resulted in the formation of the XL9—HLA-DQA1, C2—HLA-DQA1, and C2—HLA-DQB1 3C interactions at comparable frequencies to those observed in Laz388. No interactions were observed with the control fragments following 5azaC treatment. Thus, DNA methylation in this system also blocks XL9 and C2 function.
Figure 7. Interactions between the XL9 and C2 insulator elements with HLA-DQ genes are restored in Laz221 cells following inhibition of DNA methylation.
A) Schematic restriction map showing the relevant portions of the HLA-DQ locus and HLA-DRB1 gene. Vertical bars show the positions of the EcoRI sites in the region. The positions of the PCR primers representing the restriction fragment assessed in the 3C assays are depicted by arrowheads and designated by the name below. The position of the insulators XL9 and C2 are represented by ovals. For these 3C assays chromatin was prepared from Laz388, Laz221, and Laz221 cells treated with 5azaC. B) 3C assay in which XL9 served as the anchor region to test interactions between the XL9 restriction fragment and the indicated restriction fragment containing the HLA-DRB1 or -DQA1 promoter. C) 3C assays in which C2 served as the anchor primer to test interactions between the C2 restriction fragment and other restriction fragments containing either the HLA-DQA1 or -DQB1 promoters. The data from three independent 3C chromatin preparations was averaged and plotted with respect to the relative amount of 3C product. Control restriction fragments are indicated as the P12, P19, CL4 and CL5 primer-positions.
Discussion
The pre B cell leukemia derived cell line Laz221 was isolated from the patient who was diagnosed with acute lymphocytic leukemia in 1978 23. A normal B cell line Laz388 was also established from the same patient 26. Nearly, a decade later it was found that these cells differentially expressed MHC-II genes 24, 25. At that time, the differential HLA-DQ expression was postulated to be due to a deficiency in Laz221 that was specific to the expression of the HLA-DQB gene 24, 27. With the current understanding that all MHC-II isotypes are coordinately regulated by a common set of factors 4, 10 and the recent identification of insulator elements that contribute to the expression of the locus, we sought to reinvestigate this cell line and its autologous partner Laz388 to determine the molecular basis of the differential expression of the HLA-DQ genes. The results showed that the HLA-DQ subregion of the MHC was hypermethylated in Laz221 compared to Laz388 cells. DNA hypermethylation disabled three separate mechanistic events necessary for the expression of an MHC-II gene. Importantly, inhibition of DNA hypermethylation within the HLA-DQ subregion restored HLA-DQ gene expression and each of the biochemical pathways associated with HLA-DQ gene regulation.
The first of these events was that DNA methylation of the HLA-DQA1 and HLA-DQB1 proximal promoter X-Y box sequences prevented the assembly of the RFX and CIITA enhanceosome complex that is required for MHC-II gene expression 7, 10. Without the binding of these factors to the proximal promoter regions, the transcription of these genes could not occur. Thus, in a gene specific manner, the DNA methylation of the proximal promoters created a situation that mimicked the phenotype of an RFX-subunit-deficient bare lymphocyte patient’s B cells 36, 37 where the critical factors failed to assemble 38. The observations that RFX and CIITA were bound at HLA-DRB1 in Laz221 cells, and that 5azaC reverted the binding of these factors to HLA-DQA1 provides evidence that the chromatin samples and assay system used in these ChIP experiments are sufficiently controlled.
Second, DNA methylation of XL9 and C2 resulted in chromatin that could not bind CTCF. This effectively removed CTCF from being able to participate in the activation of the neighboring MHC-II genes. A surprise was that in addition to XL9 and C2 being hypermethylated, we found that the XL9 region contained high levels of the repressive histone modification H3K9me3 and was associated with HP-1, the heterochromatin protein that recognizes this mark 39, 40. This suggests the possibility that this region is packaged in a heterochromatic state and that the histone modification is playing a role in the silencing of the region. The heterochromatin mark did not spread to the HLA-DQ gene promoter regions, suggesting that it was highly localized. Third, the loss of both CIITA and CTCF binding to their target sites had the net effect of eliminating the ability of these sites to interact with each other. Thus, the long-distance chromatin loops that form between the CTCF sites and the HLA-DQ genes did not occur. In addition to XL9 and C2, the MHC-II region has 8 other CTCF sites in the locus 15. These CTCF sites interact with each other, as well as the neighboring MHC-II gene promoters. Although large in scale (~150–200 kb), the frequency of interactions is dependent on the distance between the sites. CTCF binding is required for these sites to interact with each other and CIITA is required for additional interactions between the CTCF sites and MHC-II gene promoter regions 15. Thus, the DNA methylation and heterochromatin events at the two insulators that border the HLA-DQ subregion, disrupt the normal architecture of the locus.
