Abstract
Drosophila melanogaster larvae usually react against eggs of the parasitoid wasp Leptopilina boulardi by surrounding them with a multicellular melanotic capsule. The genetic determinism of this response has been studied previously using susceptible (non-capsule-forming) and resistant (capsule-forming) strains. The results suggest that differences in their encapsulation response involve a single gene, resistance to Leptopilina boulardi (Rlb), with two alleles, the resistant one being dominant. Rlb confers specific protection against Leptopilina boulardi and is thus probably involved in parasitoid recognition. Recent studies have localized this gene on the right arm of the second chromosome and our aim was to precisely determine its genetic and molecular location. Using strains bearing deletions, we demonstrated that resistance to Leptopilina boulardi is conferred by the 55C; 55F3 region and that the 55E2–E6; F3 region is particularly involved. A physical map of the 55C; 56A region was then constructed, based on a set of overlapping cosmid and P1 phage clones. Using single and double digests, cross hybridization of restriction fragments, and location of genetically mapped genes and STSs, a complete, five-enzyme restriction map of this 830-kb region was obtained.
Insects possess a complex immune system, with both humoral and cellular components to protect themselves from a foreign entity. Bacterial invasion for example induces the production of an array of peptides with antimicrobial or antifungal activity as well as the activation of the prophenol oxydase cascade (Hoffmann 1996; Hoffmann and Reichart 1997). In the case of parasitism by a larger parasite, an encapsulation response is provoked that involves the deposition of eumelanin resulting from the prophenol oxydase system activation as well as the adhesion of numerous hemocytes around the egg of the parasite (Carton and Nappi 1997). No experimental data have clearly identified the components of the melanotic encapsulation response that kill the parasite, even if production of cytotoxic radicals is probably involved (Nappi et al. 1995), but the presence of the capsule always indicates the failure of parasitism. Despite the recent advances in understanding these immunological processes and the demonstration that the immune response is not completely aspecific (Lemaitre et al. 1997), little is known about how insects recognize a parasite and which factors underlie the specificity of the response.
Drosophila melanogaster is parasitized by several wasps including Leptopilina boulardi (Cynipidae) and Asobara tabida (Braconidae). In both of these systems, virulent (immune-suppressive) and avirulent (non-immune-suppressive) strains of parasites have been obtained. Virulent strains always succeed in escaping or suppressing the host immune response, whereas parasitoids from avirulent strains are encapsulated by some of the hosts. The use of an avirulent strain has allowed selection of two isofemale lines of D. melanogaster with different immune responses to L. boulardi (Carton and Boulétreau 1985). Flies from the resistant (R) strain encapsulate eggs of the parasite within 24 hr after infestation, whereas flies from the susceptible (S) strain do not exhibit the encapsulation response. Experiments using R and S strains have demonstrated that induction of the phenoloxydase cascade by parasitism occurs only in the R strain (Nappi et al. 1991). The resistance gene(s) thus act at an early phase in the immune response. Parasitism does not induce the production of antibacterial peptides in the R strain, which indicates that the antibacterial and the antiparasite immune responses are controlled by different pathways (Cousteau et al. 1996). Very interestingly, comparative infestation experiments with L. boulardi and A. tabida have shown that host resistance is highly specific (Carton and Nappi 1997). The S strain susceptible to L. boulardi is able to encapsulate the eggs of A. tabida, demonstrating that it is not immune-incompetent but simply unable to recognize L. boulardi (Vass et al. 1993). The molecular mechanisms sustaining this specificity are currently completely unknown. Characterization of the resistance gene(s) and determination of its function would greatly help in understanding how insects recognize the nonself.
In the systems studied, the resistance of D. melanogaster to avirulent parasitoids has been demonstrated to be autosomal and monogenic. Two major resistance genes have been described that confer resistance to L. boulardi—resitance to L. boulardi (Rlb) (Carton and Nappi 1997) and A. tabida–resistance to A. tabida (Rat) (Orr et al. 1997; Benassi et al. 1998), respectively. These genes are dominant and both localized on the second chromosome of D. melanogaster. The genetics of resistance to Leptopilina has been further studied using the S and R strains and Rlb has been localized in the region 55–56 (Carton and Nappi 1997). It should be noted that the 55A–56A region of the Drosophila genome contains several genes involved in immunological processes. The prophenol-oxydase-encoding gene, A1, has been located in 55A (Fujimoto et al. 1995), the immune deficiency (imd) gene (Lemaitre et al. 1995) in 55CD, and the gene encoding the diptericin antibacterial peptide in 56A (Wicker et al. 1990).
This paper reports genetic experiments using strains bearing deletions, which allowed us to obtain a more precise localization of Rlb. We demonstrated that deletion of the 55C; 55F3 region and especially of the E2–E6; F3 region affects the resistant phenotype considerably. Further characterization of the 55C–56A region was then undertaken by taking advantage of the existence of two D. melanogaster genomic libraries, based on cosmid and P1 phages. A complete, fine restriction map of the Rlb-containing region was obtained that constitutes an essential tool for the molecular cloning of the Rlb gene.
RESULTS
Genetic Localization of Rlb
As mentioned above, classical genetic studies have shown that Rlb is completely dominant. Nevertheless, using deficiency (Df) strains bearing deletions in the 55–56 region of the second chromosome [Df(2R) strains], we observed that, unexpectedly, when the R allele of the Rlb gene (including its regulatory regions) is facing a deletion, the resistant phenotype is disturbed. The important implication of this phenomenon for our studies is that deletions can be useful for localizing the dominant Rlb gene. To perform this localization, five D. melanogaster strains bearing deletions were used.
Individuals of each deleted strain [Df(2R)/CyO] were crossed with individuals of the R strain, homozygous for Rlb. F1 hybrids [Df(2R)/R and CyO/R] were then infested by L. boulardi as were control individuals from the deleted strains. The number of individuals used in each experiment is reported Figure 1. The controls were used to confirm the infestation ability of the parasitoid and the susceptible phenotype of the deleted strains. The encapsulation ability was determined by dissecting larvae and recording melanotic encapsulation, and the F1 encapsulation rates (ER) were calculated as stated by Carton et al. (1992). At the larval stage, when infestation occurs, CyO/R individuals cannot be distinguished from Df(2R)/R individuals. The ER were then corrected, considering that half of the F1 progenies consist of CyO/R-resistant (capsule-forming) larvae.
Figure 1.
