Abstract
Conspecific pollen precedence can be a strong reproductive barrier between polyploid and diploid species, but the role of genome multiplication in the evolution of this barrier has not been investigated. Here, we examine the direct effect of genome duplication on the evolution of pollen siring success in tetraploid Chamerion angustifolium. To separate the effects of genome duplication from selection after duplication, we compared pollen siring success of synthesized tetraploids (neotetraploids) with that of naturally occurring tetraploids by applying 2x, 4x (neo or established) or 2x + 4x pollen to diploid and tetraploid flowers. Seed set increased in diploids and decreased in both types of tetraploids as the proportion of pollen from diploid plants increased. Based on offspring ploidy from mixed-ploidy pollinations, pollen of the maternal ploidy always sired the majority of offspring but was strongest in established tetraploids and weakest in neotetraploids. Pollen from established tetraploids had significantly higher siring rates than neotetraploids when deposited on diploid (4xest = 47.2%, 4xneo = 27.1%) and on tetraploid recipients (4xest = 91.9%, 4xneo = 56.0%). Siring success of established tetraploids exceeded that of neotetraploids despite having similar pollen production per anther and pollen diameter. Our results suggest that, while pollen precedence can arise in association with the duplication event, the strength of polyploid siring success evolves after the duplication event.
Keywords: polyploidy, neotetraploid, pollen size, pollen precedence, fireweed
1. Introduction
Polyploidy, or whole genome multiplication, is considered an important mechanism of plant speciation. It has occurred recurrently throughout the diversification of plants [1–3] and is associated with 15 per cent of all speciation events in flowering plants and 23 per cent in ferns [4]. Reproductive isolation between polyploids and diploids is commonly attributed to strong postzygotic isolation, which arises as a direct result of differences in chromosome number [5]. However, polyploids also differ phenotypically from diploids in ways that enhance assortative mating and prezygotic reproductive isolation [6,7]. For example, polyploids may differ from diploids with respect to spatial distribution [8–10], flowering time [11], pollinator visitation [12,13] and pollen precedence [14,15]. Where multiple reproductive barriers have been quantified for the same species, prezygotic mechanisms contribute most to total isolation [7].
The mechanism by which prezygotic barriers evolve in polyploids is not known. Phenotypic differences between polyploids and their progenitors are presumed to arise in concert with genome duplication [16]. However, most reports of phenotypic divergence are unable to distinguish the effects of genome duplication from selection operating afterwards. Recent studies involving synthesized polyploids (neopolyploids) suggest that genome duplication may not account for the full difference in reproductive phenotypes between naturally occurring diploids and polyploids. For example, time of flowering in synthesized tetraploids of Chamerion angustifolium [17] and Dactylis glomerata [18] is more similar to the diploid progenitors than established tetraploids. This raises the question of the timing of divergence in reproductive traits relative to the duplication event and the role of selection in polyploid speciation.
Conspecific gamete precedence [19] is an important yet poorly understood mechanism of assortative mating in plants. It operates after pollination when conspecific male gametes have a siring advantage over heterospecific gametes. This phenomenon has been documented within numerous plant genera [20–22] although the strength of conspecific pollen precedence often differs between species (i.e. is asymmetrical). The underlying mechanisms that favour rapid growth of conspecific pollen over heterospecific are not fully understood but involve pollen–pistil interactions [23] or traits intrinsic to the pollen that influence resources and directionality of pollen tubes. The only polyploid system in which post-pollination, prezygotic barriers have been examined is C. angustifolium and, in that case, tetraploids have a uniform siring advantage over diploids regardless of maternal ploidy [15]. This pattern is consistent with there being an intrinsic attribute of pollen, such as size or energy reserves, that uniformly enhances pollen germination and tube growth.
Chamerion angustifolium L. Holub (Onagraceae) is a herbaceous perennial with a circumpolar distribution, most often occurring in open and disturbed habitats. In North America and Asia individuals are either diploid (2n = 2x = 36) or autotetraploid (2n = 4x = 72) [11,24]. The strength of reproductive isolation between ploidy states is comparable to many recognized species [25]. Compared with diploids, tetraploids have delayed flowering, receive a disproportionate number of bee visits in mixed-ploidy populations and exhibit a pollen siring advantage in mixed-ploidy pollinations [15]. This siring advantage makes C. angustifolium a useful model for studying the evolution of homoploid pollen precedence and, more generally, conspecific pollen precedence. The degree to which the siring advantage is caused by genome duplication itself or to selective divergence subsequent to the duplication is unknown.
Here, we examine the role of genome duplication in the evolution of post-pollination, prezygotic barriers in polyploid C. angustifolium. We examine the siring success of pollen from diploids and tetraploids in single and mixed-ploidy experimental pollinations. To differentiate between the effects of genome duplication and selection on siring success, we compare established tetraploids (4xest) with experimentally synthesized neotetraploids (4xneo), which have experienced little selection. We ask four specific questions: (i) is the siring rate of established tetraploids and neotetraploids greater than that of diploids?, (ii) does pollen precedence depend on the ploidy of the maternal plant?, (iii) is the pollen siring rate of neotetraploids the same as that of established tetraploids?, and (iv) are differences in siring success between neotetraploids and established tetraploids related to differences in pollen size or pollen production? If genome duplication is responsible for pollen precedence in tetraploids, then neotetraploids should exhibit the same pollen characteristics and siring rate as established tetraploids when in competition with pollen from diploids.