The loss of CTCF binding to XL9 raises the issue of how HLA-DRB1 can be expressed as its closest CTCF site is silenced. In addition to XL9, HLA-DRB1 interacts with the CTCF site that is upstream of the HLA-DRA gene, termed C1 15. Thus, it has a choice of which site it can use. The data presented here suggests that the loss of one available CTCF site does not prevent expression of an MHC-II gene if another site can be used. Thus, the system has redundancies built in. Intriguingly, in Raji cells, HLA-DQA1 interacts with only XL9 and C2 15, for which both of these sites are inactive in Laz221.
Transcriptional insulators are often complex elements that in addition to having the ability to block an enhancer from activating a downstream target gene, they can also prevent the encroachment of heterochromatin into a region 41. This latter activity is attributed to barrier or boundary function of the elements. More recently the activities associated with the β-globin HS4 insulator, which exhibited all of these activities, have been dissected and individual functions, enhancer blocking vs boundary function, were found to be the result of separate definable elements 42. This raises the question as to whether the CTCF binding sites within the MHC-II locus have multiple activities. XL9 mediates strong enhancer blocking activity 12; however, barrier activity was not assessed for XL9 or any of the other MHC-II CTCF elements. In the results presented here, the differential DNA hypermethylation did not appear to spread outside of the HLA-DQ region. Because CTCF cannot bind to its sites, it is reasonable to postulate that it does not contribute to such an activity. This assessment is consistent with the fact that the zinc finger protein VEZF1 was found to be responsible for preventing the spread of heterochromatin in the HS4 insulator 42.
DNA methylation of MHC-II genes has been observed previously and correlated with silencing of the locus 43–45. In these studies, the HLA-DR region was methylated as determined by Southern blots and methylation sensitive restriction enzymes, and HLA-DQ genes were not assessed. In other studies, differential HLA-DR and –DQ expression was observed, but the molecular basis was not established 25, 46–48. Thus, there is a long history of methylation as a mechanism to control MHC-II expression. Because MHC-II gene products are typically co-expressed, this raises the question of how both alleles received the same epigenetic mark to silence the HLA-DQ subregion. We pose that there are two possibilities. The first is that the Laz221 leukemic line may represent a developmental snap shot of B cell development. In such a scenario, the HLA-DQ region would be normally silenced and activated at a later developmental cell stage. Several reports support this notion. HLA-DQ expression was reported to lag behind HLA-DR and HLA-DP during early fetal development 46. Additionally hematopoegenic progenitors were found to express HLA-DR and HLA-DP but not HLA-DQ 48. Lastly, in a similar vein, it is possible that the entire locus is controlled by DNA methylation events. In embryonic and pluripotent stem cells, HLA-DRA was not expressed or inducible by IFNγ, unless the cells were pretreated with 5azaC 49. Bisulfite sequencing showed that the HLA-DRA locus was hypermethylated in these cells.
A second possibility is that a strong selection event led to the silencing of both HLA-DQ alleles and that DNA methylation was the successful strategy to silence the loci. In diffuse large B cell lymphoma (DLBCL), MHC-II expression is often lost through a variety of mechanisms some of which include the reduction of CIITA expression but others remain unknown 50–53. In these cancers, the loss of MHC-II expression is associated with a poorer prognosis than those lymphomas expressing MHC-II 51. Fewer tumor infiltrating CD8 T cells were associated with MHC-II negative DLBCLs than MHC-II positive, suggesting a connection between MHC-II positive cells and tumor surveillance mechanisms. The DNA methylation events found associated with Laz221 cells represent one mechanism by which MHC-II genes and a particular isotype can be silenced; thus, it would not be surprising if other lymphomas/leukemias were ultimately identified that displayed MHC-II silencing events that occurred through a DNA methylation and/or the loss of an MHC-II specific insulator function.
Materials and Methods
Cell culture
The Laz221 and Laz388 cell lines 23, 26 originally derived from the peripheral blood of a patient with acute lymphocytic leukemia. These two cell lines were obtained from Dr. G. Nepom (Benaroya Institute, WA) and were grown in RPMI-HEPES supplemented with 10% fetal bovine serum (Hyclone, Inc., Logan, UT), 2 mM L-glutamine, penicillin /streptomycin 50 U/ml and 1mM Sodium pyruvate. Laz221 cells were grown in the absence or presence of 5 µM 5’-deoxyazacytadine (5azaC) (Sigma-Aldrich, Cat#A2385). 5azaC was added at 8 hour intervals to the growth media for 2 days.
