Abstract
Kalirin-7 (Kal7), a multi-functional Rho GDP/GTP exchange factor (GEF) for Rac1 and RhoG, is embedded in the post-synaptic density at excitatory synapses, where it participates in the formation and maintenance of dendritic spines. Kal7 has been implicated in long-term potentiation, fear memories and addiction-like behaviors. Using liquid chromatography and tandem mass spectroscopy, we identified sites phosphorylated by six PSD-localized kinases implicated in synaptic plasticity and behavior, sites phosphorylated when myc-Kal7 was expressed in non-neuronal cells and sites phosphorylated in mouse brain Kal7. A site in the Sec14p domain phosphorylated by calcium/calmodulin dependent protein kinase II, protein kinase A and protein kinase C, was phosphorylated in mouse brain, but not in non-neuronal cells. Phosphorylation in the spectrin-like repeat region was more extensive in mouse brain than in non-neuronal cells, with a total of 20 sites identified. Sites in the pleckstrin homology domain and in the linker region connecting the GEF domain to the PDZ binding motif were heavily phosphorylated in both non-neuronal cells and in mouse brain and affected GEF activity. We postulate that the kinase convergence and divergence observed in Kal7 identify it as a key player in integration of the multiple inputs that regulate synaptic structure and function.
Keywords: phosphorylation, CaMKII, CKII, kinase convergence, spectrin repeat, plasticity, dendritic spine, synaptogenesis, Rho-GEF
Introduction
In the mammalian forebrain, the vast majority of excitatory neurons synapse onto dendritic spines1. Dendritic spines show rapid plasticity in both their size and number in response to various stimuli2–4, a feature that is dependent on rearrangement of the actin cytoskeleton which makes up the core of each spine5,6. Rho-guanine nucleotide exchange factors (RhoGEFs) are critical regulators of the actin cytoskeleton. In the mouse and human genomes there are ~60 Rho-GEFs and these proteins are known to play important roles in cell signaling, actin rearrangement and protein localization7. Dysfunctions in Rho-GEF signaling pathways are associated with various mental retardation syndromes and schizophrenia8,8–11. Previous studies have shown that about a dozen Rho-GEFs are localized to the postsynaptic density12, a subcellular placement that puts these proteins in position to participate in changes that occur in response to synaptic stimulation.
Kalirin-7 (Kal7) is an190kDa Rho-GEF that is a product of the extensively spliced Kalrn gene. Linkage studies have identified roles for Kalrn in early onset coronary artery disease13, schizophrenia14,15, and Alzheimers disease16. While there are many isoforms of Kalirin, Kal7 accounts for the majority of Kalrn gene products in the adult mammalian brain. Kal7 is localized almost exclusively to the PSD and has a C-terminal PDZ-binding motif known to interact with PSD-9517. Over the past decade a number of studies have demonstrated that Kal7 is essential for dendritic spine formation and maintenance in cultured primary neurons18,19. Additionally, Kal7 has been shown to interact with a number of other principal components of the PSD including the scaffolding molecules DISC-1 and AF-6, as well as with the enzyme iNOS and the glutamate receptor subunit NR2B20–24. Mice with a constitutive genetic deletion of the exon unique to the Kal7 isoform show a significant decrease in dendritic spine density in the hippocampus, as well as deficient long-term potentiation (LTP) and focal learning impairments25. When given repeated doses of cocaine, the Kal7 knockout mice show abnormal dendritic spine plasticity in the nucleus accumbens and have aberrant locomotor sensitization and conditioned place preference responses26.
Despite its clear role in multiple signaling pathways, little is known about how the catalytic activity, protein/protein and protein/lipid interactions of Kal7 are regulated. As might be expected for a component of the PSD, phosphorylation of Kal7 affects its function. A single site (Thr1590) in the unstructured region that links the GEF domain of Kal7 to its PDZ binding motif is phosphorylated by the proline-directed kinase Cdk5, increasing its catalytic GEF activity; Kal7 that cannot be phosphorylated at this site (T1590A) produces dendritic spines of altered morphology27. Calcium influx through NMDA receptors localized to the PSD triggers changes in spine morphology and function, many of which result from activation of calcium/calmodulin dependent protein kinase II (CaMKII)28,29. Based on indirect evidence, Xie et al. suggested that CaMKII-mediated phosphorylation of Thr95 in the Sec14p domain of Kal7 was essential for Kal7 to exhibit GEF activity30.