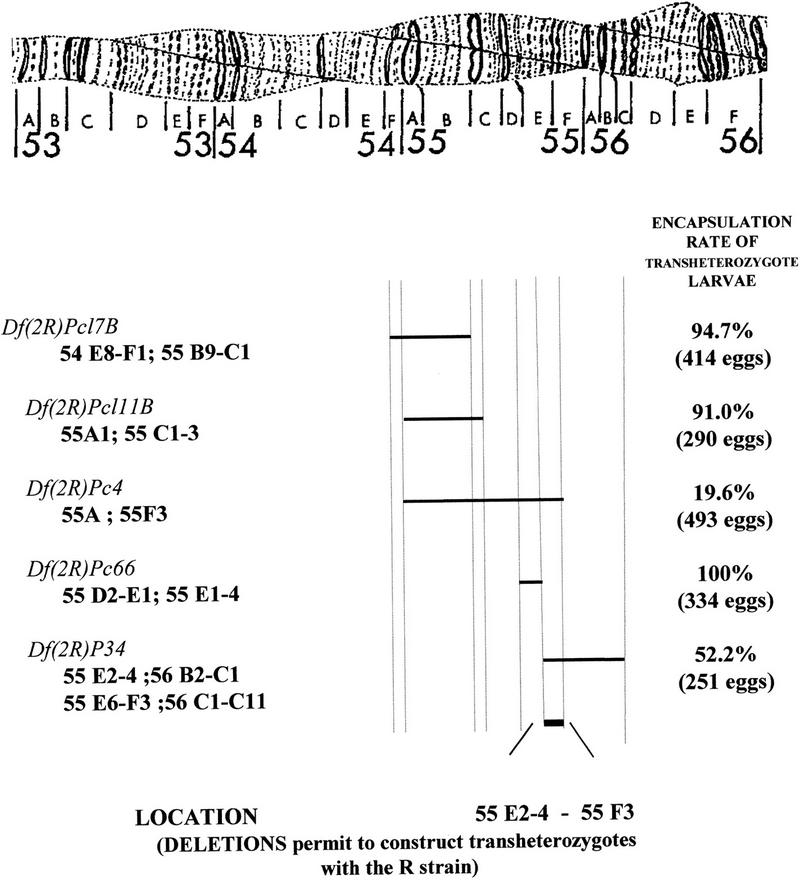
In situ localization of the regions deleted in the Df(2R) strains and corresponding encapsulation rates in trans-heterozygote larvae. The localization of the limits of the deletions in the different strains Df(2R) is provided. For the Df(2R)P34 strain, data have changed in course of our experiments and both old and new localizations are given. Different results were found for the Df(2R)Pc66 strain and the choice of the given limits is discussed in the text. The encapsulation rates of the R × Df(2R) progenies were calculated as the number of encapsulated eggs divided by the total number of eggs. This total number of recovered eggs is indicated for each F1. The ER of transheterozygote larvae were obtained using the assumption that half of the F1 progenies are of CyO/R genotype and encapsulate parasite eggs. These results demonstrate that deletion of the 55E2–6; 55F3 region has an important effect on the encapsulation ability.
The localization of the right and left limits of the five deletions were obtained in the FlyBase databank. These data have sometimes be specified or changed in the course of our experiments: For example, the deleted region in Df(2R)Pc4 considered previously as 55A1–55F was further specified as 55A1–55F3. When in doubt, the different data were taken into account.
Encapsulation rates recorded for the Df(2R)Pc4, Df(2R)Pc17B, Df(2R)Pc66, Df(2R)P34, and Df(2R)P111B strains were, respectively, 4.8% (126 individuals), 0% (249 individuals), 0.02% (124), 3.9% (152 individuals), and 0% (32 individuals). As these results do not differ significantly from the ER usually obtained in the S strain (Carton et al. 1992), the deleted strains were considered as susceptible subsequently.
The length of the region involved in the resistance phenomenon was first defined using the Df(2R)Pc4 strain: The encapsulation rate of Df(2R)/R heterozygous individuals fell under 20% as compared with 80%–95% found in the R homozygous strain or in the F1 between the R and S strains (Fig. 1; Carton et al. 1992). This demonstrated that the 55A1; 55F3 region is involved in resistance to L. boulardi. This region was then restricted to 55C1; 55F3 using the Df(2R)Pc17B and Df(2R)Pc111B strains. The corrected ER obtained in their F1 progenies with the R strain were of the same order as the ER of the R strain itself (94.7% and 91%, respectively; Fig. 1). These results showed that deletions of the 54E8–F1; 55B9–C1 [Df(2R)Pc17B] or the 55A1; 55C1–C3 [Df(2R)Pc111B] region do not impair the resistant phenotype. On the contrary, the corrected ER was impaired greatly (52%) when hybrids between the R strain and Df(2R)P34 were infested. The region deleted in this strain had been first located in 55E2–4; 56B2–C1 but recent FlyBase data report a 55E6–F3; 56C1–C11 localization. In combination with the previous results, this allowed us to conclude that deletion of the E2–E6 F3 region has a great impact on the ability of Drosophila to recognize and encapsulate Leptopilina eggs. Finally, F1 hybrids between the R strain and the Df(2R)Pc66 strain were infested. The corrected encapsulation rate was 100%. This deletion had been first localized to the 55D2–E1; 55E3–E4 region and is now reported to be in the 55D2–E1; 56B2 position. Nevertheless, other experiments reported only a slight alteration of the D region in the Pc66 strain (Georgel, pers. comm.), and we considered subsequently a 55D2–E1; 55E1–E4 localization. As a consequence, our main conclusion is that the E2–E6; F3 region is clearly involved in resistance. Nevertheless, the ER is significantly higher in R/P34 hybrids (52%) as compared with R/Pc4 hybrids (19%), which indicates that another region, located on the left side of the Df(2R)Pc66 deletion (55C1–C3; 55D2–E1) or on its right side (55E1–E4) could also have an impact on the resistance phenotype.
Mapping of the 55C–55F Region
Covering of the Region
The genome of D. melanogaster is studied by the European Drosophila Genome Program (EDGP) using cosmid clones (Siden-Kiamos et al. 1990; Kafatos et al. 1991) and by the Berkeley Drosophila Genome Program (BDGP) using P1 phage clones (Smoller et al. 1991; Hartl et al. 1994; Kimmerly et al. 1996). The results on the genetic localization of Rlb interested us in the mapping of the 55C–55F region. Two contigs of P1 clones were available in this region, otefin (55BC) and diptericin (55F–56AB) but the region between the two was not covered. Furthermore, some of the clones had not been localized, the position of others had to be confirmed, and the limits of the complete region had to be determined (Rubin 1996).
Cosmid Map
We first attempted to recover the complete 55C–55F region using the cosmid library. Thirty-one cosmids predicted to cover this region or not localized yet were obtained from I. Siden Kiamos [Foundation for Research and Technology—Hellas (FORTH) Heraklion, Crete]. Their DNA was digested with BsrgI and NotI enzymes and transferred. BsrgI and NotI cut, respectively, at the positions 91 and 5144 in Lorist6 (5156 bp) and a 5-kb restriction fragment containing almost the entire vector is obtained (Cross and Little 1986; Gibson et al. 1987). This fragment is thus the only one common to all the clones. The blots were hybridized with radioactively labeled DNA of each clone and cross-hybridizations were analyzed. Cosmids containing repeated DNA were detected by high-stringency hybridization of the Southern blots with radioactively labeled Drosophila genomic DNA. Several bands were detected for five cosmids (86B6, 152H3, 65H8, 195C7, 128B5) which indicated that they contain repeated DNA. This result was taken into account during the analysis. Finally, four contigs containing, respectively 2, 4, 11, and 5 cosmids were determined but the complete region could not be obtained (Fig. 2). The use of the P1 library was then necessary to confirm cosmid overlaps and solve remaining gaps.