Flow cytometry
Cells were harvested, washed twice with cold PBS, and incubated with phycoerythrin (PE)-conjugated anti-human HLA-DR antibody or unconjugated anti-human HLA-DQ antibody (BD bioscience Pharmingen, San Diego, CA) followed by staining with a PE-conjugated anti–mouse IgG secondary antibody (Southern Biotech). A FACScalibur (BD Biosciences) flow cytometer was used to analyze the expression level of human MHC-II surface expression. MHC-II positive cells were visualized using the FL2 channel. Control stained samples were processed in parallel using only the secondary antibodies indicated in the figures.
Quantitative RT-PCR
Total RNA was isolated from Laz388 and Laz221 cells using the RNeasy RNA isolation kit (QIAGEN, Valencia, CA) according to the company’s protocol. cDNA synthesis was performed using Superscript II reverse transcriptase (Invitrogen, Inc., Carlsbad, CA) with 2 µg of total RNA in a volume of 20 µl in PCR II buffer containing 5 mM MgCl2 (Applied Biosystems, Foster City, CA). After reverse transcription, sample volumes were increased to 200 µl with TE buffer and 3 µl of the cDNA was used for quantitative PCR assays. GAPDH primers were used in each of the experiments to normalize the data between samples. PCR primers are listed in Supplemental Table 1. The data presented represent the average of three independent experiments and are plotted with their standard deviation. The student’s T test was used to determine statistical significance.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described previously 8, 12, 14, 22. For immunoprecipitations, 5–10 µg of anti-CTCF (Upstate, Cat no. 06-917), anti-RFX5 28, anti-CIITA 8, or anti-T-cell receptor (non-specific control) antibodies were used. Protein A beads (60 µl) were used to isolate the chromatin-antibody bound complexes. After extensive washing, the immunoprecipitated chromatin was eluted in 1% SDS and incubated overnight at 65°C to reverse the crosslinks. The DNA was purified and used as a template in real-time PCR analyses. Immunoprecipitated DNA samples were quantitated by real-time PCR using a 5-point genomic DNA standard curve and an I-cycler (Biorad Laboratories, Inc.) as previously described 22. SYBR-Green incorporation quantitative PCR reactions contained 5% DMSO, 1 X SYBRgreen (Bio Whittaker Molecular Applications), 0.04% gelatin, 0.3% Tween 20, 50 mM KCl, and 20 mM Tris [pH 8.3], 3 mM MgCl2, 0.2 mM dNTP, and 100 nM of each primer. Primer sequences are listed in Supplemental Table 1. All ChIP experiments were performed at least three times from independent preparations of chromatin. The data were averaged and plotted as a percentage of input chromatin with standard error bars as indicated.
Quantitative methylated DNA immunoprecipitation (MeDIP) assay
MeDIP assays were performed as described by Weber et al. 29 with the following modifications. Genomic DNAs were purified from Laz388 and Laz221 cells using DNAzol genomic DNA isolation reagent as described by the manufacturer (Molecular Research Center, Inc., Cat no. DN-127). Genomic DNAs were sonicated in TE buffer to produce random fragments of about 600 bp. 4 µg of fragmented DNA for each experimental sample was denatured and incubated for 2 hr with 5 µg of anti-5-methylcytidine antibodies (Eurogenetec, Inc.; Cat no. BI-MECY-0100) in a final volume of 500 µl of IP buffer (10 mM sodium phosphate (pH 7.0), 140 mM NaCl, 0.05% Triton X-100). Thirty µl of Dynabeads (M-280 sheep antibody to mouse IgG; Dynal Biotech, Inc.) were used to isolate the 5-methycytidine bound DNA complexes. After extensive washing and treating with proteinase K, the immunoprecipitated DNA was extracted with phenol-chloroform, and recovered by ethanol precipitation. To quantitate the samples, real-time PCR was carried out with input and the immunoprecipitated DNAs as described in the above section for ChIP assays.
Quantitative chromosome conformation capture (3C) assay
3C assays 30 were performed with modifications described previously 22, 30. Here, 1×107 cells were isolated, cross-linked with formaldehyde to a final concentration of 1% for 10 minutes at room temperature and glycine was added to 125 mM to stop the crosslinking reaction. Nuclei were purified and digested overnight with EcoRI at 37°C. Following heat inactivation of the enzyme, the samples were diluted ~40:1 and then ligated with T4 DNA ligase. The DNA was purified and then used as a substrate for real-time PCR using the primers described in 22 and shown in Supplemental Table 1. All restriction sites assessed by 3C were equally accessible to restriction enzyme digestion as determined by PCR amplification across the sites following digestion. Standard curve templates for the 3C products were generated in vitro by restriction enzyme cleaving and religating the bacterial artificial chromosome (BAC) RP11-257P24 (purchased from Children’s Hospital Oakland Research Institute), which encodes the HLA-DRB1 and HLA-DQA1 subregion and includes XL9 and C2. All primer combinations are listed in Supplemental Table 1), and were tested previously 22 had a greater than 90% PCR efficiency, and produce a single amplicon on cleaved/religated BAC DNA. Data are presented as crosslinked frequency and represent an average derived from three independent biological replicates. Standard error is provided for the dataset. Student’s T tests were used to determine if observed differences were significant.