The PSD is home to at least 50 protein kinases, and Tiam-1 and β-Pix, other Rho-GEFs localized to the PSD, are known to be regulated by phosphorylation12,31–34. Advancements in phosphoproteomic and mass spectroscopic techniques have begun to unearth the depth and breadth of the phosphorylation events that occur at the PSD35. Hundreds of phosphorylation events occur within the PSD in response to neurotransmitter stimulation; single proteins can serve as phosphorylation hubs, being phosphorylated as many as 58 times by 23 different kinases36. We used liquid chromatography and tandem mass spectrometry to identify Kal7 as a potential phosphorylation hub. Using recombinant myc-Kal7, we identified sites phosphorylated by protein kinase A, protein kinase C, CaMKII, casein kinase II and Fyn.
Analysis of myc-Kal7 expressed in non-neuronal cells and endogenous Kal7 isolated from mouse brain revealed 39 sites of phosphorylation, many conserved in the single Kalirin/Trio homologues found in D. melanogaster and C. elegans. Based on direct analysis, Thr95 was neither phosphorylated in vivo nor a CaMKII site in Kal7. While the functional consequences of the multiple phosphorylation events identified remain to be determined, it is clear that Kal7 is a heavily phosphorylated target of numerous PSD kinases.
Experimental Methods
Cell culture and transfection
pEAK Rapid cells (Edge Biosystems, Gaithersburg, MD) were maintained in DMEM:F12 medium containing 200 U/ml penicillin G, 20 μg/ml streptomycin sulfate, 25 mM HEPES, and 10% fetal bovine serum. Cells were fed with serum-free medium for 2 hours before transfection. A vector encoding His6-myc-tagged full-length rat Kal727,37 was mixed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in Opti-MEM (Life Technologies) and added to the cells for 6 hours at 37°C to allow for transfection. After six hours, cells were returned to growth medium for 24–48 hours. Cells were stimulated with the cyclic AMP analog 8-Br-cAMP (500μM), the PKC agonist phorbol myristate acetate (PMA) (1μM) and the calcium ionophore A23187 (10μM) for 30 minutes in the presence of two phosphatase inhibitors, calyculin A (50nM) and fenvalerate (100nM). Cells were then extracted in lysis buffer containing 1% SDS, 50mM NaF and 2mM Na orthovanadate; after heating at 95°C for five minutes, samples were centrifuged in a benchtop centrifuge for 15 minutes to remove insoluble particulates. To prepare Kal7 for phosphorylation by known kinases, cells were not stimulated and were harvested in SDS lysis buffer without phosphatase inhibitors (see below).
Preparation of cortical/striatal tissue for immunoprecipitation
Male C57BL/6 mice (2–6 months) were sacrificed and striata and prefrontal cortices were rapidly dissected and weighed on ice. Samples were then sonicated into 15 volumes of lysis buffer containing 1% SDS, 50mM Tris [pH 7.4], 130mM NaCl, 5mM EDTA, 50mM NaF, 1mM PMSF, protease inhibitor cocktail and phosphatase inhibitor cocktails I & II (Calbiochem). Samples were heated at 95°C for five minutes and then centrifuged for 15 minutes in a benchtop centrifuge to remove insoluble components. Following dilution into buffer containing NP-40 (see below), samples were used for immunoprecipitation.
Immunoprecipitation
For isolating Kal7 from cultured cells or neurons, we used a Kal7 monoclonal antibody (20D8, specific to the amino terminal portion of the final 20 amino acids of Kal7)25 crosslinked to agarose beads using the Pierce Direct IP kit (Pierce, Rockford, IL). Crosslinking was performed according to the manufacturer’s specifications. For immunoprecipitation from cortex, 4–5mg of total protein (as determined by BCA) was used for each sample; for cultured cells, protein from two wells of a six well plate was used. Before immunoprecipitation, SDS lysates were incubated with 0.5 volumes of 15% NP-40 for 20 minutes at 4°C. Samples were then diluted with 5 volumes of TES-mannitol (TM) buffer containing 50mM NaF and 2mM Na orthovanadate. Immunoprecipitations were performed overnight with continuous agitation. In the morning, beads were centrifuged and washed twice with TM + 1% TX-100 (TMT) and twice with TM. Samples were then heated at 95°C in 40μl of 1X Laemmli sample buffer and fractionated on polyacrylamide gels.