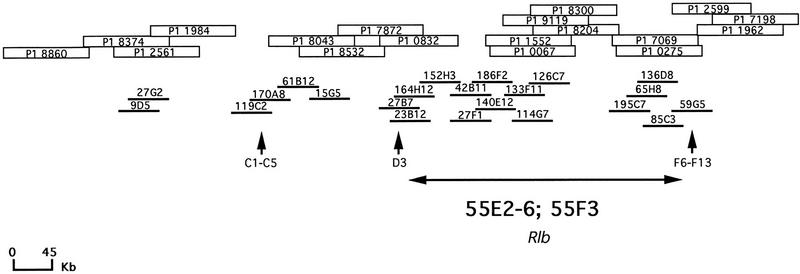
Figure 2.
Localization of P1 and cosmid clones in a 55C–56A contig: Thirty-one cosmid and thirty-three P1 clones supposed to be located in 55C–56A or not localized yet, were analyzed by cross-hybridization experiments to obtain a contig of this region. Four contigs (2, 4, 11, and 5 clones) were obtained with the cosmids and three contigs with the P1s (4, 4, and 10 clones). Cosmids are represented by thick lines and P1s by empty boxes. Both P1 and cosmid libraries proved necessary to determine a complete contig. Vertical arrows indicate in situ localization of three of the clones, 119c2 (55C1–2/C3/C4–5), 27B7 (55D1–2/D3), and 59G5 (55 F6–F13) which allowed to demonstrate that the entire 55C–55F region had been recovered. Finally, an estimated localization of the 55E2–6; 55F3 region, based on BDGP in situ data is given.
In Situ Localization of Cosmids
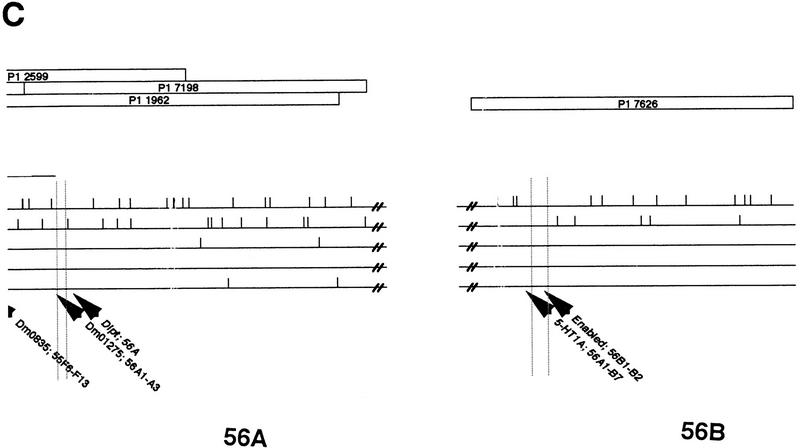
Five cosmids, chosen to define the limits of the 55C–55F region, were hybridized on Drosophila polytene chromosomes. Their localization was compared with data from the EDGP. As expected, 119C2, 27B7, and 59G5 hybridized in 55C1–2/C3/C4–5, 55D1–2/D3, and 55 F5–F11/56A, respectively, which indicated the limits of the entire 55C–55F region (Figs. 2 and 3). On the contrary, the 55G2 clone, localized in 56A1–B7 by the EDGP program, was found to hybridize in 55F1–2/F3. Sequence data found on a 55G2T STS in the EMBL databank (accession no. Z5065) were analyzed with a BLAST program (Infobiogen Bisance, Dessen et al. 1990). The results demonstrated clearly that 55G2 shares homologies with a P1 clone localized in 52E1–E8 (DS03910) and with sequences of the retrotransposon Burdock (EMBL accession no. U89994). Therefore, the contradictory results are most probably caused by differences between strains of D. melanogaster concern ing the presence of this transposable element. This confirms the high rate of D. melanogaster middle repetitive DNA as a source of false positives in clone localization. The 55G2 clone was not further considered. A last clone, 75A9 had been located both in 90E and 55C but only the first localization (90E) was confirmed.
Figure 3.
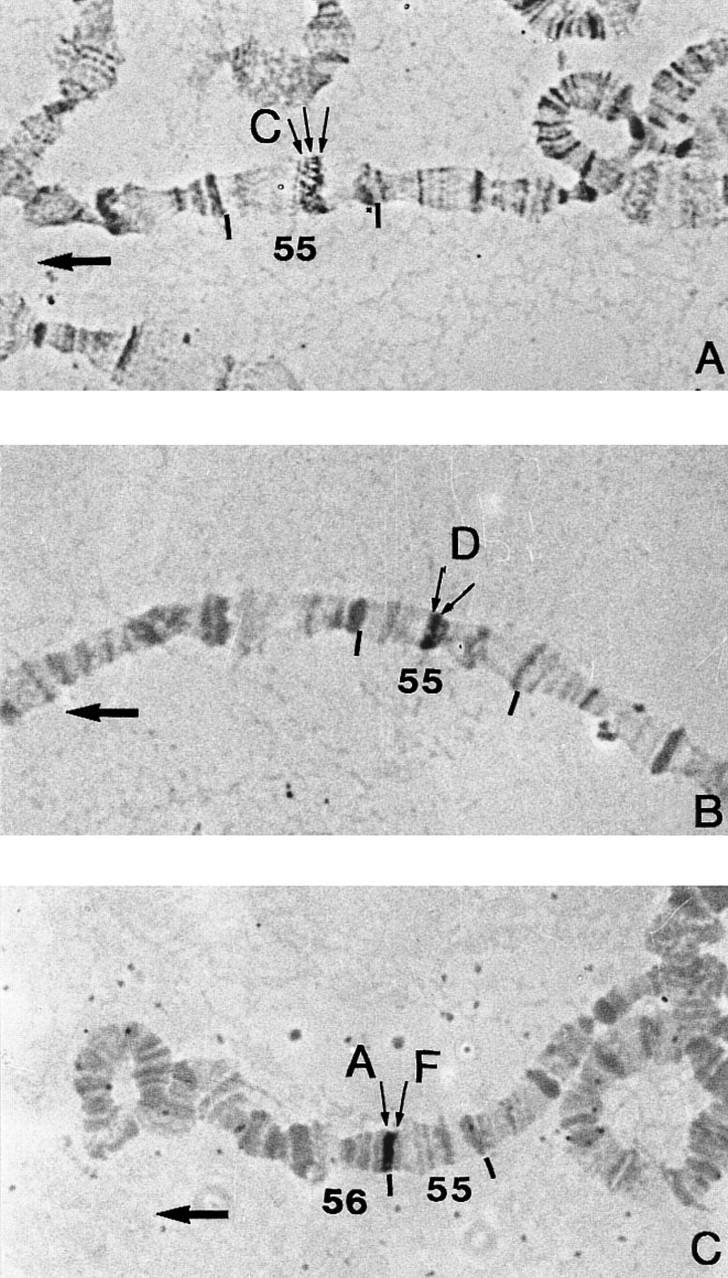
In situ hybridization with biotinylated cosmid probes to Drosophila melanogaster 2R chromosomes (Canton S strain). Thin arrows denote the probe insertions on sections 55 and 56. Thick arrows point to the tips of the 2R chromosomes (at the left). Cosmid 119c2 hybridizing on bands 55C1–2, 55C3, and 55C4–5. Cosmid 27B7 hybridizing on bands 55D1–2 and 55D3. Cosmid 59G5 hybridizing on bands 55F5–11 and 56A.