Supplementary Material
Acknowledgements
We thank Dr. C. Scharer for aiding with the development of the MeDIP assay and his comments of the work. We thank and acknowledge the technical support for this project from Royce Butler. This work was supported by NIH grant GM47310.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Germain RN, Braunstein NS, Brown MA, Glimcher LH, Lechler RI, McCluskey J, et al. Structure and function of murine class II major histocompatibility complex genes. Mt Sinai J Med. 1986;53(3):194–201. [PubMed] [Google Scholar]
- 2.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375(6534):802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 3.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274(5287):618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 4.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 5.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annual review of immunology. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 6.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr Opin Immunol. 2003;15(1):105–111. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 7.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes & development. 2000;14(9):1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 8.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2(7):652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 9.Boss JM. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9(1):107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 10.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annual review of immunology. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 11.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265(5168):106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 12.Majumder P, Gomez JA, Boss JM. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J Biol Chem. 2006;281(27):18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- 13.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol. 2003;4(2):132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 14.Gomez JA, Majumder P, Nagarajan UM, Boss JM. X box-like sequences in the MHC class II region maintain regulatory function. J Immunol. 2005;175(2):1030–1040. doi: 10.4049/jimmunol.175.2.1030. [DOI] [PubMed] [Google Scholar]
- 15.Majumder P, Boss JM. CTCF controls the expression and the chromatin architecture of the human major histocompatibility complex class II locus. Molecular and cellular biology. 2010 doi: 10.1128/MCB.00327-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez C, Borgel J, Court F, Cathala G, Forne T, Piette J. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochemical and biophysical research communications. 2010;392(2):129–134. doi: 10.1016/j.bbrc.2009.12.159. [DOI] [PubMed] [Google Scholar]
- 18.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16(3):453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111(2):151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 20.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32(1):1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell CM, Grinberg A, Huang SP, Chen D, Pichel JG, Westphal H, et al. A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse beta -globin locus. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14554–14559. doi: 10.1073/pnas.97.26.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. The Journal of experimental medicine. 2008;205(4):785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarus H, Barell EF, Krishan A, Livingston DM, Harris K, Schlossman SF, et al. Characterization if a unique cell line (Laz221) from human acute lymphocytic ("Null" cell) leukemia. Cancer Res. 1978;38:1362–1367. [PubMed] [Google Scholar]
- 24.Woolfrey AE, Andersen LC, Shewey L, Chung J, Nepom GT. Analysis of differential HLA-DQB expression in autologous B cell lines. J Leukoc Biol. 1993;53(6):697–706. doi: 10.1002/jlb.53.6.697. [DOI] [PubMed] [Google Scholar]
- 25.Pesando JM, Graf L. Differential expression of HLA-DR, -DQ, and -DP antigens on malignant B cells. J Immunol. 1986;136(11):4311–4318. [PubMed] [Google Scholar]
- 26.Hercend T, Meuer S, Reinherz EL, Schlossman SF, Ritz J. Generation of a cloned NK cell line derived from the "null cell" fraction of human peripheral blood. J Immunol. 1982;129(3):1299–1305. [PubMed] [Google Scholar]
- 27.Woolfrey AE, Nepom GT. Differential transcription elements direct expression of HLA-DQ genes. Clinical immunology and immunopathology. 1995;74(2):119–126. doi: 10.1006/clin.1995.1018. [DOI] [PubMed] [Google Scholar]
- 28.Moreno CS, Rogers EM, Brown JA, Boss JM. Regulatory factor X, a bare lymphocyte syndrome transcription factor, is a multimeric phosphoprotein complex. J Immunol. 1997;158(12):5841–5848. [PubMed] [Google Scholar]
