Abstract
The phytohormone abscisic acid (ABA) plays a central role in plant development and in plant adaptation to both biotic and abiotic stressors. In recent years, knowledge of ABA metabolism and signal transduction has advanced rapidly to provide detailed glimpses of the hormone's activities at the molecular level. Despite this progress, many gaps in understanding have remained, particularly at the early stages of ABA perception by the plant cell. The search for an ABA receptor protein has produced multiple candidates, including GCR2, GTG1, and GTG2, and CHLH. In addition to these candidates, in 2009 several research groups converged on a novel family of Arabidopsis proteins that bind ABA, and thereby interact directly with a class of protein phosphatases that are well known as critical players in ABA signal transduction. The PYR/PYL/RCAR receptor family is homologous to the Bet v 1-fold and START domain proteins. It consists of 14 members, nearly all of which appear capable of participating in an ABA receptor–signal complex that responds to the hormone by activating the transcription of ABA-responsive genes. Evidence is provided here that PYR/PYL/RCAR receptors can also drive the phosphorylation of the slow anion channel SLAC1 to provide a fast and timely response to the ABA signal. Crystallographic studies have vividly shown the mechanics of ABA binding to PYR/PYL/RCAR receptors, presenting a model that bears some resemblance to the binding of gibberellins to GID1 receptors. Since this ABA receptor family is highly conserved in crop species, its discovery is likely to usher a new wave of progress in the elucidation and manipulation of plant stress responses in agricultural settings.
Keywords: Abiotic stress, abscisic acid, Bet v 1-fold, drought, PP2C, PYR/PYL/RCAR, salinity, SnRK, signal–receptor, START domain
Introduction
Abscisic acid (ABA) is a sesquiterpenoid molecule made by organisms across kingdoms, including fungi, plants, and metazoans (Oritani and Kiyota, 2003; Bruzzone et al., 2007). It was first discovered and is best understood in angiosperms, where it acts as a hormone to regulate diverse processes including seed development, dormancy, germination, and seedling growth (Finkelstein et al., 2002). ABA is also a central regulator of plant adaptation to biotic (Fujita et al., 2006) and abiotic stressors (Zhu, 2002). ABA protects plants under abiotic stress, particularly dehydration and salinity, through mechanisms including the production of osmoprotective proteins and metabolites, and the regulation of stomatal conductance (Zhu, 2002). Many details of ABA biosynthesis, transport, catabolism, and ABA-mediated effects on global transcription and metabolism have been elucidated (Nambara and Marion-Poll, 2005; Christmann et al., 2006; Verslues and Zhu, 2007; Urano et al., 2009). Genomic resources of Arabidopsis, in particular, have enabled the identification of key components of ABA signal transduction, including protein kinases, phosphatases, transcription factors, RNA processing factors, proteasome components, chromatin remodelling proteins, and histone deacetylases that mediate epigenetic regulation (Hirayama and Shinozaki, 2007; Chinnusamy and Zhu, 2009; Cutler et al., 2010). However, many significant gaps in our understanding of ABA signal transduction still exist.
One active area of ABA research has been the quest for protein receptors that directly bind the hormone and trigger signalling events leading to ABA's distinctive effects on plant physiology. Genetic redundancy has complicated the search for ABA receptors. Moreover, receptor searches have led to multiple—in some cases controversial—candidate proteins (McCourt and Creelman, 2008). Recently, a combination of chemical genetic and proteomic approaches led to significant breakthroughs in the receptor hunt. In this review, we focus on the convergence of separate research groups upon a novel class of ABA-binding proteins in Arabidopsis—the PYR/PYL/RCAR family—that bear unequivocal hallmarks of ABA receptors. These rapid developments have opened a wide doorway for exploring the structural basis of ABA perception and signal transduction in great detail. The emerging picture reveals a remarkable, surprisingly simple, assemblage of key proteins that form an ABA signal–receptor complex, with an arrangement that bears some resemblance to perception mechanisms for other plant hormones, but that also presents a distinctive model likely to establish new paths of research. The PYR/PYL/RCAR family is highly conserved in other plant species, and its discovery could lead to the development of novel means for manipulating stress tolerance in crops.
Dodging redundancy
Independent, complementary approaches led to the first published reports that identified PYR/PYL/RCAR proteins as ABA receptors. One strategy employed yeast two hybrid (Y2H) screening to search for interacting partners of Arabidopsis protein phosphatases that are negative regulators of ABA signalling (Ma et al., 2009). In an alternative, innovative approach, a chemical genetic screen of small synthetic molecules that perturb seed germination in Arabidopsis identified the receptor family through a forward genetic analysis (Park et al., 2009).
In the latter approach, the Cutler group identified one small molecule, termed pyrabactin, that specifically inhibits seed germination and cell expansion (Park et al., 2009). Pyrabactin's effects on global gene transcription in the seed are highly correlated with those of ABA but not with other germination inhibitors. This correlation is much weaker in seedlings, indicating that the chemical is a highly selective ABA agonist that mimics only a subset of the hormone's activities during plant development. A forward genetic screen for mutants resistant to pyrabactin identified the PYRABACTIN RESISTANCE 1 (PYR1) locus; map-based cloning identified several mutations in PYR1, which is a founding member of the PYR/PYL/RCAR protein family. Thirteen PYR1-LIKE (PYL) proteins encoded in the Arabidopsis genome were identified by sequence analysis and designated PYL1 through PYL13. The specificity of pyrabactin for a seed germination phenotype and the relatively high expression level of PYR1 in the seed, compared with other family members, may explain why a pyrabactin-insensitive mutant could be detected, bypassing the functional redundancy that has hindered other forward genetic screens for ABA receptors.
In a paper that was published simultaneously with that of the Cutler group, the Grill group used the Y2H method to screen the Arabidopsis proteome for interactors with ABI2, a type 2C serine threonine protein phosphatase (PP2C) (Ma et al., 2009). PP2Cs are found across kingdoms, but comprise an exceptionally large and diverse family in plants (Schweighofer et al., 2004). Some members of clade A of this family are known for their central roles in negative regulation of ABA signalling, including the well-studied proteins ABI1 and ABI2 (ABA INSENSITIVE 1 and 2), HAB1 (HOMOLOGY TO ABA INSENSITIVE 1), and AHG1 and AHG3 (ABA-HYPERSENSITIVE GERMINATION 1 and 3) (Koornneef et al., 1984; Leung et al., 1994, 1997; Meyer et al., 1994; Saez et al., 2004; Yoshida et al., 2006; Nishimura et al., 2007). By using the Y2H method with ABI2 as bait, the Grill group discovered a protein called REGULATORY COMPONENT OF ABA RESPONSE 1 (RCAR1), identical to PYL9, leading to the identification of the other 13 members of the PYR/PYL/RCAR family (Ma et al., 2009). The simultaneous reports of PYR/PYL and RCAR proteins have created two sets of nomenclature for this family (Table 1). This report from the Grill group was followed by other publications in which researchers used similar approaches and added details about the PYR/PYL/RCAR receptor family.
Table 1.