2. Material and methods
(a). Parent plants
Diploid plants were derived from 46 seed families collected in two populations from the Canadian Rocky Mountains: Marmot Basin (lat. 52°48.176′ N, long. 118°04.700′ W) and Rampart Creek (lat. 52°02.498′ N, long. 116°51.835′ W). Established tetraploids (4xest) came from 45 seed families collected at Rampart Creek and Moose Meadows (lat. 51°11.571′ N, long. 115°44.515′ W).
Neotetraploid plants (4xneo) were generated by treating diploids with colchicine. Diploid seeds were collected from four populations within a 250 km radius: Coleman Clearcut (lat. 52°18.540′ N, long. 114°36.784′ W), Mount Norquay (lat. 51°12.240′ N, long. 118°04.700′ W), Continental Divide (lat. 51°13.673′ N, long. 116°02.910′ W), and Fortress Mountain (lat. 50°49.004′ N, long. 115°11.670′ W). Seeds were germinated in Petri dishes on moist filter paper under 16 h, 24°C days and 8 h, 22°C nights. Four-day old seedlings were bathed in 0.02 per cent (m/v) colchicine for 18 h, rinsed with deionized water and planted into 5 : 1 Sunshine mix (Sun Grow, Vancouver, British Columbia) and Turface (Beacon Athletics, Middleton, WI, USA) in 2.8 l pots. Leaf tissue was collected from treated plants and screened for DNA content using flow cytometry (see below). Plants with twice the DNA content as known diploids were considered tetraploid, and crossed in randomly selected pairs to produce nine independent families of first generation neotetraploid seed.
Seeds of diploids, established tetraploids and neotetraploids were grown to flowering in a greenhouse. Seeds were first germinated in Petri dishes, as before. After 10 days, seedlings were transplanted into 2.8 l pots. In total, we transplanted 60 diploid plants from 31 families, 46 neotetraploid plants from nine families and 55 established tetraploid plants from 42 families. Two months after planting, the ploidy of all parent plants was confirmed using flow cytometry. Leaves were finely chopped with a razor blade in Galbraith's buffer [26], passed through a 30 µm filter and then centrifuged for 6 min at 1000 rpm. The supernatant was removed and the pellet resuspended in 0.5 ml Galbraith's buffer with 50 µg ml−1 of the nucleic acid specific fluorochrome, propidium iodide, and 50 µg ml−1 of RNase. Samples were stained for at least 20 min. DNA content was estimated using a BD Biosciences FACSCalibur Flow Cytometer (BD Biosciences, San Jose, USA). The FL2 detector (585/42nm) was used to measure relative fluorescence, and the parameter FL2-area (integrated fluorescence) was used to quantify DNA content. Over 1000 nuclei were measured per sample. Mean fluorescence and coefficients of variation (CV) for each sample were measured using Modfit LT software (Verity Software House, Inc.). Chamerion angustifolium plants of known ploidy were run as an external reference to assign ploidy to each sample. Previous research [17,27] indicates that DNA content of 2x and 4x plants are sufficiently distinct to identify ploidy reliably using an external standard. Over the duration of this study 68.3, 69.1 and 24.4 per cent of diploids, established tetraploids and neotetraploids, respectively, developed to flowering.
(b). Experimental pollinations
We pollinated flowers of diploid, neotetraploid and established tetraploid plants with pollen from either a single-ploidy donor (2x, 4xneo or 4xest) or a 1 : 1 diploid-tetraploid mixture (2x + 4xest or 2x + 4xneo) (table 1). Donor and recipient plants were randomly chosen from the available pool of flowering plants each day. Recipient flowers were emasculated prior to pollination and pollinated with four anthers worth of pollen (two anthers per pollen donor for mixed pollinations). Each of the nine single donor cross types was replicated with a minimum of five different parental combinations, while the four mixed-ploidy crosses were replicated with a minimum of 13 independent parental combinations for a total of 135 crosses (table 1). Fruits were collected when mature and stored in dust-free silica gel at 4°C.
Table 1.
Observed proportions of diploid (2x), triploid (3x), tetraploid (4x), pentaploid (5x) and hexaploid (6x) offspring from single-ploidy and mixed-ploidy pollinations of diploid (2x), established tetraploid (4xest) and neotetraploid (4xneo) Chamerion angustifolium. Expected ploidy levels in the offspring based on the assumption of reduced gamete production are indicated in italic.