P1 Map
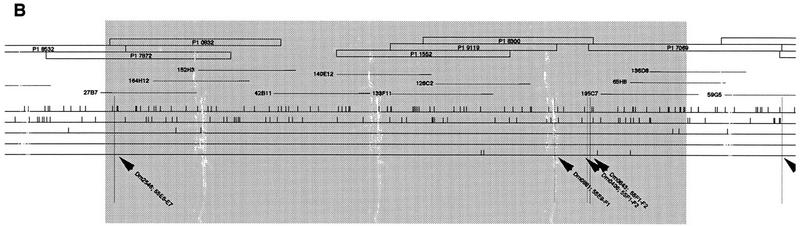
Thirty-three P1 clones believed to cover the 55C–56A region were obtained from M. Ashburner and C. McKimmie (University of Cambridge, UK). Their DNA was digested with SfiI and NotI enzymes and transferred. SfiI and NotI cut at the 39 and 15978 positions, respectively in the Ad10SacBII vector and a 16-kb restriction fragment containing almost the entire vector was obtained (Pierce et al. 1992; Sternberg 1990, 1994). This fragment is thus common to all the clones with the exception of the P1 8374, 2561, 1552, and 2599 which correspond to the pNS582 Ad10tet14 vector. Radioactively labeled DNA of each P1 or cosmid clone was hybridized on Southern blots of all the P1 clones and Southern blots of all the cosmid clones. Controls showing hybridization patterns of the two vectors (Lorist6 and Ad10SacBII) onto these blots were also realized. Cross hybridizations were finally analyzed to determine the P1 contigs. As shown in Figure 2, the three gaps found in the cosmid map were solved with P1 clones, and the cosmid contigs were confirmed. Nevertheless, the 55C–55F region could not be covered completely using only the P1 library as three contigs (4, 4, and 10 P1) were detected. The two gaps were solved with cosmid clones. The 55BC–56A contig, made of P1 and cosmids is represented Figure 2.
Restriction Map of the Region
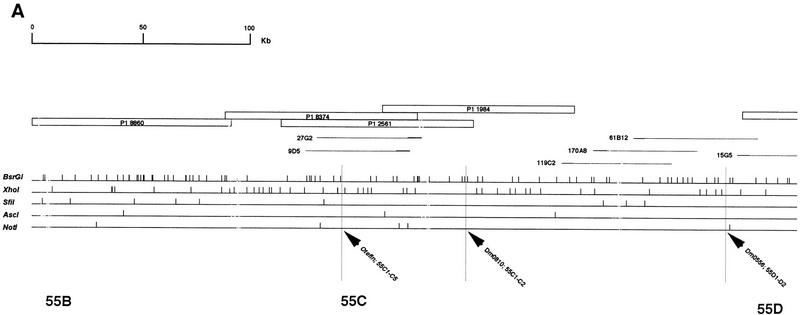
The restriction mapping of the region with five enzymes was realized to obtain a chromosomal walk, and with the objective of future subcloning and sequencing of the region. The DNA extracted from each cosmid and P1 clone located in the region was digested with NotI, BsrgI, XhoI, AscI, and SfiI and double-digested with NotI–BsrgI, NotI–XhoI, XhoI–BsrgI, AscI–XhoI, and XhoI–SfiI. Migrations were done on both 0.7% (or 0.5%) and 1.2% agarose gels. Southern blots were hybridized with radioactively labeled DNA of the vector and the clone itself and the restriction map of each clone was drawn. Genomic DNA fragments inserted into the Lorist cosmid vector ranged from 35 to 45 kb and fragments inserted into the Ad10SacBII P1 vector from 80 to 85 kb. Southern blots of each clone were then hybridized with probes of other clones located in the same part of the map. This allowed us to detect cross-hybridizing fragments between the clones. When necessary, fragments corresponding to the ends of inserted DNA were used as probes on Southern blots of other clones to detect small cross hybridizations. The complete restriction map of the 55BC–56A region finally drawn from covering of individual maps is ∼830 kb long and contains 357 restriction sites (Fig. 4).
Figure 4.
Physical map of the 55B–56AB region of Drosophila melanogaster 2R chromosome. The contig of P1 and cosmid clones that were used in restriction mapping is represented at the top. Empty boxes correspond to P1 clones and thin lines correspond to cosmid clones. The thicker part of the lines represent the cosmid vector, Lorist 6. The restriction map of the region is given below the corresponding clones. Each horizontal line corresponds to a restriction enzyme and vertical traits indicate restriction sites. A total of 357 restriction sites were placed in the 55B–56A region. The precise localization of STSs and gene fragments on the restriction map is pointed out by high vertical traits. otefin and diptericin were found in the contig, whereas 5-HT1A and enabled probes hybridized on the 7626 P1 in 56B. The limits of the different parts of the contig (55B, 55C…) were estimated using BDGP data regarding clones and STS in situ localization. The 55E2–E6; 55F3 region which when deleted greatly impairs the resistant phenotype, is represented by shading. Its size is probably overestimated because of the inaccuracy of in situ localization of the clones and of the limits of the deletions.
Genes and STS Localization
The 55C–56A region was analyzed further by localizing STSs and genes predicted to be in the region. FlyBase databank localization of the STSs Dm0406, Dm0810, Dm0556, Dm2546, Dm0881, Dm0843, Dm406, Dm0835, and Dm1275 was, respectively, 55C1–2, 55D1–D2, 55E6–E7, 55E9–F1, 55F1–F2, 55F1–F2, 55F6–F13, and 56A1–A3. Fragments corresponding to these STSs were obtained by PCR experiments, radioactively labeled, and hybridized on Southern blots of all the clones located in the same part of the map. As shown on Figure 4, the chromosomal walk was confirmed by the fine localization of these STSs. Nevertheless, a clear discrepancy was sometimes recorded between in situ localization data of STSs and clones.