Nomenclature and corresponding Arabidopsis accession numbers for the 14 members of the PYR/PYL/RCAR family of ABA receptors
| Nomenclature |
Locus accession no. | |
| PYR/PYLa | RCAR | |
| PYR1 (1,2) | RCAR11 | At4g17870 |
| PYL1 (3,4,5) | RCAR12 | At5g46790 |
| PYL2 (4,5) | RCAR14 | At2g26040 |
| PYL3 | RCAR13 | At1g73000 |
| PYL4 | RCAR10 | At2g38310 |
| PYL5 | RCAR8 | At5g05440 |
| PYL6 | RCAR9 | At2g40330 |
| PYL7 | RCAR2 | At4g01026 |
| PYL8 | RCAR3 | At5g53160 |
| PYL9 | RCAR1 | At1g01360 |
| PYL10 | RCAR4 | At4g27920 |
| PYL11 | RCAR5 | At5g45860 |
| PYL12 | RCAR6 | At5g45870 |
| PYL13 | RCAR7 | At4g18620 |
Crystal structure solved by: (1) Nishimura et al., 2009; (2) Santiago et al., 2009a; (3) Miyazono et al., 2009; (4) Melcher et al., 2009; (5) Yin et al., 2009.
The Rodriguez group performed a Y2H screen for interacting partners of HAB1, and identified PYL5, PYL6, and PYL8 proteins (Santiago et al., 2009b). In another protein-based approach, the Schroeder group screened for ABI1-interacting proteins in planta by affinity purification and mass-spectrometry, using YFP-tagged ABI1 expression lines of Arabidopsis. This method identified nine of the 14 PYR/PYL/RCAR proteins (Nishimura et al., 2010). After identifying PYR1 in the forward genetic screen, the Cutler group conducted a Y2H screen of protein interactors with PYR1, and identified HAB1 (Park et al., 2009). This study also detected physical interactions between multiple PP2Cs and several members of the PYR/PYL/RCAR family. Physical interactions of PYR/PYL/RCAR with PP2Cs have been further supported in planta with other co-immunoprecipitation experiments and with bimolecular fluorescence complementation (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009b). These lines of evidence add weight to the in vitro reconstitution assays and crystallographic studies described later in this review.
Manipulations of PYR/PYL/RCAR expression have confirmed their central role in ABA signal transduction and adaptation to abiotic stress. Since the pyr1 mutant responds normally to ABA, it appeared possible that functional redundancy from other family members could mask PYR1’s role in ABA signal transduction. Triple and quadruple mutants with genotypes pyr1pyl1pyl4 and pyr1pyl1pyl2pyl4, respectively, showed markedly reduced ABA sensitivity in seed germination and seedling growth (Park et al., 2009). The quadruple mutant also showed reduced sensitivity to ABA-mediated stomatal closure (Nishimura et al., 2010) and ABA-mediated transcriptional responses (Park et al., 2009). Transgenic over-expression of either PYL1 or PYL4 restored ABA sensitivity in the triple mutant (Park et al., 2009). Suppression of RCAR1 through RNA interference in protoplasts lowered ABA-induced transcriptional responses, while over-expression of RCAR1 in stably transformed lines enhanced ABA responses at the level of germination, seedling growth and stomatal aperture (Ma et al., 2009). Similarly, over-expression of PYL5 and PYL8 enhanced ABA responses and resistance to water-deficit stress (Santiago et al., 2009b; Saavedra et al., 2010).
Helix-grip fold receptors
The 14-member PYR/PYL/RCAR family is part of the ancient, ubiquitous Bet v 1-fold superfamily, named for a major allergen in pollen of white birch (Betula verrucosa) (Iyer et al., 2001; Radauer et al., 2008). A central feature of this superfamily is the presence of a seven-stranded β-sheet and two α-helices enfolding a long, carboxy-terminal α-helix, which collectively form a helix-grip fold structural motif. The helix-grip fold creates a large cavity that can bind hydrophobic ligands including lipids and hormones (Iyer et al., 2001; Radauer et al., 2008). Some members of the Bet v 1-fold superfamily have been shown to bind cytokinin and brassinosteroid hormones in vitro (Markovic-Housley et al., 2003; Pasternak et al., 2006; Fernandes et al., 2008), so it is not surprising that PYR/PYL/RCAR proteins bind ABA, as described below. Other Bet v 1-fold members are known as pathogenesis-related proteins (class PR-10) that play roles in defence against microbes and in abiotic stress tolerance (Liu and Ekramoddoullah, 2006). Bet v 1-fold proteins have been classed together with the large, ubiquitous START (steroidogenic acute regulatory—StAR—related lipid transfer) domain superfamily (Ponting and Aravind, 1999; Iyer et al., 2001). START domain-containing proteins have also been implicated in plant adaptation to both biotic and abiotic stressors (Yu et al., 2008; Cao et al., 2009; Fu et al., 2009). However, in a recent phylogenetic analysis many Bet v 1-fold proteins were found to differ from START domain proteins with respect to the numbers and relative positions of β-strands and α-helices that create the helix-grip fold; some Bet v 1-fold proteins of plants, including PYR/PYL/RCAR members from Arabidopsis, were segregated into a subfamily of polyketide cyclase-like proteins (Radauer et al., 2008).
PP2C inhibition linked to ABA-receptor binding
The significance of PYR/PYL/RCAR interactions with PP2Cs is supported by multiple lines of evidence that link the ABA-binding properties of these receptors with their suppression of phosphatase activity and the release of ABA signal transduction from PP2C-mediated inhibition. PYR/PYL/RCAR proteins strongly inhibit the phosphatase activity of PP2Cs in vitro, an effect that is ABA dose-dependent (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009b; Szostkiewicz et al., 2010). Site-directed mutations identify critical residues for these interactions. For example, PYR1 protein containing either of two of the amino acid substitutions (PYR1S152L or PYR1P88S) that gave rise to pyrabactin resistance in planta failed to inhibit HAB1 phosphatase activity, and displayed a reduced ABA-dependent interaction with HAB1 in Y2H tests (Park et al., 2009). Conversely, the ABI2G168D mutation, which confers the dominant abi2 phenotype, abolished the inhibitory effect of PYR1 on phosphatase activity (Park et al., 2009), and also blocked the physical interaction between ABI2 and RCAR1 (Ma et al., 2009). A similar result was observed with the interaction between RCAR1 and ABI1G180D, the mutation causing the dominant abi1 phenotype (Ma et al., 2009). Likewise, the HAB1G246D mutation abolished its physical interaction with PYL5 in vivo (Santiago et al., 2009b). Furthermore, this same study reported that over-expression of PYL5 and HAB1 in the same plant removed the ABA-insensitivity of HAB1-overexpressing plants, consistent with PYL5 antagonism of HAB1 phosphatase activity.
The dependence of PYR/PYL/RCAR proteins on ABA to inhibit PP2Cs is paralleled by observations that the binding of ABA to PYR/PYL/RCAR proteins is greatly enhanced by the presence of PP2Cs (Ma et al., 2009; Santiago et al., 2009b). Interestingly, the evidence for PP2C-mediated enhancement of ABA binding comes from experiments in which a structural variant of ABA was compared with the natural form; these comparisons provide insights into the stereoselectivity of plant responses to the molecule. Synthetic ABA can exist as two stereoisomers, S-(+) and R-(–), although only the S-(+) form occurs naturally. In some experimental contexts, the non-natural R-(–) form has been shown to be bioactive (Lin et al., 2005; Nambara et al., 2002; Sondheim et al., 1971; Walker-Simmons et al., 1992). These and other studies, including transcriptional profiling in response to ABA structural analogues (Huang et al., 2007), indicated that multiple ABA receptors with differing specificities may exist.