Gel electrophoresis and staining
For isolation of Kal7, samples were analyzed on 4–15% polyacrylamide gels (Bio-Rad, Hercules, CA). For samples from transfected cells, gels were stained with Coomassie Brilliant Blue R-250. For samples immunoprecipitated from brain, proteins were visualized using the SilverSNAP Kit (Pierce, Rockford, IL). For all samples, the Kal7 band was excised using a sterile scalpel, placed into a sterile microfuge tube and frozen on dry ice.
Protein digestion and TiO2 enrichment
Gel bands were washed with 250μl 50% acetonitrile/50% water for 5 minutes followed by 250μl 50mM ammonium bicarbonate/50% acetonitrile/50% water for 30 minutes, and 10mM ammonium bicarbonate/50% acetonitrile/50% water for 30 minutes. After washing, the gel was dried using a Speedvac and rehydrated with 0.1μg modified porcine trypsin (Promega) in 15 μl 10mM ammonium bicarbonate. Samples were digested at 37°C for 16 hours.
The digest was next acidified with 0.5% TFA, 50% acetonitrile. Top Tips (Glygen Corp.) were prepared by washing with 3 × 40μl 100% acetonitrile, followed by 3 × 40μl 0.2M sodium phosphate pH 7.0, and 3 × 40μl 0.5% TFA, 50% acetonitrile. Washes were spun through into an eppendorf tube at 2,000 rpm for 1 minute. Acidified digest supernatants were loaded into Top Tips, spun at 1,000 rpm for 1 minute, and then 3,000 rpm for 2 minutes. Gel pieces were rinsed with 40μl 0.5% TFA, 50% acetonitrile, with the supernatant transferred to the Top Tip and the spin repeated. The Top Tip was then washed with 40μl 0.5% TFA, 50% acetonitrile and the spin repeated. The flow through from these washes were saved and analyzed by LC-MS/MS as below. Phosphopeptides were eluted from the TopTip with 3 × 30μl 28% ammonium hydroxide. Both the flow through and eluted fractions were dried using a vacuum centrifuge, and then dried again from 40 μl of water. Samples were dissolved in 3μl 70% formic acid, vortexed, diluted with 7μl 50mM sodium phosphate, pH 7.8, spun and transferred to LC-MS/MS vials from which 5μl was injected.
Liquid chromatography/mass spectrometry analysis (LC-MS/MS) - was performed on a Thermo Scientific LTQ Orbitrap XL equipped with a Waters nanoACQUITY UPLC system, a Waters Symmetry® C18 180μm × 20mm trap column and a 1.7 μm, 75 μm × 250 mm nanoACQUITY™ UPLC™ column (35ºC) for peptide separation. Trapping was done at 15μl/min, 99% Buffer A (100% water, 0.1% formic acid) for 1 min. Peptide separation was performed at 300nl/min with Buffer A: 100% water, 0.1% formic acid and Buffer B: 100% CH3CN, 0.075% formic acid. A linear gradient (51 minutes) was run with 5% buffer B at initial conditions, 50% B at 50 minutes, and 85% B at 51 minutes. MS was acquired in the Orbitrap using 1 microscan, and a maximum inject time of 900 followed by six data dependent MS/MS acquisitions in the ion trap. Multistage activation was used for neutral losses of 98.0, 49.0, 32.7 and 24.5 amu. External calibration of the mass spectrometer was performed as per the manufacturer with a solution of caffeine, MRFA and Ultramark 1621.
Identification of phosphorylated peptides
The LC-MS/MS data were searched using the Mascot Distiller and Mascot search algorithms (Matrix Science, www.matrixscience.com). The Mascot Distiller program combines sequential MS/MS scans from profile data that have the same precursor ion. Charge states of +2 and +3 were preferentially located with a signal to noise ratio of 1.2 or greater and a peak list was generated for database searching against the NCBInr database, mouse taxonomy and/or the IPI mouse database. The Mascot significance score match is based on a MOWSE score and relies on multiple matches to more than one peptide from the same protein. Parameters used for searching were a significance threshold of p<0.05, a peptide tolerance of ±20ppm, MS/MS fragment tolerance of ±0.6 Da, 1–3 missed cleavage sites and peptide charges of +2 or +3. Variable modifications included propionamide, methionine oxidation, phosphoserine, phosphothreonine and phosphotyrosine. Normal and decoy database searches were run against the mouse Kal7 sequence (NCBI Reference Sequence: NP_001157740.1).