According to FlyBase data, a few genes localized in the 55C–56A region or near this region have been cloned and sequenced. Fragments of these genes were cloned and sequenced following amplification with specific primers. Two fragments (222 and 780 bp) were obtained for otefin (55C1–C13), a 395-bp fragment was obtained for diptericin (56A1–A3), a 195-bp fragment was obtained for enabled (56B), a 270-bp fragment was obtained for coracle (56C), a 170-bp fragment was obtained for 5HT1-A (56AB), a 250-bp fragment was obtained for 5HT1-B (56AB), and a 1021-bp fragment was obtained for three rows (55A1–A4). All these DNA fragments were radioactively labeled and used as probes on Southern blots of cosmid and P1 clones. The precise localization of otefin, diptericin, 5HT1-A, and enabled is reported Figure 4. The three rows fragment was found to be located on the 128B5 cosmid which is not part of the studied region but is most probably located in 55A. No hybridization was detected for 5HT1-B and coracle.
The localization of the diptericin and otefin genes on the 1962 and 2561 P1 clones confirmed the limits of the 55C–56A contig. 5HT1-A and enabled are located outside this region, in 56B.
DISCUSSION
Genetic Localization of Rlb
D. melanogaster resistance to parasitism by L. boulardi is determined by a major gene, Rlb, located on the second chromosome, and whose expression is completely dominant. Nevertheless, preliminary experiments designed to localize Rlb demonstrated that some deletions in a heterozygous form are able to disturb the otherwise Rlb-dominant phenotype, that is, the encapsulation ability. No molecular hypothesis explaining this phenomenon has been proposed, yet but it might be reminiscent of the transvection effects recorded in Drosophila. Transvection as “the complementation of heteroalleles allowed by their proximity” was first described by Lewis (1954) for the Ultrabithorax (Ubx) locus. Where alleles subject to transvection have been characterized, it appeared that one allele carried a mutation in the regulatory region of the gene, and the other in its coding region. It is thus likely that transcription of the intact coding region on one homolog is activated by the intact enhancer on its pairing partner (Bender et al. 1983). When the heteroalleles are no longer able to interact, as is the case following chromosomal rearrangements, the transvection is disrupted. In the case of Ubx, it was demonstrated recently that such rearrangements reduce the expression of both alleles and not only the one encoding the active product (Goldsborough and Kornberg 1996). This result suggests strongly that somatic pairing is not only involved in the rescue of gene expression from mutant alleles, but may also play a role in the normal expression of this gene. This phenomenon might occur by a variety of molecular mechanisms and Henikoff (1997) refers to the ability of a gene to sense its paired state by the umbrella term of “trans-sensing.” For example, the occurrence under certain circumstances of physical interactions between enhancer and promoter sequences located on homologous chromosomes has been pointed out by Geyer et al. (1990).
Our working hypothesis is that such somatic pairing might have a role in the expression of Rlb. Chromosomal deletions including the Rlb region could prevent the usual short range pairing between the Rlb alleles (including regulatory regions) and result in a dramatic reduction in the level of expression of the Rlb allele. The lowered level of the Rlb+ allele product would be responsible for the reduced rate of encapsulation in the R/Df(2R) heterozygous strains.
Whatever its molecular underlying, this phenomenon allowed us to precisely localize the region involved in resistance. The five strains bearing deletions that we used were first demonstrated to be susceptible to parasitism by Leptopilina. Then, the encapsulation rate of the hybrids R/Df(2R) were obtained and compared, taking into account the fact that half of the F1 progenies must be of CyO/R genotype and therefore of resistant phenotype. An important number of individuals has been tested (Fig. 1) and the differences recorded between the ERs were high enough to show immediately if a deletion has an impact on the resistance phenotype or not.
When using deletions in genetic studies, the accuracy of in situ localization of their limits is of great importance. The limits of the deletions that we used were found in the FlyBase databank but some of them had been refined or changed in the course of our experiments. When determining the Rlb-containing region, we used all the known data to keep it as large as possible. The ERs were as high as in the R strain for three of the four deleted strains: thus, the 55A; 55C1–3 and 55D2–E1; 55E1–4 regions seem not to be involved in the resistance phenotype. On the contrary, the 55E2–6; 55F3 region is likely to be implied since the R/Df(2R)P34 individuals showed an ER of only 52%. The fact that resistance might not be determined completely by this region is suggested by the comparison of this ER (52%) with the ER (19%) of individuals heterozygous for the 55A–55F3 deletion (Fig. 1). The 55C1–C3; 55D2 region could be responsible for this effect. Nevertheless, the right limit of the Df(2R)Pc66 deletion and the left limit of the Df(2R)P34 are not defined precisely. Part of the 55E2–E6 region may not be included in the Pc66 or Pc34 deletions and play a role in determining the resistant phenotype.
The detailed interpretation of these data will remain difficult until the trans-sensing effects will be better understood. For example, the ER of R/Df(2R)Pc4 individuals (19%) is not in the same range as the ER of the S strain (0%–5%) but this could result from either the way deletions affect the expression of Rlb on the homologous chromosome or it could indicate that another region outside the 55A–55F3 part of the chromosome is also involved in the resistance phenomenon. Finally, resistance to Leptopilina appears to be determined in a great part by the 55E2; 55F3 region even if the involvement of the 55C1–C3; 55D2–E1 region can not be excluded completely.
Physical Mapping of the 55C–55F Region
To map this region we used a cosmid library (EDGP) and a P1 phage library (BDGP) as well as numerous data classified in FlyBase including in situ localization data. For instance, the diptericin P1 contig was known to cover 55F–56A and the otefin contig was located in 55B–C. Both cosmid and P1 clones, located in the region or not localized yet, had to be used to obtain the complete 55C–55F region. Having identified the clones containing repeated DNA, we used direct cross-hybridization and restriction mapping to determine the contigs. The main limitation of this bottom-up mapping technique is the difficulty to detect small overlaps between clones. Nevertheless, unique restriction sites were available in the vectors that allowed us to separate them from the inserted DNA and comparison of the restriction maps allowed us to determine clone overlaps with good accuracy. To solve remaining ambiguities, the ends of some clone inserts were prepared as probes and used in hybridization experiments.
Four contigs covering the 55C–55F region were obtained with the cosmids and the gaps (75, 30, and 45 kb) were further solved using P1 phages. Using the P1 library, three contigs were determined in the 55BC–56A region. Again, the two gaps (75 and 25 kb) were covered by cosmid clones. Therefore, the limit of the bottom-up technique was not responsible for the nonrecovering of the complete region from a given library. The fact that some gaps remain in the cosmid map as well as in the P1 map of the 55C–56A region can be explained in two ways. First, even if all the clones located in this region have been a priori provided, other clones corresponding to the missing regions may be contained in the libraries. Nevertheless, the fact that different regions are missing in each library seems more likely. The cosmid library represents four times the genome of D. melanogaster (Siden Kiamos et al. 1990) but instability has been described for some inserts that are replicated by multicopy replicons and this could account for the failure to recover certain genomic segments in cosmid cloning systems (Smoller et al. 1991). One of the advantages of the P1 vector is supposed to be the avoidance of this instability with the use of a one-copy replicon and the framework map obtained by Hartl et al. (1994) included 85% of the euchromatic genome. Nevertheless, it can be noticed that in their chromosome 2R map, no clones were obtained for part of the 55A band.