Strong inhibition of ABI1 and ABI2 phosphatase activities was observed with the addition of both RCAR1 and S-ABA while R-ABA, the non-natural form, was relatively ineffective at inhibition (Ma et al., 2009). Similarly, Santiago et al. (2009b) measured an 8-fold stronger inhibition of HAB1 phosphatase activity by PYL5 in the presence of S-ABA, compared with the R form. Isothermal titration calorimetry experiments found that the apparent binding affinity of RCAR1 to S-ABA was enhanced approximately 10-fold by the addition of ABI2, from a KD of 660 nM to 64 nM (Ma et al., 2009). Santiago et al. (2009b) report a similar result with this same technique, observing an apparent KD of 38 nM S-ABA for PYL5 in the presence of the catalytic core of HAB1 phosphatase. These results show that ABA binding to PYR/PYL/RCAR proteins is intimately tied to PP2C inhibition, raising the possibility that the two proteins could serve as co-receptors. Interestingly, Park et al. (2009) found differences in ABA-dependence and in stereoselectivity among PYR/PYL/RCAR family members with respect to interactions with PP2C proteins. In Y2H experiments, PYL2, PYL3, and PYL4 required the presence of either ABA stereoisomer for their interaction with HAB1 in yeast, while PYL5 through PYL12 (excluding PYL8) were shown to interact constitutively with HAB1. Twelve out of the 14 members (excluding PYL8 and PYL13) interacted with HAB1 in the presence of the natural S-ABA, whereas all these 12 except PYR1 and PYL1 could interact in the presence of the non-natural R-ABA.
Defining core components of ABA signalling
The discovery of PYR/PYL/RCAR proteins has coincided with recent insights on the regulation of ABA-responsive genes by serine-threonine kinases and PP2Cs, providing tools that define a minimal set of factors required for ABA signalling in vitro. Three members of a plant-specific kinase group, called subfamily 2 SNF1-related kinases (SnRK2s), play central roles in Arabidopsis as positive regulators in ABA signal transduction (Mustilli et al., 2002; Yoshida et al., 2002; Fujii et al., 2007). This was demonstrated recently by the extreme ABA-insensitivity of the snrk2.2/2.3/2.6 triple mutant (Fujii and Zhu, 2009; Fujita et al., 2009). The phenotype of this mutant suggests that phosphorylation of transcription factors and other substrates by SnRK2s may be a general requirement for ABA function at all stages of plant development. Transcription factors that are activated through phosphorylation by SnRK2s, and thereby promote ABA signalling, include basic leucine zipper (bZIP) proteins called ABFs/AREBs (Furihata et al., 2006; Johnson et al., 2002).
A transfection assay of protoplasts of the snrk2.2/2.3/2.6 triple mutant background was developed to test the roles of several factors involved in ABA signal transduction, including an ABA-dependent ABF (ABF2), SnRK2s, PP2Cs, and PYR/PYL/RCARs (Fujii et al., 2009). In these protoplasts, the co-transfection of DNAs encoding ABF2 and any of the three SnRK2s induced the transient expression of a luciferase (LUC) reporter driven by the ABA-responsive RD29B promoter, in an ABA-dependent manner. LUC expression was blocked by the co-transfection of DNA encoding PP2C proteins ABI1 or HAB1, while the additional co-transfection with almost any of the PYR/PYL/RCAR members restored RD29B-LUC expression. In this same study, bimolecular fluorescence complementation analysis demonstrated physical interactions between ABI1 and SnRK2s in vivo, which were localized in the cytoplasm and in the nucleus of tobacco cells; these results were supported by Y2H experiments. In vitro kinase assays indicated that SnRK2.6 (also named OST1; Mustilli et al., 2002) undergoes autophosphorylation and can phosphorylate an ABF2 fragment. In these assays, PP2Cs were shown to inhibit both SnRK2-mediated phosphorylation of ABF2 and SnRK2 auto-phosphorylation, effects that could be blocked by the addition of different PYR/PYL/RCAR proteins. Yeast three hybrid assays established that PYR/PYL/RCAR protein PYL8 or PYL5 can disrupt physical interactions between SnRK2.6 and any of the PP2C proteins ABI1, ABI2 or HAB1, in an ABA-dependent manner.
The isolation, reassembly and manipulation of these signalling components thus demonstrated their interactions in vitro, in yeast and in protoplasts, showing that this minimal set of factors can reconstitute an ABA signal transduction pathway. Collectively, these results point to a model of a signal–receptor complex, in which PYR/PYL/RCAR proteins bind to ABA and thereby disrupt the physical interaction between PP2Cs and their target SnRK2s. In this way, the target kinases are released from intermolecular inhibition and are able to phosphorylate transcription factors such as ABF2, leading to the activation of ABA-responsive gene expression (Fig. 1).
Fig. 1.
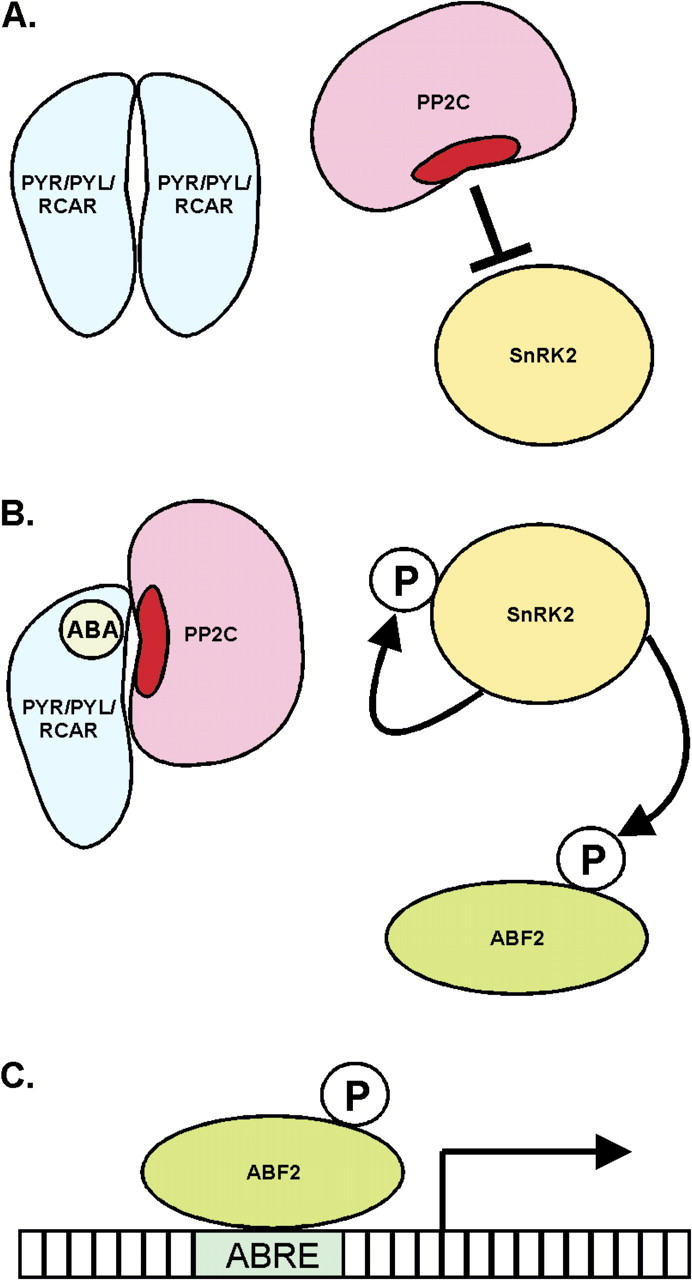
Schematic view of the ABA signal–receptor complex, including a PYR/PYL/RCAR homodimer (A) and hormone-bound protomer (B); a PP2C phosphatase, with the active site indicated in dark red; a SnRK2 kinase; and the ABF2 transcription factor. In the absence of ABA (A), the receptor forms a homodimer, while the PP2C inhibits both autophosphorylation of the SnRK2 and phosphorylation of ABF2. In the presence of ABA (B), a receptor protomer engulfs the hormone within a pocket, allowing the receptor to bind the PP2C and cover the phosphatase active site. This permits the autophosphorylation of the SnRK2 and phosphorylation of its ABF2 substrate. In its phosphorylated, active state (C), ABF2 binds to an ABA-responsive element (ABRE) in the promoter of ABA-responsive genes, activating transcription.