Generation of Kal7 expression vectors
Point mutations of selected phosphorylation sites in rat His6-myc-Kal7 [Thr95/Ala (T95A); Ser487-Thr495/Ala-Ala (S3-ST/AA); Thr1519-Ser1520/Ala-Ala (PH-TS/AA); Thr1519-Ser1520/Glu-Asp (PH-TS/ED); Tyr1653/Phe (Y1653F)] were introduced into the His6-myc-Kal7 vector using the QuickChange point mutation kit (Stratagene, La Jolla, CA) as described27. The constructs encoding the inactivated GEF (ND/AA) and Kal7 lacking the Sec14p domain (ΔSec14p) were created previously37.
Rac activation assays
pEAK Rapid cells were transfected as described above with full length Kal7 or one of the mutant Kal7 constructs. Twenty-four hours after transfection, cells were scraped into MLB [25mM HEPES [pH 7.4], 150mM NaCl, 1% NP-40, 10mM MgCl2, 5mM EDTA, 2% glycerol, and protease inhibitor cocktail38] with 10μg/ml of GST-Pak-CRIB. Samples were briefly sonicated and centrifuged at 1000 × g for 5 minutes at 4°C to remove insoluble particulates. A sample of this supernatant was saved to be measured as the input fraction and the remainder was incubated with 15μl of glutathione Sepharose 4B resin for 1 hour at 4°C. Beads were then pelleted and washed twice with 1X MLB; bound Rac was eluted by heating at 95°C for 5 min in 30μl of 1X Laemmli buffer. Input and bound samples were analyzed on polyacrylamide gels; Rac1 was detected with monoclonal antibody from Millipore (Billerica, MA).
Dephosphorylation of His6-myc-Kal7 and phosphorylation by known kinases
His6-myc-Kal7 expressed in pEAK Rapid cells was left on the antibody beads, which were exposed to Lambda phosphatase or calf intestinal phosphatase (CIP) (both from New England Biolabs, Ipswich, MA). These phosphatases were chosen because they have high specific activities for phosphorylated Ser/Thr and Tyr, respectively (http://www.neb.com/nebecomm/tech_reference/proteintools/phosphatases.asp). Samples to be treated with Lambda phosphatase were washed with PMP buffer (50mM HEPES, 100mM NaCl, 2mM DTT, 0.01% Brij 35 [pH 7.5]) and resuspended in 1X PMP supplemented with 1mM MnCl2 and 2μl of Lambda phosphatase. Samples to be treated with CIP were washed with NEB Buffer 3 (50mM Tris-HCl, 100mM NaCl, 10mM MgCl2, 1mM DTT [pH 7.9]) and then resuspended in NEB Buffer 3 with 2μl of CIP. All samples had 1μM PMSF and protease inhibitor cocktail added for the phosphatase treatment, which was carried out for 60 minutes at 31°C with constant agitation. EDTA (50mM final concentration) was added to stop the reaction; after 5 min, the beads were washed twice with TMT and once with TM. One set of samples was boiled into sample buffer immediately after phosphatase treatment to identify sites that were not dephosphorylated.