To confirm the limits of the recovered region, we used two complementary approaches: in situ mapping of some of the clones and localization of known STSs and genes on the physical map. The right limit of the region was confirmed by in situ localization of 59G5 in 55F5–F13/56A, and of the diptericin gene (56A) on P1 1962. The localization of 27B7 in 55D1–2/D3 and of the STSs Dm0810, Dm0556, Dm2546, Dm0881, Dm0406, Dm0843, Dm0835, and Dm1275 gave confirmation of different parts of the map. Finally, the left limit of the covered region was obtained with in situ localization of 119C2 in 55C1–2/C3-4/C5 and of the otefin gene (55C1–C13) on the 27G2 and 9D5 cosmids. FlyBase localization of the P1 8860 at the left end of our contig is 55B. Occurrence of false localization of clones because of middle repetitive DNA was demonstrated for the cosmid 55G2.
The 55C–56A region has been mapped with three rarely cutting enzymes, AscI, NotI, SfiI, and two frequently cutting enzymes, XhoI and BsrgI. A total of 357 restriction sites was obtained. The size of P1 inserts varied from 80 to 85 kb, whereas the cosmid inserts size ranged from 35 to 45 kb, which did not differ from the expected 85 and 35 kb (Siden Kiamos et al. 1990; Smoller et al. 1991). Because of the number of BsrgI fragments, the relative position of a few neighbored BsrgI–BsrgI fragments remained uncertain. Nevertheless, the opportunity to test the restriction maps arose recently with the publication by the BDGP of sequence data regarding six P1 clones located in 55F–56A (1552, 9119, 8204, 7069, 2599, and 1962) and three P1 clones in 55BC (8374, 2561, and 8860). Only one inversion between two BsrgI–BsrgI fragments was recorded.
The complete restriction map is reported Figure 4 with the precise localization of the diptericin, otefin, 5HT1-A, and enabled gene fragments. The size of the mapped 55BC–56A region is ∼830 kb. According to Sorsa (1988), the total DNA contents of the band 55 is 1316 kb (with 284 kb in 55C, 103 kb in 55D, 298 kb in 55E, and 220 kb in 55F). The 56A band would represent 93 kb and the complete 55C–56A region would then cover ∼900 kb. The 70-kb difference could be explained partly by the fact that the 56A region is not recovered completely in our contig but on the other part, we also included part of the 55B region in our map. It is then likely that the DNA content in each band had been slightly overestimated previously.
Based on our physical map, we estimated the maximal size of the E2; F3 region as ∼300 kb. The expected number of genes in the Drosophila genome ranges from 12,000 (Miklos and Rubin 1996) to 43,000 (Louis et al. 1997), this last number probably being overestimated. As the size of the euchromatic genome is 1.2 × 108 bp, one to three genes should be found every 10 kb and the region could contain 30–90 genes. Nevertheless, the size of this region is probably overestimated because of the limits of in situ hybridization data and because of the possible discrepancy between the localization of the clones and the localization of the limits of the deletions. Our first aim will be to determine which of the clones are included in which deletion.
To carry on the cloning project of Rlb, we have the choice between a genetic and a physical approach. For instance, a genetic study was chosen by Collins et al. (1997) for the pen1 gene responsible for the encapsulation of malarial parasites. In our case, some sequences of the 55E–55F region are available. Our future efforts will be based on the analysis of these sequences and on the characterization of the regions of the contig expressed at the larval stages when parasitism occurs. The potential function of the genes determined will be considered, taking into account the recent advances underlining the homologies between the immunological responses of mammals, insects, and plants (Baker et al. 1997; Dushay and Eldon 1998; Vilmos and Kurucz 1998). Depending on the number of clones finally retained and the estimated number of candidate genes, it will be possible to decide if a genetic strategy involving the use of microsatellites markers will be or not necessary.
Whatever the final strategy, this physical mapping represents the first step toward the cloning of an insect resistance gene to its parasitoid, a type of gene that has never been cloned to date.
METHODS
Origin of Insect Strains
The origin of the avirulent strain of L. boulardi (Gif stock no. 486) and the resistant D. melanogaster strain (strain 940), as well as their rearing conditions have been described elsewhere (Carton et al. 1992). The Df(2R)Pc4, Df(2R)Pc17B, Df(2R)P34, Df(2R)Pc66, and Df(2R)Pc111B fly strains were obtained from the Bloomington Stock Center.
Bioassay Procedures
The working hypothesis was that chromosomes bearing the resistant allele could display lower resistance levels when combined with Df chromosomes missing the Rlb gene (or its regulatory regions). To test this hypothesis, crosses were performed between each deleted strains [Df(2R)] and the resistant R strain. Hybrid progenies as well as individuals from the deleted strains were submitted to parasitism by exposing batches of 50 second-instar larvae hosts to L. boulardi females. Infestation and rearing of infested larvae were conducted as described in Carton et al. (1992). Dissection of infested larvae were carried out 3 days after infestation to determine the encapsulated or nonencapsulated status of the parasitoid egg. The encapsulation rate ER was calculated as the ratio of the encapsulated egg number to the recovered egg number. Superparasitized larvae were included in the counts, as encapsulation rate do not vary if calculated with monoparasitized or superparasitized larvae. The ER of transheterozygous individuals bearing the deletion was calculated considering that they represent only 50% of the F1 progeny. Any significant change from a 100% value could be interpreted as a role of the deleted region in this character.
Origin of Clones and Experimental Procedures
Cosmids were provided by I. Siden Kiamos and originated from a library made of OregonR adult DNA (Siden Kiamos et al. 1990). P1 phages were provided by C. MacKimmie and M. Ashburner and originated from a library made of y, cn bw sp strain adults DNA (Smoller et al. 1991). In situ localization of P1 clones was found in Hartl et al. (1994) or provided by the BDGP. In situ localization of cosmid clones was provided by the EDGP. Data on the contigs assembled by STS mapping come from Kimmerly et al. (1996). Data concerning the STSs and gene sequences were found in the FlyBase databank. The P1 sequences were obtained from the BDGP databank.
Unless otherwise indicated, all molecular procedures were performed as described by Ausubel et al. (1994).
Cosmid and P1 DNA Extraction
Cosmid clones were grown in 2YT-kanamycin broth as indicated in Siden-Kiamos et al. (1990). P1 clones were grown in LB-kanamycin broth for 3 hr, then supplemented with 5 ml of 0.1 m IPTG and shaken for another 3 hr, as reported in FlyBase. Clone and vector DNA was isolated by an alkaline extraction procedure followed by CsCl-gradient separation. Standard techniques including purification by ultracentrifugation on ethidium bromide–cesium chloride gradients were used for extraction of genomic DNA.