Receptor activity involved in a rapid ABA response
Several lines of evidence suggest that SnRK2s, in particular SnRK2.6, also directly regulate the most immediate responses to ABA, such as the production of reactive oxygen species (ROS) and the regulation of plasma membrane anion channels. SnRK2.6 has been shown to interact with and phosphorylate AtRBOHF (RESPIRATORY BURST OXIDASE HOMOLOG F), a NADPH oxidase involved in the ABA-triggered production of second messenger ROS (Kwak et al., 2003; Sirichandra et al., 2009). SLAC1 (SLOW ANION CHANNEL-ASSOCIATED 1) is a recently identified S-type anion channel on the plasma membrane of guard cells; its anion efflux activity causes membrane depolarization leading to stomatal closure (Vahisalu et al., 2008; Negi et al., 2008). These studies reported that Arabidopsis mutants deficient in SLAC1 fail to close stomata in response to CO2 and ABA. Geiger et al. (2009) recorded currents on whole Xenopus oocytes expressing SLAC1, SnRK2.6 and/or PP2Cs to show that S-anion channel activity, driven by SLAC1, could be activated only in the presence of SnRK2.6. Moreover, the co-expression of ABI1 or ABI2 inhibited the activation of SLAC1 by SnRK2.6. Furthermore, it was shown that SnRK2.6 can interact with SLAC1 and phosphorylate the N-terminal, cytoplasmic region of this channel protein. No phosphorylation could be detected when ABI1 protein was added to the reaction mixture, indicating that ABI1 inhibits SnRK2.6-mediated channel activation (Geiger et al., 2009). Similar results were obtained with the protein phosphatase PP2CA in place of ABI1 (Lee et al., 2009).
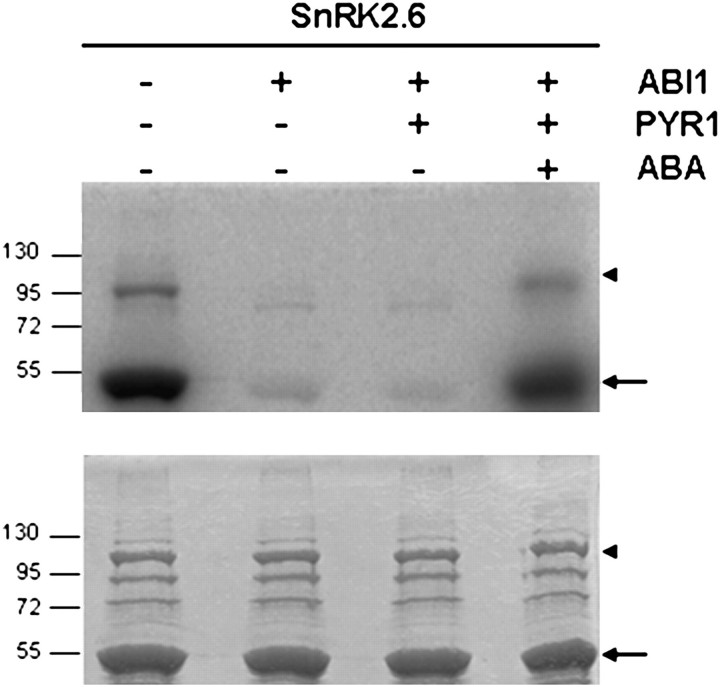
It is therefore conceivable that PYR/PYL/RCAR proteins control, through SnRK2.6 and related SnRK2s, not only the activation of gene expression but also the activity of proximate effectors of an ABA response. To test this hypothesis, the in vitro reconstitution assay developed by Fujii et al. (2009), was used but with the N-terminal fragment of SLAC1 (SLAC N) as a final substrate instead of ABF2 (Fig. 2). As expected from previous reports (Geiger et al., 2009; Lee et al., 2009), when SnRK2.6 was incubated with SLAC N the kinase was capable of phosphorylating the substrate. When SnRK2.6 was pretreated with ABI1, phosphorylation of the substrate was significantly reduced, and the addition of His-PYR1 in the absence of ABA could not restore SLAC N phosphorylation by SnRK2.6. However, the addition of 100 μM (±)-ABA to the pretreatment of SnRK2.6 with ABI1 and PYR1 could restore the ability of SnRK2.6 to phosphorylate SLAC N (Fig. 2). Our results suggest that the PYR/PYL/RCAR family of proteins controls not only the transcriptional response to ABA but also the transport activity of channels, thereby regulating the most immediate responses to stress signals mediated by ABA. Interestingly, a similar result was recently reported by Geiger et al. (2010), using RCAR1 (PYL9) instead of PYR1.
Fig. 2.
Regulation of SLAC1 phosphorylation status by the ABA-dependent PYR1, ABI1, SnRK2.6 signalling cascade. GST-SLAC1 N-terminal fragment (SLAC N, Met1 to Phe188) was incubated in the presence of [γ-32P]ATP with MBP-SnRK2.6 pretreated without (–) or with (+) GST-ABI1, His-PYR1, and 100 μM (±)-ABA. Bands of SLAC N fragment and MBP–SnRK2.6 are indicated by an arrow and an arrowhead, respectively. Coomassie-stained SDS-PAGE (bottom) and autoradiogram of the gel (top) are shown. Recombinant proteins and reaction conditions were as described previously (Fujii et al., 2009; Park et al., 2009). When SLAC N was incubated with SnRK2.6 not treated with GST-ABI1, both SnRK2.6 autophosphorylation and SLAC N phosphorylation bands were visualized (first lane). SnRK2.6 pre-treated with GST-ABI1 was unable to phosphorylate SLAC N (second lane). In the absence of ABA, the addition of His-PYR1 to the pretreatment of SnRK2.6 with GST-ABI could not restore SnRK2.6 phosphorylation activity on SLAC N (third lane). However, when ABA was added to the pretreatment reaction, SLAC N phosphorylation was recovered (fourth lane).