| maternal ploidy | pollen donor ploidy | offspring ploidy |
no. of seeds | no. of crosses | ||||
|---|---|---|---|---|---|---|---|---|
| 2x | 3x | 4x | 5x | 6x | ||||
| 2x | 2x | 0.947 | — | 0.053 | — | — | 57 | 8 |
| 2x | 4xest | — | 1.000 | — | — | — | 39 | 8 |
| 2x | 4xneo | 0.071 | 0.714 | 0.190 | — | 0.024 | 42 | 6 |
| 4xest | 2x | — | 0.684 | 0.316 | — | — | 57 | 10 |
| 4xneo | 2x | — | 0.857 | 0.095 | 0.048 | — | 42 | 7 |
| 4xest | 4xest | — | 0.026 | 0.974 | — | — | 78 | 10 |
| 4xneo | 4xneo | — | — | 1.000 | — | — | 57 | 8 |
| 4xest | 4xneo | 0.022 | — | 0.978 | — | — | 45 | 5 |
| 4xneo | 4xest | — | — | 1.000 | — | — | 45 | 5 |
| 2x | 2x−4xest | 0.585 | 0.341 | 0.057 | 0.016 | — | 123 | 13 |
| 2x | 2x−4xneo | 0.817 | 0.183 | — | — | — | 153 | 15 |
| 4xest | 2x−4xest | — | 0.029 | 0.929 | 0.036 | 0.007 | 140 | 14 |
| 4xneo | 2x−4xneo | 0.056 | 0.208 | 0.736 | — | — | 144 | 16 |
(c). Seed set
We measured seed set for five replicates of each single pollen-donor cross type and 10 replicates for each mixed-ploidy treatment as the percentage of ovules that developed into mature seeds per fruit. Seeds were classified as mature when they were plump and had a fully developed seed coat [28]. Total ovule number was measured as the sum of mature seed, immature (aborted) seed and unexpanded ovules (which includes mostly unfertilized ovules).
(d). Offspring ploidy
Offspring ploidy was determined by estimating DNA content of seeds using flow cytometry. Seeds were rehydrated for a minimum of 1 h in deionized water. For efficiency, two seeds from the same mixed-ploidy cross (i.e. same fruit) or three seeds from the same single-ploidy cross were chopped simultaneously in 40 µl LBO1 buffer [29] following procedures above. When multiple fluorescence peaks were identified from a single-ploidy cross, the peak corresponding to an unexpected ploidy was attributed to one seed, and the expected ploidy to two seeds. Seed ploidy was calculated for each sample as the DNA content for the seed, divided by the daily average 1C value (DNA content of a single chromosome set). In total, ploidy was determined for 1023 seeds from all crosses (table 1).
Seed ploidy from single donor crosses was used to determine the expected distribution of offspring ploidies for the mixed-ploidy pollinations. Assuming that all gametes are reduced (i.e. half the somatic number), then between-ploidy crosses should yield strictly triploid offspring. However, previous studies have found triploid and tetraploid offspring in 2x × 2x (maternal parent is listed first) crosses [30], probably reflecting a low rate of unreduced gametes. The expected ploidy distributions in offspring from mixed-ploidy pollinations, under random siring success, were calculated as the mean of the observed ploidy distributions of single-ploidy crosses (table 1). For example, the expected proportion of 2x offspring in the 2x × (2x + 4x) cross treatment is equal to the mean frequency of 2x offspring from 2x × 2x and 2x × 4x crosses.
The deviation between observed and expected offspring ploidy distributions was tested with a G goodness-of-fit test with Williams' correction [31]. Offspring with ploidies other than 2x, 3x and 4x were pooled into a separate ‘rare ploidy’ group. When no rare ploidies were expected (because no rare ploidies were observed in the corresponding single-ploidy crosses), the ‘rare’ ploidy class was not included in the G-test and the relative frequencies for the remaining categories were recalculated. The distribution of offspring ploidy in crosses with 4xest pollen was compared with that of 4xneo pollen using a contingency analysis.
Measured siring rates based on seed ploidy may deviate from the actual siring patterns at the time of fertilization due to differential mortality of embryos formed through within- and between-ploidy crosses. We estimated the corrected frequency of offspring of a given ploidy at fertilization by adjusting seed frequencies using [32],
| 2.1 |
where b is the rate of between-ploidy fertilization, bs is the rate of between-ploidy mating measured at the seed stage and wb is the relative seed fitness of triploids based on single donor crosses. To calculate b for the 2x × 2x + 4xneo cross, relative triploid fitness was based on seed set in 2x × 4xneo crosses relative to that of 2x × 2x crosses (i.e. bs = 0.18). Subsequently, recalculated siring rates of 2x and 4x were adjusted to sum to one within each pollination. This correction was applied to the observed and expected siring rates in the mixed pollen-donor pollinations to better estimate siring rate isolated from the confounding effect of high triploid embryo mortality. In these corrected data ‘rare’ ploidy categories were excluded.
(e). Pollen diameter and number
Pollen diameter and number of viable pollen grains per anther were estimated for 19 diploid, 18 established tetraploid, and 13 neotetraploid plants using a Multisizer 3 Coulter Counter (Beckman Coulter Inc.). Mature anthers were collected and stored in 0.2 ml of 70 per cent ethanol at 4°C. After evaporating the ethanol, dehisced anthers were suspended in 7 ml of Coulter Counter diluent and 0.1 ml tween. Samples were vortexed for more than 10 s to separate pollen from anther sac and filament tissue. Two 1 ml subsamples from each 7 ml sample were run. Visual inspection on a brightfield microscope indicates that viable C. angustifolium pollen grains have a size distribution of 60–105 µm [13]. Particle counts and size distributions within this range were subtracted from a null distribution acquired by running sterile anthers with no visible pollen production. Pollen number and diameter from the two subsamples were averaged and then multiplied by 7 to estimate pollen number per anther.