Obtaining Contigs and Restriction Mapping
Standard techniques were used for DNA digestion with restriction enzymes (1 μg of DNA in each lane), gel electrophoresis (0.5% or 0.7% and 1.2% Seakem agarose) and Southern blotting onto Nylon+ (ICN products) membranes. The probes were random-primed labeled with [α-32P]dATP (ICN products) using the Klenow fragment of DNA polymerase I (Promega) and used at a concentration of 106 cpm/ml of hybridizing solution. Hybridizations were carried out at 65°C in 0.5 m at pH 7.2 Na2HPO4-NaH2PO4, 7% SDS, and 1 mm EDTA. The final washing was done in 0.2× SSC, 0.1% SDS at 65°C. Hybridized filters were autoradiographed with Fuji RX films at −80°C. Restriction maps were drawn using Plasmid Artist TM 1.13 software.
Fragments corresponding to the end of clone inserts were obtained by standard fragment extraction procedures (Ausubel et al. 1994), quantified, radioactively labeled, and used as probes.
Localization of STS and Gene Fragments
Fragments corresponding to STS DM0881, 0843, 0556, 0835, 1275, 2546, 0810, and 0406 were obtained by PCR experiments using the primers and amplification conditions described in FlyBase.
Fragments of genes located in the region were obtained by PCR using 300 ng of D. melanogaster genomic DNA. The DNA was dissolved in 10 mm Tris-HCl at pH 9, 3 mm MgCl2, 50 mm KCl, 0.1% TritonX-100, 150 mm each dATP, dCTP, dGTP, and dTTP, 0.1 mm of each oligonucleotide, in a 100-μl reaction volume with 5 units of Taq Polymerase (Promega). Each PCR was carried out in a programable thermal controller (Perkin-Elmer) for 30 cycles. The cycle was: denaturing at 94°C for 1 min, annealing for 1 min, and extending at 72°C for 30 sec. At the end of the thirtieth cycle, the heat denaturation step was omitted and extension was allowed to proceed at 72°C for 3 min. Primers containing restriction sites (EcoRI, BamHI) were used to obtain the following fragments: a 1065-bp three rows fragment (NTc1 5′-GGAATTCCGACGGCACTGCATATTGG-3′ and NTc2 5′-CGGGATCCCGCAATCAATAGACGTTT-3′ TA 61°C), a 395-bp diptericin fragment (NDipt1 5′-GGAATTCCCTGCAGCAAAGGTATCA-3′ and NDipt2 5′-CGGGATCCCGAAGCTTAGAAATTCGGA-3′ TA 57°C), a 195-bp enabled fragment (Nena1 5′-GGAATTCCCTGCAGCAGTTCAAGCTC-3′ and Nena2 5′-CGGGATCCCGGATCCTTCTTTTCTGC-3′ TA 60°C), a 270-bp coracle fragment (Ncora1 5′-GGAATTCCGAGACGCCCACATCCG-3′ and Ncora2 5′-CGGGATCCCGTAGTCGCCCATCTCCG-3′ TA 64°C), a 170-bp 5HTA-1 fragment (NHTA1 5′-GGAATTCCCTGCAGCGTATCGAGCA-3′ and NHTA3 5′-CGGGATCCCGTCGACGATGGATGCGTT-3′ TA 61°C), a 250-bp 5HTB-1 fragment (NHTB1 5′-GGAATTCCTGCAGAACAGTGATCGGAG-3′ and NHTB2 5′-CGGGATCCCGGATCCGGGTTATGCAAAAT-3′ TA 60°C). The primers Ote1 5′-GAGACGCCCACAGATCCG-3′ and Ote1b 5′-CGTAGTCGCCCATCTCCG-3′ were used to obtain a 220-bp otefin fragment and the Ote1 and Ote2 5′-AGTGCGACCCTTGTAGCG-3′ primers were used to obtain a 627-bp otefin fragment. In both cases, the TA was 52°C. The DNA fragments were eluted using a Promega PCR Prep Kit, and the concentration of each was estimated on agarose gels. The otefin fragments were subcloned with the pGEM-T vector (Promega) and the other fragments with a M13mp18 phage vector digested with BamHI and EcoRI. Each DNA fragment was sequenced, for both strands, in a Li-Cor automatic sequencer, using a Sequitherm Excel II long-read sequencing kit (Epicentre Technology) and labeled universal and reverse IRD41 primers in the conditions described by the suppliers. Sequences were analyzed with Infobiogen Bisance programs (Dessen et al. 1990).
STS and gene DNA fragments were labeled with 32P by random priming and used at a concentration of 106 cpm/ml of hybridizing solution. Hybridizations and washings were carried out as described previously.
In situ hybridization
In situ hybridization of biotinylated probes (Boehringer kit) to salivary gland polytene chromosomes was adapted from Engels et al. (1986). This technique permits identification of the sites of homologous DNA.
Acknowledgments
We thank I. Siden-Kiamos and C. MacKimmie for providing the cosmid and P1 clones, P. Georgel for personal communication on the Df(2R)Pc66 limits, and E. Huguet and J.M. Drezen for helpful comments on the manuscript. This work was supported by grants from the CNRS (UPRES-A 6035), the Ministère de Enseigment National de la Recherche et de la Technologie (MENRT) (Génome et Interactions durables). We are grateful for the assistance provided by European Community (EC) (grant no. AIR36CT9-1433) and the French CNRS-INRA program Biodiversité. M. Hita is supported by an EC grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL poirie@univ-tours.fr; FAX 33247366966.