Crystal structures: pockets, gates, and latches
The discovery of PYR/PYL/RCAR proteins led to five crystallographic studies, published in a wave of reports in late 2009 (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009). Collectively, these groups derived crystal structures for three proteins of the 14-member family (Table 1). These studies support the role of PP2Cs as intimate players, if not co-receptors, in ABA perception. All five reports confirm the importance of the helix-grip fold in ABA binding, as anticipated from previous bioinformatic analyses. This structure forms a large pocket that, in the absence of ABA, remains open to the solvent. Bound ABA is completely contained within the pocket, with the carboxylic acid portion of the hormone in contact with a Lys residue at the innermost region. Upon binding, the PYR/PYL/RCAR protein undergoes a conformational shift that covers the hormone from the solvent. In all proteins studied, the β-strands of the helix-grip fold, the three α-helices, and two loops joining β-strands β3–β4 and β5–β6 were deemed critical for the formation of the binding pocket and the structures covering bound ABA from the exterior. ABA is buried by the folding over of the two loops that connect the two pairs of β-strands. These loops consist of highly conserved residues that serve as ‘lids’ (Nishimura et al., 2009; Miyazono et al., 2009; Santiago et al., 2009a; Yin et al., 2009) or, more descriptively, as a ‘gate’ and ‘latch’ corresponding to the β3–β4 and β5–β6 loops, respectively (Melcher et al., 2009; Fig. 1). The functional importance of residues within the binding pocket and within the gate and latch structures was confirmed by various in vitro experiments, and by transfecting snrk2.2/2.3/2.6 triple mutant protoplasts with PYL2 containing site-directed mutations in these domains (Melcher et al., 2009).
The folding of the β3–β4 loop forms a hydrophobic surface with which an individual PP2C (in these cases, ABI1 or HAB1) can interact. A highly conserved Trp residue in the PP2C protein inserts between the β3–β4 ‘gate’ and the β5–β6 ‘latch’ loops to interact, through a water-mediated H-bond, with the ketone group of ABA's cyclohexene ring, causing the PP2C protein to serve as a molecular ‘lock’ that stabilizes the closed position of the PYR/PYL/RCAR protein loops (Melcher et al., 2009). This interaction is observed in four of the five structural studies, but is interpreted somewhat differently by different groups. Melcher et al. (2009) consider this interaction as evidence that PP2Cs serve as ‘co-receptors’ that sense binding of ABA with PYR/PYL/RCAR proteins, while Yin et al. (2009) consider the PYR/PYL/RCAR protein as the sole receptor and eschew the co-receptor terminology, presumably because the hormone is buried completely by PYR/PYL/RCAR residues and interacts only indirectly, through a water molecule, with the PP2C. The binding of a PP2C protein to the ABA-bound PYR/PYL/RCAR covers the active site of the phosphatase, which explains the mechanism by which the PYR/PYL/RCAR acts as a competitive inhibitor of PP2C activity.
Another finding reported in four of the five crystal structure papers (Melcher et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009) is the formation of a homodimer between PYR/PYL/RCAR proteins in the absence of a PP2C protein. The homodimer interface encompasses the same region that participates in the interaction with PP2Cs (Fig. 1). The model arising from these observations is that the homodimer can exist with zero or one protomer binding ABA and, upon binding of an additional ABA molecule to the other protomer, the homodimer dissociates and allows the binding of a PP2C protein to an ABA-bound PYR/PYL/RCAR protein, forming a hormone–heterodimer complex.
Since all members of the PYR/PYL/RCAR family have highly conserved amino acid sequences in critical parts of the helix-grip fold structure, it appears likely that they generally share properties of ABA-binding and interaction with PP2Cs. One interesting exception may be PYL13, which has a Gln residue instead of the Lys common to all the other family members at a critical position. It is this Lys residue that contacts the hormone's carboxylic acid group at the pocket interior. Mutations that replace the Lys residue (K59 in PYR1; K64 in PYL2) eliminate ABA and PP2C binding to recombinant proteins (Melcher et al., 2009; Nishimura et al., 2009; Yin et al., 2009), consistent with the lack of evidence for ABA or PP2C binding with PYL13. The relation between PYL13 and its other family members remains unclear. At least two other residues in PYL13 differ at important locations, compared with all other paralogues: the Leu residue within the SGLPA ‘gate’ is replaced with Asp; and Glu/Asp in the GG(E/D)HRL ‘latch’ is replaced with Asn. Minor structural differences among PYR/PYL/RCAR proteins, within the binding pocket and at other critical domains, are likely to play roles in the different strengths of interaction that have been observed between the receptors and ABA stereoisomers, and between the receptors and different PP2C proteins. If one considers the family diversity of the PYR/PYL/RCARs, the PP2Cs and the SnRK2s, it is clear that the ABA signal–receptor complex could be subjected to a highly sophisticated level of combinatorial control. This diversity of structure might enable the functional plasticity that plants require throughout their development, and across the environmental conditions they encounter.
Ma et al. (2009) grouped the PYR/PYL/RCAR family into three subfamilies, based on amino acid sequence similarity. It is presently unclear whether these groupings relate to any functional specialization. Gene expression patterns vary among family members, according to the Arabidopsis eFP Brower (Winter et al., 2007; http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi), but these patterns do not appear to correlate, in any obvious way, with phylogenetic groupings within the family, nor do they correlate with the ABA-dependency of their interactions with PP2C proteins in yeast (Park et al., 2009). It remains to be seen whether patterns can be identified for the differences in properties among individual family members—at the level of ABA binding affinity, interaction with other components of the ABA signal–receptor complex(s), transcriptional control, protein turnover, subcellular localization, or cell-type specificity. It is possible that some of the PYR/PYL/RCARs bind to ABA biosynthetic intermediates and/or ABA metabolites.
Comparisons with receptors for other hormones
The discovery of ABA receptors coincides with other exciting developments in the field of phytohormone perception. Protein receptors for other phytohormones have been identified in recent years (Santner and Estelle, 2009), and interesting similarities and differences can be seen between these and the PYR/PYL/RCAR proteins that perceive ABA. The binding pocket and the gate–latch–lock structures of the PYR/PYL/RCAR-PP2C complex bear some resemblance to the binding features the gibberellin (GA) receptors GID1A and GID1 (gibberellin insensitive dwarf 1) of Arabidopsis and rice (Oryza sativa), respectively (Murase et al., 2008; Shimada et al., 2008). In both studies, the hydrophilic carboxylate portion of both the GA3 and the GA4 structural variants was shown to be directed at the inner, or bottom, part of the binding pocket of either receptor, while the hydrophobic, aliphatic rings are positioned near the pocket entrance. An N-terminal extension of GID1A (Murase et al., 2008) or GID1 (Shimada et al., 2008), which in each case contains α-helices, serves as a ‘lid’ that encloses the GA molecule when it enters the receptor pocket. In both studies, this movement of the lid appeared to alter the surface of the receptor, allowing it to interact with the Arabidopsis DELLA protein GAI (Murase et al., 2008), or with the rice DELLA homologue SLR1 (Shimada et al., 2008), which are negative regulators of GA hormone signalling (Peng et al., 1997; Ueguchi-Tanaka et al., 2007). These negative regulators are, therefore, analogous to the PP2Cs that inhibit ABA signalling. As with the PP2C proteins, the DELLA protein in the Arabidopsis GA-GID1A–GAI complex does not directly contact the receptor-bound hormone (Murase et al., 2008). Whether an analogous situation exists in rice was not explicitly reported by Shimada et al. (2008).