Each sample was then washed once with the appropriate kinase buffer and resuspended in 100μl of the same buffer (detailed below) plus 200μM ATP, phosphatase inhibitor cocktails I & II (Tocris Bioscience, Ellisville, MO), 1μM PMSF, and protease inhibitor cocktail. Incubations were performed for 45 minutes at 31°C with agitation. Casein kinase II (CKII; 1000 units/reaction; New England BioLabs, Ipswich, MA) was used in 20mM Tris-HCl, 50mM KCl, 10mM MgCl2 [pH 7.5]. The catalytic subunit of protein kinase A (PKA; 5000 units/reaction; New England Biolabs, Ipswich, MA) was used in 50mM Tris-HCl, 10mM MgCl2 [pH 7.5]. Calcium/calmodulin dependent protein kinase II (CaMKII; 1000 units/reaction; New England Biolabs, Ipswich, MA) was pre-activated by incubating 1000 units of enzyme in CaMKII buffer (50mM Tris-HCl, 10mM MgCl2, 2mM DTT, 0.1mM Na2EDTA [pH 7.5]) containing 200μM ATP, 1.2μM calmodulin and 2mM CaCl2 for 10 minutes at 31°C. The activated kinase was then added to the immunoprecipitated protein. Protein kinase C (PKCa; ~5000units/ reaction; Sigma-Aldrich, St. Louis, IL) was used in 50mM Tris-HCl, 10mM MgCl2 [pH 7.5]. Fyn (~7000 units/reaction; Sigma-Aldrich, St. Louis, IL) was used in 50mM Tris-HCl, 10mM MgCl2, 2mM DTT, 0.1mM Na2EDTA [pH 7.5]. After phosphorylation, the beads were pelleted, washed once with TMT and once with TM and Kal7 was eluted into 1X Laemmli buffer by heating at 95°C for 5 min.
Results and Discussion
Purified recombinant Kal7 is phosphorylated by kinases of multiple classes
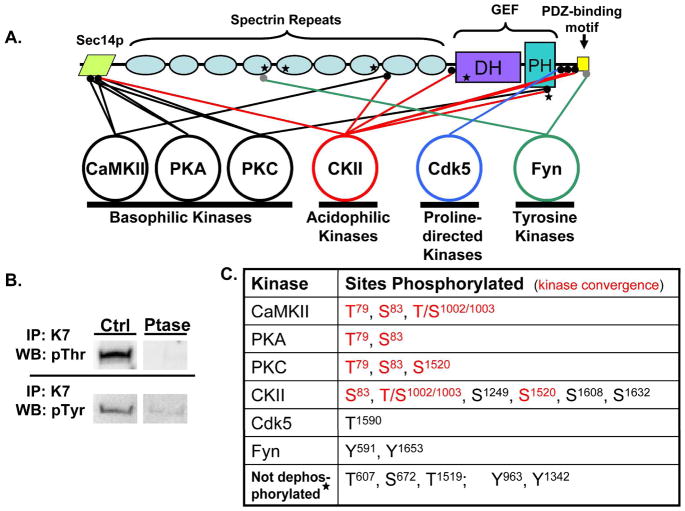
Kal7 is a complex molecule with multiple functional domains (Fig. 1A). At its N-terminus is a Sec14p domain that is known to interact with phosphatidylinositol-3-phosphate and play an essential functional role37. The Sec14p domain is followed by nine spectrin-like repeat regions that have been shown to interact with a variety of other proteins including PAM (peptidylglycine a-amidating monooxygenase), Arf6, HAPIP, iNOS and DISC-120,23,39,40. While the spectrin-repeat region is not necessary for GEF activity, it plays an essential role in the spine formation observed when exogenous myc-Kal7 is expressed in neurons. The spectrin-repeats are followed by tandem DH-PH domains characteristic of Rho-GEFs7. At the extreme C-terminus of Kal7 is a PDZ-binding motif which binds to PSD-95, AF-6 and a number of other PDZ-domain containing proteins22. The final 20 amino acids of Kal7, which are encoded by a separate 3′-terminal exon, are unique to Kal7 and ΔKal7, an N-terminally truncated isoform. The PDZ-binding motif is connected to the preceding GEF domain by an unstructured 60 amino acid linker region.
Figure 1.