- 29.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 30.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10(6):1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 31.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282(46):33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 32.Spektor TM, Rice JC. Identification and characterization of posttranslational modification-specific binding proteins in vivo by mammalian tethered catalysis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(35):14808–14813. doi: 10.1073/pnas.0907799106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nature reviews. 2009;10(11):805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic acids research. 2010 doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor S, Jones P. Mechanism of action of eukaryotic DNA methyltransferase *1: Use of 5-azacytosine-containing DNA. Journal of Molecular Biology. 1982;162(3) doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- 36.Steimle V, Durand B, Barras E, Zufferey M, Hadam MR, Mach B, et al. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes & development. 1995;9(9):1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 37.Nagarajan UM, Peijnenburg A, Gobin SJ, Boss JM, van den elsen PJ. Novel mutations within the RFX-B gene and partial rescue of MHC and related genes through exogenous class II transactivator in RFX-B-deficient cells. J Immunol. 2000;164(7):3666–3674. doi: 10.4049/jimmunol.164.7.3666. [DOI] [PubMed] [Google Scholar]
- 38.Kara CJ, Glimcher LH. In vivo footprinting of MHC class II genes: bare promoters in the bare lymphocyte syndrome. Science. 1991;252(5006):709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- 39.Dialynas GK, Vitalini MW, Wallrath LL. Linking Heterochromatin Protein 1 (HP1) to cancer progression. Mutation research. 2008;647(1–2):13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438(7071):1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 41.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes & development. 2002;16(3):271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 42.Dickson J, Gowher H, Strogantsev R, Gaszner M, Hair A, Felsenfeld G, et al. VEZF1 elements mediate protection from DNA methylation. PLoS genetics. 2010;6(1) doi: 10.1371/journal.pgen.1000804. e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reitz MS, Jr, Mann DL, Eiden M, Trainor CD, Clarke MF. DNA methylation and expression of HLA-DR alpha. Molecular and cellular biology. 1984;4(5):890–897. doi: 10.1128/mcb.4.5.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrington MN, Salter RD, Cresswell P, Ting JP. Evidence for methylation as a regulatory mechanism in HLA-DR alpha gene expression. Immunogenetics. 1985;22(3):219–229. doi: 10.1007/BF00404481. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda H, Redford A, Magalong D. Allele-specific methylation in the 5'-regulatory region of class II DQ beta genes in the human major histocompatibility complex (MHC): relationship to autoimmune disease susceptibility. Dis Markers. 1992;10(1):7–18. [PubMed] [Google Scholar]
- 46.Edwards JA, Durant BM, Jones DB, Evans PR, Smith JL. Differential expression of HLA class II antigens in fetal human spleen: relationship of HLA-DP, DQ, and DR to immunoglobulin expression. J Immunol. 1986;137(2):490–497. [PubMed] [Google Scholar]
- 47.Falkenburg JH, Fibbe WE, Goselink HM, Van Rood JJ, Jansen J. Human hematopoietic progenitor cells in long-term cultures express HLA-DR antigens and lack HLA-DQ antigens. The Journal of experimental medicine. 1985;162(4):1359–1369. doi: 10.1084/jem.162.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linch DC, Nadler LM, Luther EA, Lipton JM. Discordant expression of human Ia-like antigens on hematopoietic progenitor cells. J Immunol. 1984;132(5):2324–2329. [PubMed] [Google Scholar]
- 49.Suarez-Alvarez B, Rodriguez RM, Calvanese V, Blanco-Gelaz MA, Suhr ST, Ortega F, et al. Epigenetic mechanisms regulate MHC and antigen processing molecules in human embryonic and induced pluripotent stem cells. PloS one. 2010;5(4) doi: 10.1371/journal.pone.0010192. e10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cycon KA, Rimsza LM, Murphy SP. Alterations in CIITA constitute a common mechanism accounting for downregulation of MHC class II expression in diffuse large B-cell lymphoma (DLBCL) Experimental hematology. 2009;37(2):184–194. doi: 10.1016/j.exphem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Rimsza LM, Roberts RA, Miller TP, Unger JM, LeBlanc M, Braziel RM, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood. 2004;103(11):4251–4258. doi: 10.1182/blood-2003-07-2365. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson ST, Fernandez DR, Murphy SP, Braziel RM, Campo E, Chan WC, et al. Decreased major histocompatibility complex class II expression in diffuse large B-cell lymphoma does not correlate with CpG methylation of class II transactivator promoters III and IV. Leukemia & lymphoma. 2009;50(11):1875–1878. doi: 10.3109/10428190903297531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rimsza LM, Chan WC, Gascoyne RD, Campo E, Jaffe ES, Staudt LM, et al. CIITA or RFX coding region loss of function mutations occur rarely in diffuse large B-cell lymphoma cases and cell lines with low levels of major histocompatibility complex class II expression. Haematologica. 2009;94(4):596–598. doi: 10.3324/haematol.2008.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.