Aside from these similarities with GA receptors, in other respects, the PYR/PYL/RCAR receptor model of competitive inhibition of an inhibitor is unique. For example, this model diverges from a common theme for other hormone receptors, wherein hormone binding triggers the ubiquitination and proteolysis—via the 26S proteasome—of negative regulators of the hormones’ signal transduction pathways. This is the case for the GA-sensing GID1 family proteins that promote the proteolysis of DELLA transcriptional repressors (Griffiths et al., 2006), the auxin-sensing TIR1 and AFBs, F-box proteins that promotes the proteolysis of Aux/IAA transcriptional repressors (Dharmasiri et al., 2005), and the jasmonate-sensing COI1, another F-box protein that promotes the proteolysis of JAZ transcriptional repressors (Chini et al., 2007; Thines et al., 2007; Yan et al., 2009). Despite this distinction, ABA sensitivity in plants is known to be regulated by the 26S proteasome (Stone et al., 2006; Zhang et al., 2005). It is likely that one or more proteins of the ABA receptor–signal complex and its downstream targets are regulated by polyubiquitination and proteolysis.
Other putative ABA receptors
Lines of evidence for different sites of ABA perception, at the plasma membrane and within the cytoplasm, have been reported (Hamilton et al., 2000; Levchenko et al., 2005). Two separate groups have published reports of Arabidopsis proteins, localized at the cell periphery, that bind to ABA and meet other criteria consistent with roles as ABA receptors. The first report (Liu et al., 2007) identified a G-protein-coupled receptor (GPCR) homologue, GCR2, as a protein that interacts with GPA1. GPA1 is the sole, canonical G-protein α subunit encoded by the Arabidopsis genome (Jones and Assmann, 2004). Liu et al. (2007) reported that this association was disrupted specifically by the naturally occurring S-ABA, and that the binding properties of the hormone to GCR2 suggested a single binding site, with a KD (dissociation constant) value of 20.1 nM, consistent with a supposed physiological range for ABA. The authors presented a model in which the binding of ABA to GCR2 disrupts the Gαβγ heterotrimeric complex, leading to the activation of downstream ABA effectors. Consistent with this model, they reported ABA-related phenotypes for loss-of-function and overexpression of GCR2, at the level of seed germination, seedling growth, and stomatal aperture.
However, GCR2 has been controversial with respect to the reproducibility of ABA-related phenotypes (Gao et al., 2007; Guo et al., 2008), the unexpected ABA hyposensitivity of a GPCR mutant, based on previous studies of G-protein signalling during ABA responses (Chen et al., 2008), ABA binding properties (Risk et al., 2009), and, on the grounds that GCR2 lacks the prototypical seven transmembrane domains of GPCRs, and is, instead, homologous to mammalian lanthionine synthetase C (LanC)-like proteins (Johnston et al., 2007; Illingworth et al., 2008). Moreover, the apparent ease with which recombinant GCR2 was solubilized for in vitro analysis was one basis for questioning its identity as a transmembrane protein (Chen, 2008). Interestingly, the human LanC-like protein LANCL2 was recently shown to be required for ABA binding to the membranes of human granulocytes and rat insulinoma cells, where endogenous ABA is implicated in the control of inflammatory and diabetic responses (Sturla et al., 2009). Nevertheless, no published reports to date have independently reproduced the results of Liu et al. (2007) in Arabidopsis, nor in any other plant species.
In a more recent study (Pandey et al., 2009), a bioinformatic analysis of the Arabidopsis genome led the authors to focus on a pair of genes encoding unusual products with predicted features of both GPCRs and G-proteins. These authors reported that the GPCR-type G proteins GTG1 and GTG2 have nine predicted transmembrane domains, are localized at the plasma membrane, have specific GTP-binding and GTPase activities that are altered through interactions with GPA1, and can both bind specifically to the natural S-ABA stereoisomer in a saturable manner with KD values of approximately 20 nM, similar to the value reported for GCR2 by Liu et al. (2007). Pandey et al. (2009) also reported that an ABA hyposensitive phenotype in seed germination and seedling growth required a gtg1gtg2 double mutant, which could be complemented by introducing either of the wild-type genes, suggesting redundancy of the GTG function. The double mutant was also hyposensitive to ABA with respect to stomatal closure, although it displayed a normal phenotype with respect to ABA inhibition of stomatal opening. The model presented by Pandey et al. (2009) portrays an unusual type of G-protein signalling, in which the GTP-bound Gα protein ‘turns off’ signalling while the GDP-bound form allows ABA binding to the GTG receptor and the initiation of a signalling cascade. Curiously, this mode of guanine nucleotide regulation is the opposite of the conventional model for signalling by G-proteins. These authors were unable to show evidence for at least one binding site per GTG protein molecule, a result they attributed to non-optimal conditions for isolating pure and intact proteins for binding assays.
Another putative ABA receptor protein in Arabidopsis is CHLH (Mg-chelatase H subunit), a chloroplast protein involved in chlorophyll biosynthesis that has also been termed ABAR, for ABA receptor (Shen et al., 2006; Wu et al., 2009). This protein's role in the cell had previously been shown to extend beyond chlorophyll biosynthesis, since CHLH is the same as GUN5 (GENOMES UNCOUPLED 5), a regulator of plastid-to-nucleus retrograde signalling (Mochizuki et al., 2001). ABA binding by CHLH has been demonstrated by more than one technique, with KD values in the order of 30–40 nM (Shen et al., 2006; Wu et al., 2009). Experiments in which CHLH levels were modulated, through over-expression, RNAi, or through insertional and point mutations, showed ABA-related phenotypes at the levels of seed germination, seedling growth, and stomatal movements (Shen et al., 2006; Wu et al., 2009). Binding experiments with potential agonists and antagonists of the hormone showed that the affinity of CHLH is specific to S-ABA (Wu et al., 2009). The chloroplast is the site of initial steps in ABA biosynthesis, and it is possible that one branch of ABA perception and signal transduction is localized within this source organelle. Like GCR2, GUN5/CHLH has been controversial, in this case because a barley homologue failed to bind ABA or show ABA-related phenotypes when mutated (Muller and Hansson, 2009). Moreover, a major unanswered question, acknowledged by Wu et al. (2009), is the mechanism by which CHLH—a wholly unexpected type of receptor for ABA—transmits a signal upon ABA binding.