3. Results
(a). Seed production
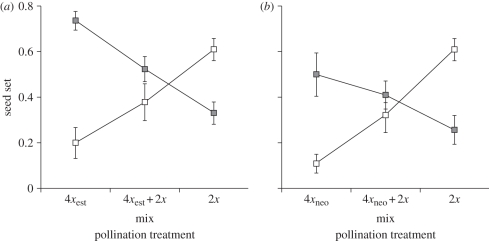
On average, seed set did not differ among pollination treatments (4x, 2x, 2x + 4x) and was weakly significantly different between maternal ploidy (2x < 4x) and between types of tetraploid (4xest > 4xneo) (table 2; figure 1). The maternal ploidy × pollination treatment interaction was strongly significant (table 2), reflecting increased seed set in diploids, and decreased seed set in tetraploids, with increasing proportions of diploid pollen (figure 1). The three-factor interaction was not significant (table 2).
Table 2.
Analysis of variance (ANOVA) of seed set for pollinations between diploid and established tetraploids and diploid and neotetraploids of Chamerion angustifolium. The independent variables are: maternal cytotype (2x or 4x), pollination treatment (2x, 4x or 2x + 4x mixture), and tetraploid type (4xest or 4xneo).
| source of variation | d.f. | d.f. error | F | p |
|---|---|---|---|---|
| maternal ploidy | 1 | 68 | 4.36 | 0.04 |
| pollination treatment | 2 | 68 | 0.71 | 0.50 |
| tetraploid type | 1 | 68 | 5.06 | 0.03 |
| maternal ploidy × pollination treatment | 2 | 68 | 23.78 | <0.001 |
| maternal ploidy × tetraploid type | 1 | 68 | 1.18 | 0.28 |
| pollination treatment × tetraploid type | 2 | 68 | 0.64 | 0.53 |
| maternal ploidy × pollination treatment × tetraploid type | 2 | 68 | 0.11 | 0.90 |
Figure 1.
Mean (±s.e.) seed set in (a) diploid (open) and established tetraploid (black) and (b) diploid (open) and neotetraploid (grey) Chamerion angustifolium flowers after pollination with either tetraploid pollen (4xest or 4xneo), diploid–tetraploid pollen mixtures or diploid pollen.
In a separate ANOVA of all possible cross combinations between established tetraploids and neotetraploids, the effect of maternal tetraploid type and the interaction between maternal and paternal type on seed set was not significant (p = 0.27). The difference between tetraploid types as pollen donors approached significance (4xest > 4xneo; p = 0.05).
(b). Siring rates
The majority of offspring produced in experimental pollinations were of the ploidy predicted when all gametes are reduced (table 1). In the within-ploidy pollination treatments, 95–100% of the resulting offspring were of the predicted ploidy. Between-ploidy pollinations produced more variable results. Offspring from 2x × 4xest pollinations were all triploid, as expected, whereas the frequency of triploids in 2x × 4xneo, 4xest × 2x, and 4xneo × 2x crosses was 71, 68 and 86 per cent, respectively. Over all, 6.8 per cent of seeds had a ploidy consistent with unreduced gametes.
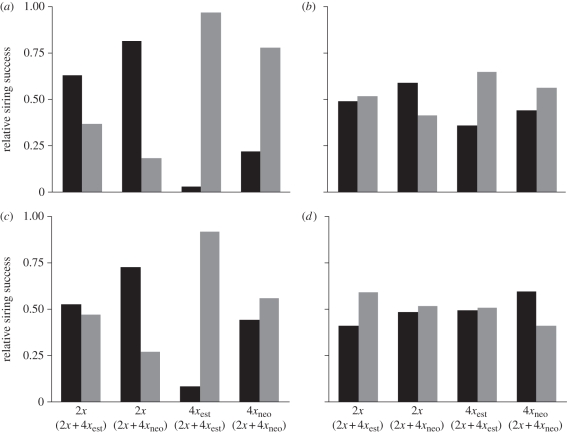
In mixed-ploidy pollinations, the siring success of tetraploid (neo or established) and diploid plants deviated significantly from random expectations, in both uncorrected and corrected data (table 3). Based on the uncorrected distributions of offspring ploidy, pollen of the maternal ploidy (homoploid pollen) had the highest siring rate regardless of maternal ploidy (table 1). For 2x × 2x + 4xest pollinations, pollen from diploids sired 59 per cent of seed; for 2x × 2x + 4xneo mixtures, pollen from diploids sired 82 per cent of seed. Established tetraploids sired 93 per cent of seed in 4xest × 2x + 4xest crosses and neotetraploids sired 74 per cent of seeds in 4xneo × 2x + 4xneo crosses. Overall, homoploid pollen sired 77 per cent of all seeds per fruit.
Table 3.