REFERENCES
- Ausubel FM, Brent R, Kingston RR, Moore DD, Seidman JG, Smith JA, Struhl K. In: Current protocols in molecular biology. Janssen K, editor. New York, NY: J. Wiley and Sons; 1994. [Google Scholar]
- Baker B, Zambrisky P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant–microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- Benassi V, Frey F, Nappi AJ. A new specific gene for wasp cellular immune resistance in Drosophila. Heredity. 1998;80:347–352. doi: 10.1046/j.1365-2540.1998.00303.x. [DOI] [PubMed] [Google Scholar]
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Carton Y, Boulétreau M. Encapsulation ability of Drosophila melanogaster: A genetic analysis. Dev Comp Immunol. 1985;9:211–219. doi: 10.1016/0145-305x(85)90112-0. [DOI] [PubMed] [Google Scholar]
- Carton Y, Nappi JA. The Drosophila immune reaction and the parasitoid capacity to evade it: Genetic and coevolutionary aspects. Acta Oecologica. 1991;12:89–104. [Google Scholar]
- ————— Drosophila cellular immunity against parasitoids. Parasitology Today. 1997;13:218–227. doi: 10.1016/s0169-4758(97)01058-2. [DOI] [PubMed] [Google Scholar]
- Carton Y, Frey F, Nappi JA. Inheritance of cellular immune resistance in Drosophila melanogaster. Heredity. 1992;69:393–399. doi: 10.1038/hdy.1992.141. [DOI] [PubMed] [Google Scholar]
- Collins FH. Progress in the map-based cloning of the Anopheles gambiae genes responsible for the encapsulation of malarial parasites. Ann Tropical Med Parasitol. 1997;91:517–521. doi: 10.1080/00034989760888. [DOI] [PubMed] [Google Scholar]
- Cousteau C, Carton Y, Nappi A, Shokoski F, Ffrench-Constant R. Differential induction of antibacterial transcripts in Drosophila susceptible and resistant to parasitism by Leptopilina boulardi. Insect Mol Biol. 1996;5(3):167–172. doi: 10.1111/j.1365-2583.1996.tb00050.x. [DOI] [PubMed] [Google Scholar]
- Cross SM, Little PFR. A cosmid vector for systematic chromosome walking. Gene. 1986;49:9–22. doi: 10.1016/0378-1119(86)90381-1. [DOI] [PubMed] [Google Scholar]
- Dessen P, Fondrat C, Valencien C, Mugnier C. BISANCE: A French service for access to biomolecular databases. Comp Appl Biosci. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- Dushay M, Eldon D. Insights from model systems. Drosophila immune responses as models for human immunity. Am J Hum Genet. 1998;62:10–14. doi: 10.1086/301694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR, Preston CR, Thompson P, Eggleston WB. In situ hybridization to Drosophila salivary chromosomes with biotinylated DNA probes and alkaline phosphatase. Focus. 1986;8:6–8. [Google Scholar]
- Fujimoto K, Kawabata S, Iwanaga S, Ohnishi E. Nucleotide sequence of the cDNA encoding the proenzyme of phenol oxydase A1 in Drosophila melanogaster. Proc Natl Acad Sci. 1995;92:7769–7773. doi: 10.1073/pnas.92.17.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Green MM, Corces VG. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: The molecular basis of transvection in Drosophila. EMBO J. 1990;9:2247–2256. doi: 10.1002/j.1460-2075.1990.tb07395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson TJ, Rosenthal AR, Waterston RH. Lorist6, a cosmid vector with BamHI, NotI, ScaI and HindIII cloning sites and altered neomycin phosphotransferase gene expression. Gene. 1987;53:283–286. doi: 10.1016/0378-1119(87)90017-5. [DOI] [PubMed] [Google Scholar]
- Goldsborough AS, Kornberg TB. Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature. 1996;381:807–810. doi: 10.1038/381807a0. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Nurminsky DI, Jones RW, Lozovskaya ER. Genome structure and evolution in Drosophila: Applications of the framework P1 map. Proc Natl Acad Sci. 1994;91:6824–6829. doi: 10.1073/pnas.91.15.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nuclear organization and gene expression: Homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichart JM. Drosophila immunity. Trends Cell Biol. 1997;7:30–316. [Google Scholar]
- Hoffmann JA. Innate immunity of insects. Curr Opin Immunol. 1996;8:8–13. doi: 10.1016/s0952-7915(96)80098-7. [DOI] [PubMed] [Google Scholar]
- Kafatos FC, Louis C, Savakis C, Glover DM, Asbhurner M, Link AJ, Siden-Kiamos I, Saunders RDC. Integrated maps of the Drosophila genomes: Progress and prospects. Trends Genet. 1991;7:155–161. doi: 10.1016/0168-9525(91)90379-5. [DOI] [PubMed] [Google Scholar]
- Kimmerly W, Stultz K, Lewis S, Lewis K, Lustre V, Romero R, Benke J, Sun D, Shirley G, Martin C, Palazzolo M. A P1-based physical map of the Drosophila euchromatic genome. Genome Res. 1996;6:414–430. doi: 10.1101/gr.6.5.414. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart J-M, Hoffmann JA. Drosophila host defense: Differential induction of antimicrobial peptides genes after infection by various classes of microorganisms. Proc Natl Acad Sci. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat. 1954;88:225–239. [Google Scholar]
- Louis C, Madueno E, Modolell J, Omar MM, Papagiannakis G, Saunders RDC, Savakis C, Siden-Kiamos I, Spanos L, Topalis P, et al. One-hundred and five new potential Drosophila melanogaster genes revealed through STS analysis. Gene. 1997;195:187–193. doi: 10.1016/s0378-1119(97)00138-8. [DOI] [PubMed] [Google Scholar]
- Miklos GL, Rubin GM. The role of the genome project in determining gene function: Insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- Nappi AJ, Carton Y, Frey F. Parasite-induced enhancement of hemolymph tyrosinase activity in a selected immune reactive strain of Drosophila melanogaster. Arch Insect Biochem Physiol. 1991;18:159–168. doi: 10.1002/arch.940180304. [DOI] [PubMed] [Google Scholar]
- Nappi AJ, Vass E, Frey F, Carton Y. Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur J Cell Biol. 1995;68:450–456. [PubMed] [Google Scholar]
- Orr HA, Irving S. The genetics of adaptation: The genetic basis of resistance to wasp parasitism in Drosophila melanogaster. Evolution. 1997;51:1877–1885. doi: 10.1111/j.1558-5646.1997.tb05110.x. [DOI] [PubMed] [Google Scholar]
- Pierce JC, Sauer B, Sternberg N. A positive selection vector for cloning high molecular weight DNA by the bacteriophage P1 system: Improved cloning efficacy. Proc Natl Acad Sci. 1992;89:2056–2060. doi: 10.1073/pnas.89.6.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM. Around the genomes: The Drosophila Genome Project. Genome Res. 1996;6:71–79. doi: 10.1101/gr.6.2.71. [DOI] [PubMed] [Google Scholar]
- Siden Kiamos I, Saunders RDC, Spanos L, Majerus T, Trenear J, Savakis C, Louis C, Glover DM, Ashburner M, Kafatos FC. Towards a physical map of the Drosophila melanogaster genome: Mapping of cosmid clones within defined genomic divisions. Nucleic Acids Res. 1990;18:6261–6270. doi: 10.1093/nar/18.21.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller DA, Petrov D, Hartl DL. Characterization of bacteriophage P1 library containing inserts of Drosophila DNA of 75–100 kilobase pairs. Chromosoma. 1991;100:487–494. doi: 10.1007/BF00352199. [DOI] [PubMed] [Google Scholar]
- Sorsa V. Chromosome maps of Drosophila. II. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- Sternberg N. Bacteriophage P1 cloning system for the isolation, amplification and recovery of DNA fragments as large as 100 kilobases pairs. Proc Natl Acad Sci. 1990;87:103–107. doi: 10.1073/pnas.87.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. The P1 cloning system: Past and future. Mamm Genome. 1994;5:397–404. doi: 10.1007/BF00356999. [DOI] [PubMed] [Google Scholar]
- Vass E, Nappi AJ, Carton Y. Comparative study of immune competence and host susceptibility in Drosophila melanogaster parasitized by Leptopilina boulardi and Asobara tabida. J Parasitol. 1993;79:106–112. [Google Scholar]
- Vilmos P, Kurucz E. Insect immunity: Evolutionnary roots of the mammalian innate immune system. Immunol Lett. 1998;62:59–66. doi: 10.1016/s0165-2478(98)00023-6. [DOI] [PubMed] [Google Scholar]
- Wicker C, Reichhart JM, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann J. Characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J Biol Chem. 1990;265:22493–22498. [PubMed] [Google Scholar]