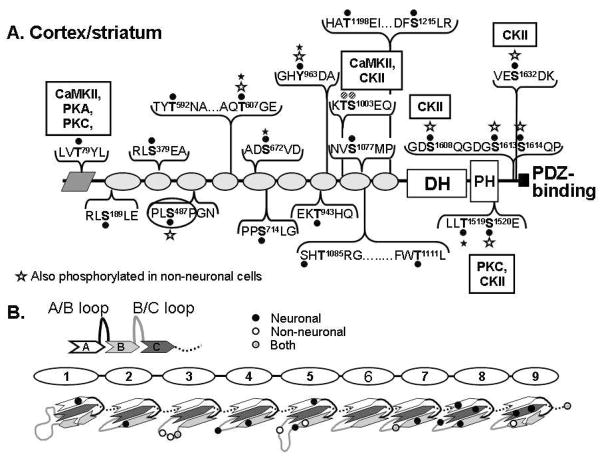
A. Purified myc-Kal7 is phosphorylated by recombinant protein kinases of different classes. Myc-Kal7 purified from transiently transfected pEAK Rapid cells was dephosphorylated as described in Methods. Recombinant protein kinases representative of four different kinase classes were then incubated with myc-Kal7 and 200 μM ATP under optimal conditions for each enzyme. LC-MS/MS analysis of tryptic peptides from each sample revealed multiple phosphorylation sites for each of the kinases except Cdk5, which had previously been shown to phosphorylate a single site in Kal727. Sites that were not dephosphorylated by phosphatase treatment are indicated by a star; Thr607 and Ser672 are consensus CKII sites (NetPhos 1.0). B. Immunoprecipitated myc-Kal7 was treated with Lambda phosphatase or CIP and then visualized with antibody specific for P-Thr or P-Tyr, respectively. Equal loading of myc-Kal7 was determined by the presence of a single Coomassie-stained band at the correct molecular weight that was not in an immunoprecipitate from non-transfected cells (not shown). C. Sites phosphorylated by each kinase are listed. Red lettering represents sites where multiple kinases converged on a single residue. It was not possible to distinguish phosphorylation of Thr1002 from phosphorylation of Ser1003.
In the adult rodent brain, Kal7 is highly localized to the PSD, a complex, 1 gigadalton molecular machine41,42. Proteomic analysis has demonstrated that more than fifty kinases are localized to the PSD24. We selected kinases known to be intricately involved in synaptic plasticity and evaluated their ability to phosphorylate purified myc-Kal7 in vitro; Ser/Thr kinases from the basophilic (CaMKII, PKA, PKC), acidophilic (CKII) and proline-directed (Cdk5) classes were selected, along with one non-receptor tyrosine kinase (Fyn). Before exposure to these recombinant kinases, myc-Kal7 was subjected to dephosphorylation by Lambda phosphatase (Ser/Thr kinases) or calf intestinal alkaline phosphatase (Tyr kinases). Although extensive dephosphorylation occurred (Fig. 1B), mass spectroscopic analysis of tryptic peptides prepared from dephosphorylated myc-Kal7 revealed continued phosphorylation of three Ser/Thr (Thr607; Ser672; Thr1519) and two Tyr (Tyr963, Tyr1342) sites (marked by a star in Fig. 1A). Data for all of the tryptic phosphopeptides identified in in vitro phosphorylated myc-Kal7 are summarized in Table 1 and representative spectra are shown in Supplementary Figs. S1 to S22
Table 1.
Sites in myc-Kal7 Phosphorylated by Recombinant Kinases
| Residue | Score | Peptide Sequence | EN/FT | M/Z | Ion Mass | Charge | |
|---|---|---|---|---|---|---|---|
| CamKII | |||||||
| 79 | 38.67 | R.KLVpTYLASVPSEDVCK.R | FT | 916.4415 | 1830.869 | 2 | |
| 83 | 33.22 | K.LVTYLApSVPSEDVCKR.G | FT | 930.4441 | 1858.874 | 2 | |
| 1002/1003 | 26.16 | K.pTpSEQVCSVLESLEQEYR.R | FT | 693.9797 | 2078.917 | 3 | |
| 95-Non | 67.48 | R.GFTVIID | FT | 534.2798 | 1066.545 | 2 | |
| PKA | |||||||
| 79 | 35.64 | R.KLVpTYLASVPSEDVCK.R | FT | 916.4418 | 1830.869 | 2 | |
| 83 | 28.25 | K.LVTYLApSVPSEDVCKR.G | FT | 930.4448 | 1858.875 | 2 | |
| PKC | |||||||
| 79 | 32.12 | R.KLVpTYLASVPSEDVCK.R | FT | 916.441 | 1830.868 | 2 | |
| 83 | 33.49 | K.LVTYLApSVPSEDVCKR.G | FT | 930.4434 | 1858.872 | 2 | |
| 1520 | 63.22 | K.LLTpSELGVTEHVEGDPCK.F | FT | 1003.952 | 2005.889 | 2 | |
| CKII | |||||||
| 83 | 30.44 | K.LVTYLASVPSEDVCKR.G | FT | 930.4428 | 1858.871 | 2 | |