Goodness-of-fit tests (with Williams correction) comparing observed seed ploidy distributions from mixed-ploidy pollinations (2x + 4xest or 2x + 4xneo) of Chamerion angustifolium to random expectations. The analyses were based on raw counts (table 1) and on data corrected for early triploid mortality.
| maternal ploidy | pollen mixture | uncorrected |
corrected |
||||
|---|---|---|---|---|---|---|---|
| d.f. | χ2 | p | d.f. | χ2 | p | ||
| 2x | 2x + 4xest | 2 | 16.77 | 0.0002 | 1 | 6.44 | 0.0112 |
| 2x + 4xneo | 1 | 36.63 | <0.0001 | 1 | 19.18 | <0.0001 | |
| 4xest | 2x + 4xest | 1 | 85.98 | <0.0001 | 1 | 106.6 | <0.0001 |
| 2x + 4xneo | 2 | 32.81 | <0.0001 | 1 | 12.61 | 0.0004 | |
After correcting for elevated mortality of triploid embryos, the siring patterns were qualitatively similar to those for uncorrected data. Regardless of maternal ploidy, homoploid pollen sired a disproportionate number of seeds (table 3). On diploid recipients, diploids sired 52.8 per cent of seed (47.2% by tetraploids) in 2x + 4xest mixtures and 72.9 per cent in 2x + 4xneo mixtures (27.1% by neotetraploids). Established tetraploids sired 91.8 per cent of seeds on established tetraploid recipients and neotetraploids sired 56.0 per cent on neotetraploid recipients (figure 2). Homoploid pollen sired 68.4 per cent of seed, overall. For 2x + 4xest pollinations, the magnitude of homoploid pollen precedence was statistically greater for established tetraploids (i.e. on 4x maternal parents) than it was for diploids (i.e. on diploid maternal parent; G = 128.5, d.f. = 2, p < 0.0001; table 3). In 2x + 4xneo crosses, homoploid pollen precedence was greater for diploids than it was for neotetraploids (G = 63.6, d.f. = 2, p < 0.0001). Across all mixed pollinations, pollen from established tetraploids had higher siring success (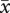 = 69.5%) than pollen from neotetraploids (
= 69.5%) than pollen from neotetraploids (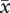 = 41.5%) or diploids (
= 41.5%) or diploids (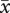 = 44.5%).
= 44.5%).
Figure 2.
Relative siring success of pollen from diploid (dark bars) and tetraploid (light bars) C. angustifolium for four different mixed-ploidy crosses (e.g. in 2x × (2x + 4x), 2x refers to ploidy of maternal plant; 2x + 4x refers to pollen parent). Tetraploids are either established tetraploids (4xest) or neotetraploids (4xneo). (a) and (b) depict observed and expected (random) siring rates, respectively, based on the raw distribution of ploidy in seeds. (c)and (d) depict observed and expected siring rates after correction for differences in mortality of 2x, 3x and 4x embryos.
Siring success of neotetraploids in 2x + 4xneo pollinations was significantly lower than the success of established tetraploids in 2x + 4xest crosses. This was true for pollen mixtures applied to diploid plants (G = 20.8, d.f. = 2, p < 0.0001) and tetraploid plants (G = 84.8, d.f. = 2, p < 0.0001).
(c). Pollen diameter and number
Mean number of pollen grains per anther for diploid (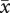 = 522.9), established tetraploid (
= 522.9), established tetraploid (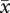 = 594.8) and neotetraploid (
= 594.8) and neotetraploid (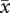 = 557.8) plants did not differ significantly (F2,45 = 0.15, p = 0.87). Pollen diameter of diploids (
= 557.8) plants did not differ significantly (F2,45 = 0.15, p = 0.87). Pollen diameter of diploids (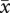 = 88.7 µm, s.e. = 2.41) was 15.2 per cent smaller than pollen of established tetraploids (
= 88.7 µm, s.e. = 2.41) was 15.2 per cent smaller than pollen of established tetraploids (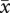 = 104.6 µm, s.e. = 3.49) and 17.3 per cent smaller than neotetraploid pollen (
= 104.6 µm, s.e. = 3.49) and 17.3 per cent smaller than neotetraploid pollen (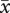 = 107.3 µm, s.e. = 5.63; F2,49 = 6.0, p = 0.0047). Pollen of established tetraploids and neotetraploids did not differ.
= 107.3 µm, s.e. = 5.63; F2,49 = 6.0, p = 0.0047). Pollen of established tetraploids and neotetraploids did not differ.
4. Discussion
In mixed-ploidy pollinations, pollen from diploid, tetraploid and neotetraploid C. angustifolium sire the majority of seeds on plants of their own ploidy. The presence of homoploid pollen precedence remains after accounting for elevated embryo mortality in triploid seed. A previous study of C. angustifolium [15] found that such variation in pollen competitive ability between diploids and tetraploids was mediated by differences in pollen tube number and pollen tube growth rate, although neotetraploids were not examined. Here, we confirm that homoploid pollen precedence occurs in neotetraploids as well as established tetraploids, suggesting that the minimum cellular and molecular mechanisms necessary for siring success are achieved immediately through the process of genome duplication. How genome duplication influences the physical and molecular matching between pollen and stylar tissues to induce homoploid pollen precedence in new polyploids is unclear. Insights from plants with self-incompatibility systems suggest that genome duplication may influence pollen tube growth loci by altering gene dosage, gene expression and allelic interactions within 2x pollen [33]. By contrast, studies of pollen size [34–36] suggest a simpler resource explanation for the success of (large) pollen from neotetraploids on neotetraploid pistils.