A critical difference between the reports of these putative ABA receptors—GCR2, GTG1/GTG2, CHLH—and those of the PYR/PYL/RCAR family is that the evidence for the former candidates has not been independently corroborated. Jones and Sussman (2009) proposed the application of strict biochemical criteria when evaluating the candidacy of a protein as a hormone receptor. It may be difficult for one or even two publications by a single research group to meet all of the desirable criteria these authors list. Perhaps the strongest case for the PYR/PYL/RCAR family is the sheer volume of data, comprising multiple lines of evidence, that has emerged from multiple groups in rapid succession. The crystallographic studies, the site-specific mutations, and the integration of PYR/PYL/RCAR function with other components of ABA signalling, through ligand binding and in vitro reconstitution assays, are the types of evidence that may need to accumulate for other candidate ABA receptors before they are widely accepted by the scientific community.
Applications in cultivated species
The challenges of genetic redundancy and multiple levels of regulation have previously hindered progress in understanding ABA perception mechanisms. Although a great number of questions about the integration of signals remain unanswered, the discovery of PYR/PYL/RCAR proteins has created a major front of progress in the elucidation of ABA function in higher plants. The complexity of this family, and of the families encoding its interacting proteins, point to a high capacity for combinatorial control by the plant to effect fine-tuning of ABA signal transduction in response to developmental and environmental cues. The great diversity among plant species in their environmental adaptations may result, to some degree, from the diversity of ABA receptor–signal complexes. Considering the abundant evidence of ABA-mediated cross-talk between biotic and abiotic stress responses (Fujita et al., 2006) it is intriguing that the Bet v 1-fold superfamily contains proteins involved in pathogen defence (e.g. PR-10) as well as ABA receptors. Future studies of this receptor family may clarify the connections between ABA-mediated responses to abiotic and biotic stressors, and lead to progress in both areas of plant stress research.
The PYR/PYL/RCAR family is well-conserved in crop species (Fig. 3), so it is likely that the modulation of these receptors and their interacting partners will enable new methods of enhancing crop tolerance to multiple types of stress. Research on these ABA receptors is already underway in non-model plant species. For example, a PP2C homologue from the beechnut tree (Fagus sylvatica L.) was shown to interact, in an ABA-dependent manner, with Arabidopsis PYR7 and PYR8, based on Y2H assays and bimolecular fluorescence complementation experiments in tobacco cells (Saavedra et al., 2010). The available genome sequences of several crops indicate levels of family diversity for the PYR/PYL/RCAR proteins similar to that of Arabidopsis (Table 2). The discovery of pyrabactin, a selective ABA agonist of PYR/PYL/RCAR proteins (Park et al., 2009), points to a potential chemical strategy for modulating ABA receptor activity in crops on a commercial scale (Cutler et al., 2010). Genetic methods of manipulating ABA perception, through conventional breeding or transgenic approaches, may also provide greater control of the hormone's function in the field. In conclusion, it appears likely that the ABA signal transduction model, as developed in Arabidopsis, will continue to create and facilitate new approaches for enhancing stress tolerance in crops.
Fig. 3.
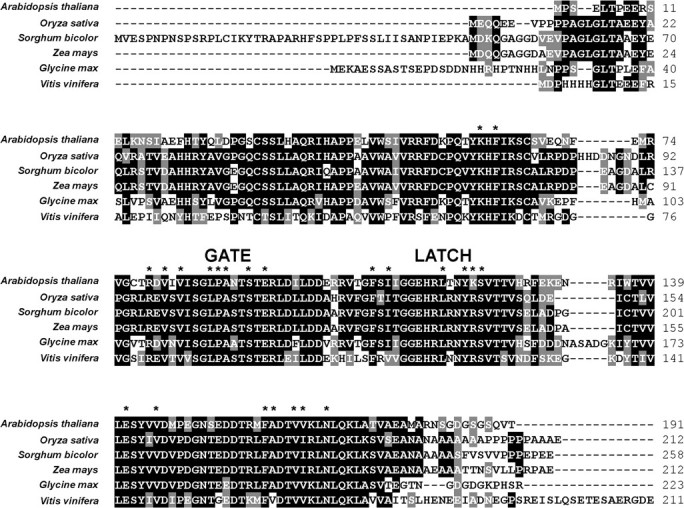
Amino acid sequence alignment of the Arabidopsis PYR1 protein with the most similar homologues in five cultivated species. The alignment was performed with the CLUSTALW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using the default settings. Asterisks indicate residues in contact with ABA hormone, according to the PYL2 crystal structure of Melcher et al. (2009). The locations of the gate and latch domains are indicated.
Table 2.
Numbers of genes homologous to PYR1 protein in seven cultivated species
| Species | Number of genes | Range of E-values | Highest % ID with PYR1 |
| Glycine max | 23 | –74 to –34 | 70.2 |
| Zea may | 20 | –56 to –37 | 61.5 |
| Populus trichocarpa | 14 | –78 to –37 | 75.1 |
| Oryza sativa | 11 | –57 to –33 | 58.7 |
| Vitis vinifera | 8 | –75 to –39 | 54.3 |
| Sorghum bicolor | 8 | –55 to –38 | 62.0 |
| Medicago truncatula | 6 | –51 to –39 | 54.1 |
Genes were identified by the BLAST algorithm with PYR1 protein as query, using the BLOSUM62 matrix and a word length equal to three.
Acknowledgments
The authors thank Viswanathan Chinnusamy for helpful comments on the manuscript and Hiroaki Fujii for advice on the in vitro reconstitution assay and for critical reading of the manuscript.