Although pollen precedence is observed in all ploidy categories, the magnitude of siring success is unequal among ploidies, as has been observed for non-polyploid species [37]. In mixed-ploidy pollinations, established tetraploids sire 91.9 per cent of seed on plants of their own ploidy, whereas neotetraploids sire 56 per cent and diploids sire 62.6 per cent. Across all pollen recipients, established tetraploids have more than 25 per cent higher siring success than either diploids or neotetraploids. The difference in siring success between established tetraploids and neotetraploids indicates that the evolution of pollen–pistil interactions and siring success is not solely the product of genome duplication. Rather, we conclude that selection after duplication is necessary for full pollen siring success to be achieved. Selection may promote the high siring success of tetraploids through direct interactions between diploids and tetraploids. Alternatively, the pollen performance of tetraploids may be a byproduct of selection for pollen that performs well on the longer styles of tetraploid pistils.
In our study, pollen from established tetraploids sired 47 per cent of the seed of diploid recipients, which is nearly as strong as the siring success of pollen from diploids (53%). This relatively high between-ploidy siring rate was similar to a previous study [15], although in that case tetraploids actually sired the majority of seeds on diploid recipients (unilateral pollen precedence). The difference in results between these studies may reflect differences in the source of plant material used in the two studies and geographical variation in pollen performance. It is noteworthy that much of the diploid and tetraploid seed in this study was collected from mixed-ploidy populations, whereas the previous study relied strictly on single-cytotype populations. It is possible that reinforcement selection in mixed-ploidy populations has strengthened homoploid pollen precedence in diploids as well as tetraploids. A controlled comparison of pollen in single and mixed-ploidy populations is necessary to test this hypothesis.
Previous studies have suggested that pollen-specific traits, such as large size, can confer a siring advantage by having increased resources [34,36]. This was also suggested in a prior study of C. angustifolium as an explanation for the advantage of tetraploids over diploids [15]. In this study, however, we find that neotetraploid pollen is also larger than pollen from diploids but has lower siring success compared with pollen from established tetraploids. This suggests that siring success arises in part through specific interactions between pollen and pistil as opposed to being simply a product of change to a style-independent pollen trait [38].
This study has implications for understanding the role of conspecific pollen precedence in the evolution of polyploid species. Establishment and persistence of tetraploid cytotypes in mixed populations is favoured by assortative mating [7,39] as it weakens the strength of minority cytotype disadvantage and the fitness costs of between-cytotype mating. Our study highlights the importance of conspecific pollen precedence for facilitating assortative mating in tetraploids [7]. Here, tetraploid pollen precedence in C. angustifolium resulted in 70 per cent of tetraploid matings being assortative. In a previous study, tetraploid siring success resulted in 44 per cent assortative mating and contributed to 4.5 per cent of the total reproductive isolation (more than 99%) between cytotypes [7].
Relative to established tetraploids, neotetraploids exhibit weaker homoploid pollen precedence and relatively weak siring success on diploid maternal plants. Without data on other barriers to hybridization in neopolyploids of other species, the magnitude of total reproductive isolation and relative contribution of conspecific pollen precedence to reproductive isolation is unclear. However, it seems probable that a number of prezygotic reproductive barriers such as ecological isolation will be weaker in neotetraploids than in established tetraploids. The question remains, what allows tetraploids to persist in sympatry with diploids and how quickly can prezygotic barriers such as strong conspecific pollen precedence evolve subsequent to the duplication? Additional research on the mechanisms of selection and the role of reinforcement on pollen siring ability and pollinator–plant interactions may shed some light on this issue.
Our results on pollen siring success mirror patterns observed for other traits such as flowering time, floral display [16–18] and drought tolerance [40], in which neotetraploids are more similar to their diploid progenitors than established tetraploids. While it does not apply to all traits [16], this pattern suggests that associations between phenotype and ploidy observed in extant populations may not represent the direct effects of genome duplication itself. Rather they reflect a much longer evolutionary process of divergence that involves selection or other population processes (e.g. drift) operating after the duplication event. This model of polyploid evolution would predict that neopolyploids may show greater phenotypic consistency than established tetraploids, and may help to explain the absence of a consistent association between polyploidy and phenotype observed in wild populations [41].
Acknowledgements
We thank M. Mucci and T. Slimmon for greenhouse support, P. Kron for assistance with flow cytometry and the Natural Sciences and Engineering Research Council of Canada, Canada Research Chair Program and Canada Foundation for Innovation for financial support to B.C.H.