References
- Bruzzone S, Moreschi I, Usai C, Guida L, Damonte G, Salis A, Scarfi S, Millo E, De Flora A, Zocchi E. Abscisic acid is an endogenous cytokine in human granulocytes with cyclic ADP-ribose as second messenger. Proceedings of the National Academy of Sciences, USA. 2007;104:5759–5764. doi: 10.1073/pnas.0609379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YJ, Wei Q, Liao Y, Song HL, Li X, Xiang CB, Kuai BK. Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Reports. 2009;28:579–588. doi: 10.1007/s00299-008-0659-x. [DOI] [PubMed] [Google Scholar]
- Chen JG. Heterotrimeric G-protein signalling in Arabidopsis: puzzling G-protein-coupled receptor. Plant Signalling and Behavior. 2008;3:1042–1045. doi: 10.4161/psb.3.12.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Current Opinion in Plant Biology. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E. Integration of abscisic acid signalling into plant responses. Plant Biology. 2006;8:314–325. doi: 10.1055/s-2006-924120. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signalling network. Annual Review of Plant Biology. 2010 doi: 10.1146/annurev-arplant-042809-112122. (In press) [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Fernandes H, Pasternak O, Bujacz G, Bujacz A, Sikorski MM, Jaskolski M. Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin. Journal of Molecular Biology. 2008;378:1040–1051. doi: 10.1016/j.jmb.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signalling in seeds and seedlings. The Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DL, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen XM, Sela HA, Fahima T, Dubcovsky J. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323:1357–1360. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S-Y, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. The Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signalling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signalling in response to water stress in Arabidopsis. Plant and Cell Physiology. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences, USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YJ, Zeng QN, Guo JJ, Cheng J, Ellis BE, Chen JG. Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. The Plant Journal. 2007;52:1001–1013. doi: 10.1111/j.1365-313X.2007.03291.x. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signalling kinase-phosphatase pair. Proceedings of the National Academy of Sciences, USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proceedings of the National Academy of Sciences, USA. 2010 doi: 10.1073/pnas.0912030107. doi/10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zeng Q, Emami M, Ellis BE, Chen J-G. The GCR2 gene family is not required for ABA control of seed germination and early seedling development in Arabidopsis. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002982. e2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DWA, Hills A, Kohler B, Blatt MR. Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proceedings of the National Academy of Sciences, USA. 2000;97:4967–4972. doi: 10.1073/pnas.080068897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends in Plant Science. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Huang DQ, Jaradat MR, Wu WR, Ambrose SJ, Ross AR, Abrams SR, Cutler AJ. Structural analogs of ABA reveal novel features of ABA perception and signalling in Arabidopsis. The Plant Journal. 2007;50:414–428. doi: 10.1111/j.1365-313X.2007.03056.x. [DOI] [PubMed] [Google Scholar]
- Illingworth CJR, Parkes KE, Snell CR, Mullineaux PM, Reynolds CA. Criteria for confirming sequence periodicity identified by Fourier transform analysis: application to GCR2, a candidate plant GPCR? Biophysical Chemistry. 2008;133:28–35. doi: 10.1016/j.bpc.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins-Structure Function and Genetics. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiology. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Temple BR, Chen J-G, Gao Y, Moriyama EN, Jones AM, Siderovski DP, Willard FS. Comment on ‘A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid’. Science. 2007;318:914c. doi: 10.1126/science.1143230. [DOI] [PubMed] [Google Scholar]
- Jones AM, Assmann SM. Plants: the latest model system for G-protein research. EMBO Reports. 2004;5:572–578. doi: 10.1038/sj.embor.7400174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Sussman MR. A binding resolution. Plant Physiology. 2009;150:3–5. doi: 10.1104/pp.109.136606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiologia Plantarum. 1984;61:377–383. [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signalling in Arabidopsis. EMBO Journal. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signalling in plant guard cells. Proceedings of the National Academy of Sciences, USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. The Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko V, Konrad KR, Dietrich P, Roelfsema MRG, Hedrich R. Cytosolic abscisic acid activates guard cell anion channels without preceding Ca2+ signals. Proceedings of the National Academy of Sciences, USA. 2005;102:4203–4208. doi: 10.1073/pnas.0500146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL, Wang HJ, Wang JS, Zaharia LI, Abrams SR. Abscisic acid regulation of heterophylly in Marsilea quadrifolia L.: effects of R-(–) and S-(+) isomers. Journal of Experimental Botany. 2005;56:2935–2948. doi: 10.1093/jxb/eri290. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Ekramoddoullah AKM. The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiological and Molecular Plant Pathology. 2006;68:3–13. [Google Scholar]
- Liu XG, Yue YL, Li B, Nie YL, Li W, Wu WH, Ma LG. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. Journal of Molecular Biology. 2003;325:123–133. doi: 10.1016/s0022-2836(02)01197-x. [DOI] [PubMed] [Google Scholar]
- McCourt P, Creelman R. The ABA receptors: we report you decide. Current Opinion in Plant Biology. 2008;11:474–478. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng L-M, Zhou XE, et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Miyazono K-I, Miyakawa T, Sawano Y, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proceedings of the National Academy of Sciences, USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AH, Hansson M. The barley magnesium chelatase 150-kD subunit is not an abscisic acid receptor. Plant Physiology. 2009;150:157–166. doi: 10.1104/pp.109.135277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T-p, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. A screen for genes that function in abscisic acid signalling in Arabidapsis thaliana. Genetics. 2002;161:1247–1255. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshi A, Nito K, et al. PYR/PYL/RACR family members are major in vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. The Plant Journal. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo R, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signalling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signalling in Arabidopsis seed. The Plant Journal. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- Oritani T, Kiyota H. Biosynthesis and metabolism of abscisic acid and related compounds. Natural Product Reports. 2003;20:414–425. doi: 10.1039/b109859b. [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G Proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak O, Bujacz GD, Fujimoto Y, Hashimoto Y, Jelen F, Otlewski J, Sikorski MM, Jaskolski M. Crystal structure of Vigna radiata cytokinin-specific binding protein in complex with zeatin. The Plant Cell. 2006;18:2622–2634. doi: 10.1105/tpc.105.037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes and Development. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. START: a lipid binding domain in StAR, HD-ZIP and signalling proteins. Trends in Biochemical Sciences. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- Radauer C, Lackner P, Breiteneder H. The Bet v. 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evolutionary Biology. 2008;8 doi: 10.1186/1471-2148-8-286. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risk JM, Day CL, Macknight RC. Reevaluation of abscisic acid-binding assays shows that G-Protein-Coupled Receptor 2 does not bind abscisic acid. Plant Physiology. 2009;150:6–11. doi: 10.1104/pp.109.135749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra X, Modrego A, Rodriguez D, Gonzalez-Garcia MP, Sanz L, Nicolas G, Lorenzo O. The nuclear interactor PYL8/RCAR3 of the Fagus sylvatica FSPP2C1 is a positive regulator of ABA signalling in seeds and stress. Plant Physiology. 2010;152:133–150. doi: 10.1104/pp.109.146381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. The Plant Journal. 2004;37:354–369. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park S-Y, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009a;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park S-Y, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. The Plant Journal. 2009b;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene L. Plant PP2C phosphatases: emerging functions in stress signalling. Trends in Plant Science. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Letters. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- Sondheim E, Galson EC, Chang YP, Walton DC. Asymmetry, its importance to action and metabolism of abscisic acid. Science. 1971;174:829–831. doi: 10.1126/science.174.4011.829. [DOI] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signalling. The Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturla L, Fresia C, Guida L, et al. LANCL2 is necessary for abscisic acid binding and signalling in human granulocytes and in rat insulinoma cells. Journal of Biological Chemistry. 2009;284:28045–28057. doi: 10.1074/jbc.M109.035329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. The Plant Journal. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. The Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. The Plant Journal. 2009;57:1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. New developments in abscisic acid perception and metabolism. Current Opinion in Plant Biology. 2007;10:447–452. doi: 10.1016/j.pbi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Walker-Simmons MK, Anderberg RJ, Rose PA, Abrams SR. Optically pure abscisic-acid analogs: tools for relating germination inhibition and gene-expression in wheat embryos. Plant Physiology. 1992;99:501–507. doi: 10.1104/pp.99.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000718. e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, et al. The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signalling: new evidence in Arabidopsis. Plant Physiology. 2009;150:1940–1954. doi: 10.1104/pp.109.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signalling by PYL proteins. Nature Structural and Molecular Biology. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-Hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signalling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiology. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signalling in Arabidopsis. Plant and Cell Physiology. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. The Plant Cell. 2008;20:1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XR, Garreton V, Chua NH. The AIP2 E3 ligase acts as a novel negative regulator of ABA signalling by promoting ABI3 degradation. Genes and Development. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]