References
- 1.Soltis D. E., Soltis P. S. 1999. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14, 348–352 10.1016/S0169-5347(99)01638-9 (doi:10.1016/S0169-5347(99)01638-9) [DOI] [PubMed] [Google Scholar]
- 2.Adams K. L., Wendel J. F. 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141 10.1016/j.pbi.2005.01.001 (doi:10.1016/j.pbi.2005.01.001) [DOI] [PubMed] [Google Scholar]
- 3.Cui L., et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16, 738–749 10.1101/gr.4825606 (doi:10.1101/gr.4825606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood T. E., Takebayashi N., Barker M. S., Mayrose I., Greenspoon P. B., Rieseberg L. H. 2009. The frequency of polyploid speciation in vascular plants. Proc. Natl Acad. Sci. USA 106, 13 875–13 879 10.1073/pnas.0811575106 (doi:10.1073/pnas.0811575106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey J., Schemske D. W. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501 10.1146/annurev.ecolsys.29.1.467 (doi:10.1146/annurev.ecolsys.29.1.467) [DOI] [Google Scholar]
- 6.Levin D. A. 1983. Polyploidy and novelty in flowering plants. Am. Nat. 122, 1–25 10.1086/284115 (doi:10.1086/284115) [DOI] [Google Scholar]
- 7.Husband B. C., Sabara H. A. 2004. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium. New Phytol. 161, 703–713 10.1046/j.1469-8137.2004.00998.x (doi:10.1046/j.1469-8137.2004.00998.x) [DOI] [PubMed] [Google Scholar]
- 8.Felber-Girard M., Felber F., Buttler A. 1996. Habitat differentiation in a narrow hybrid zone between diploid and tetraploid Anthoxanthum alpinum. New Phytol. 133, 531–540 10.1111/j.1469-8137.1996.tb01921.x (doi:10.1111/j.1469-8137.1996.tb01921.x) [DOI] [Google Scholar]
- 9.Husband B. C., Schemske D. W. 1998. Cytotype distribution at a diploid-tetraploid contact zone in Chamerion (Epilobium) angustifolium (Onagraceae). Am. J. Bot. 85, 1688–1694 10.2307/2446502 (doi:10.2307/2446502) [DOI] [PubMed] [Google Scholar]
- 10.Baack E. J., Stanton M. 2005. Ecological factors influencing tetraploid establishment in snow buttercups (Ranunculus adoneus): niche differentiation and tetraploid establishment. Evolution 59, 1936–1944 [PubMed] [Google Scholar]
- 11.Husband B. C., Schemske D. W. 2000. Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. J. Ecol. 88, 689–701 10.1046/j.1365-2745.2000.00481.x (doi:10.1046/j.1365-2745.2000.00481.x) [DOI] [Google Scholar]
- 12.Segraves K. A., Thompson J. N. 1999. Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution 53, 1114–1127 10.2307/2640816 (doi:10.2307/2640816) [DOI] [PubMed] [Google Scholar]
- 13.Kennedy B. F., Sabara H. A., Haydon D., Husband B. C. 2006. Pollinator-mediated assortative mating in mixed ploidy populations of Chamerion angustifolium (Onagraceae). Oecologia 150, 398–408 10.1007/s00442-006-0536-7 (doi:10.1007/s00442-006-0536-7) [DOI] [PubMed] [Google Scholar]
- 14.Williams J. H., Jr, Friedman W. E., Arnold M. L. 1999. Developmental selection within the angiosperm style: using gamete DNA to visualize interspecific pollen competition. Proc. Natl Acad. Sci. USA 96, 9201–9206 10.1073/pnas.96.16.9201 (doi:10.1073/pnas.96.16.9201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husband B. C., Schemske D. W., Burton T. L., Goodwillie C. 2002. Pollen competition as a unilateral reproductive barrier between sympatric diploid and tetraploid Chamerion angustifolium. Proc. R. Soc. Lond. B 269, 2565–2571 10.1098/rspb.2002.2196 (doi:10.1098/rspb.2002.2196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsey J., Schemske D. W. 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639 10.1146/annurev.ecolsys.33.010802.150437 (doi:10.1146/annurev.ecolsys.33.010802.150437) [DOI] [Google Scholar]
- 17.Sabara H. A. 2008. The evolution of reproductive isolation between diploid and tetraploid Chamerion angustifolium (Onagraceae). PhD thesis, University of Guelph, Canada [Google Scholar]
- 18.Bretagnolle F., Lumaret R. 1995. Bilateral polyploidization in Dactylis glomerata L. subsp. lusitanica: occurrence, morphological and genetic characteristics of first polyploids. Euphytica 84, 197–207 10.1007/BF01681812 (doi:10.1007/BF01681812) [DOI] [Google Scholar]
- 19.Howard D. J. 1999. Conspecific sperm and pollen precedence and speciation. Annu. Rev. Ecol. Syst. 30, 109–132 10.1146/annurev.ecolsys.30.1.109 (doi:10.1146/annurev.ecolsys.30.1.109) [DOI] [Google Scholar]
- 20.Rieseberg L. H., Desrochers A. M., Youn S. J. 1995. Interspecific pollen competition as a reproductive barrier between sympatric species of Helianthus (Asteraceae). Am. J. Bot. 82, 515–519 10.2307/2445699 (doi:10.2307/2445699) [DOI] [Google Scholar]
- 21.Carney S. E., Hodges S. A., Arnold M. L. 1996. Effects of differential pollen-tube growth on hybridization in the Louisiana irises. Evolution 50, 1871–1878 10.2307/2410745 (doi:10.2307/2410745) [DOI] [PubMed] [Google Scholar]
- 22.Diaz A., Macnair M. R. 1999. Pollen tube competition as a mechanism of prezygotic reproductive isolation between Mimulus nasutus and its presumed progenitor M. guttatus. New Phytol. 144, 471–478 10.1046/j.1469-8137.1999.00543.x (doi:10.1046/j.1469-8137.1999.00543.x) [DOI] [PubMed] [Google Scholar]
- 23.Cheung A. Y. 1995. Pollen–pistil interactions in compatible pollination. Proc. Natl Acad. Sci. USA 92, 3077–3080 10.1073/pnas.92.8.3077 (doi:10.1073/pnas.92.8.3077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosquin T. 1967. Evidence for autopolyploidy in Epilobium angustifolium (Onagraceae). Evolution 21, 713–719 10.2307/2406768 (doi:10.2307/2406768) [DOI] [PubMed] [Google Scholar]
- 25.Soltis D. E., Soltis P. S., Schemske D. W., Hancock J. F., Thompson J. N., Husband B. C., Judd W. S. 2007. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56, 13–30 [Google Scholar]
- 26.Galbraith D. W., Harkins K. R., Maddox J. R., Ayres N. M., Sharma D. P., Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051 10.1126/science.220.4601.1049 (doi:10.1126/science.220.4601.1049) [DOI] [PubMed] [Google Scholar]
- 27.Greilhuber J., Temsch E. M., Loureiro J. C. M. 2007. Nuclear DNA content measurement. In Flow cytometry with plant cells (eds Dolezel J., Greilhuber J., Suda J.), pp. 67–101 Weinheim, Germany: Wiley-VCH [Google Scholar]
- 28.Burton T. L., Husband B. C. 2000. Fitness differences among diploids, tetraploids and their triploid progeny in Chamerion angustifolium: mechanisms of inviability and implications for polyploid evolution. Evolution 54, 1182–1191 [DOI] [PubMed] [Google Scholar]
- 29.Dolezel J., Binarová P., Lucretti S. 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 31, 113–120 [Google Scholar]
- 30.Burton T. L., Husband B. C. 2001. Fecundity and offspring ploidy in matings among diploid, triploid and tetraploid Chamerion angustifolium (Onagraceae): consequences for tetraploid establishment. Heredity 87, 573–582 10.1046/j.1365-2540.2001.00955.x (doi:10.1046/j.1365-2540.2001.00955.x) [DOI] [PubMed] [Google Scholar]
- 31.Sokal R. R., Rohlf F. J. 1981. Biometry, 2nd edn. San Francisco, CA: WH Freeman [Google Scholar]
- 32.Maki M. 1993. Outcrossing and fecundity advantage of females in gynodioecious Chionographis japonica var. kurohimensis (Liliaceae). Am. J. Bot. 80, 629–634 10.2307/2445432 (doi:10.2307/2445432) [DOI] [Google Scholar]
- 33.Mable B. K. 2004. Polyploidy and self-compatibility: is there an association? New Phytol. 162, 803–811 10.1111/j.1469-8137.2004.01055.x (doi:10.1111/j.1469-8137.2004.01055.x) [DOI] [PubMed] [Google Scholar]
- 34.Anderson J. M., Barrett S. C. H. 1986. Pollen tube growth in tristylous Pontederia cordata L. (Pontederiaceae). Can. J. Bot. 64, 2602–2607 10.1139/b86-344 (doi:10.1139/b86-344) [DOI] [Google Scholar]
- 35.Cruzan M. B. 1990. Variation in pollen size, fertilization ability and post-fertilization siring ability in Erythronium grandiflorum. Evolution 44, 843–856 10.2307/2409550 (doi:10.2307/2409550) [DOI] [PubMed] [Google Scholar]
- 36.Lau T. C., Stephenson A. G. 1994. Effects of soil phosphorus on pollen production, pollen size, pollen phosphorus content and the ability to sire seeds in Curcurbita pepo (Curcubitacaea). Sexual Plant Reprod. 7, 215–220 [Google Scholar]
- 37.Ramsey J., Bradshaw H. D., Schemske D. W. 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Pharymaceae). Evolution 57, 1520–1534 [DOI] [PubMed] [Google Scholar]
- 38.Fishman L., Aagaard J., Tuthill J. C. 2008. Toward the evolutionary genomics of gemetophytic divegence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution 62, 2958–2970 10.1111/j.1558-5646.2008.00475.x (doi:10.1111/j.1558-5646.2008.00475.x) [DOI] [PubMed] [Google Scholar]
- 39.Levin D. A. 1975. Minority cytotype exclusion in local plant populations. Taxon 24, 35–43 10.2307/1218997 (doi:10.2307/1218997) [DOI] [Google Scholar]
- 40.Maherali H., Walden A. F., Husband B. C. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytol. 184, 721–731 10.1111/j.1469-8137.2009.02997.x (doi:10.1111/j.1469-8137.2009.02997.x) [DOI] [PubMed] [Google Scholar]
- 41.Levin D. A. 2002. The role of chromosomal change in plant evolution. New York, NY: Oxford University Press [Google Scholar]