| 1002/1003 | 48.81 | K.pTpSEQVCSVLESLEQEYR.R | FT | 1040.466 | 2078.917 | 2 | |
| 1247 | 35.52 | K.ALGVNTEDNKDLELDIIPApSLSDR.E | FT | 893.4325 | 2677.276 | 3 | |
| 1249 | 83.59 | K.DLELDIIPASLpSDR.E | EN | 818.8926 | 1635.771 | 2 | |
| 1520 | 43.97 | K.LLTpSELGVTEHVEGDPCK.F | FT | 1003.951 | 2005.887 | 2 | |
| 1608 | 91.63 | R.DGVEDGDpSQGDGSSQPDTISIASR.T | EN | 1236.997 | 2471.98 | 2 | |
| 1632 | 86.22 | R.TSQNTVEpSDKDGNLVPR.W | EN | 970.4376 | 1938.861 | 2 | |
| Fyn | |||||||
| 591 | 34.3 | K.RHDDFEEVAQNTpYTNADK.L | EN | 744.9745 | 2231.902 | 3 | |
| 1653 | 56.15 | R.WHLGPGDPFSTpYV. | EN | 778.3316 | 1554.649 | 2 | |
| Not dephosphorylated | |||||||
| 607 | 62.72 | K.LLEAAEQLAQpTGECDPEEIYK.A | FT | 1215.531 | 2429.048 | 2 | |
| 672 | 52.77 | K.EVLEDVCADpSVDAVQELIK.Q | FT | 1077.988 | 2153.961 | 2 | |
| 1519 | 53.79 | K.LLpTSELGVTEHVEGDPCK.F | FT | 1003.952 | 2005.889 | 2 | |
| 963 | 36.75 | K.AEALLQAGHpYDADAIRECAEK.V | FT | 1177.519 | 2353.023 | 2 | |
| 1342 | 61.96 | K.pYEQLPEDVGHCFVTWADK.F | FT | 1108.962 | 2215.91 | 2 | |
All peptides are numbered using the sequence of rat Kal7 with the N-terminus encoded by the KalrnA promoter (U88157). Peptides are grouped by the kinase used for phosphorylation. For each peptide, the sequence is shown, with cleavage sites indicated by dots and the phosphorylated residue in red. EN/FT indicates whether the peptide was detected in the TiO2 enriched (EN) or unbound (FT) fraction. M/Z, mass/charge ratio. Information for the non-phosphorylated Thr95 peptide (95-Non) is provided along with the data for CamKII. Representative spectra are shown in Supplementary Figs. S1 to S22.
Dephosphorylated myc-Kal7 was phosphorylated by each of the protein kinases selected. With the exception of Cdk5, a proline-directed kinase studied previously27, each protein kinase phosphorylated myc-Kal7 at more than one site (Fig. 1A,C). The three basophilic kinases tested each phosphorylated the same two sites in the Sec14p domain of myc-Kalirin (Thr79, Ser83). Kinase convergence of this type is observed for many targets of synaptically localized protein kinases36. Casein kinase II, an acidophilic kinase, phosphorylated one of these same sites in the Sec14p domain (Ser83). In another case of kinase convergence, casein kinase II phosphorylated a site in spectrin repeat 8 that was also phosphorylated by CaMKII (Thr/Ser1002/3) and a site in the PH domain that was also phosphorylated by PKC (Ser1520). The phenomenon of primed convergence, in which one phosphorylation event facilitates another, may also occur. Both CKII and PKC phosphorylate Ser1520 in the PH domain; the preceding residue, Thr1519, was not completely dephosphorylated by the phosphatase treatment employed. The single tyrosine kinase tested, Fyn, phosphorylated a site in spectrin repeat 4 along with the penultimate residue of the PDZ binding motif (Tyr1653).
An essential role for CaMKII activity in the activation of Rac1 in neuronal cultures was demonstrated using CaMKII inhibitors30. Based on indirect evidence, it was suggested that CaMKII phosphorylated Kal7 at a single site, Thr95, leading to activation of its GEF domain. Using purified Kal7 and purified CaMKII, we observed phosphorylation of Thr79, Ser83 and Thr/Ser1002/1003, but not of Thr95; the tryptic peptide containing non-phosphorylated Thr95 was detected repeatedly (Table 1).
Identification of myc-Kal7 phosphorylation sites used in non-neuronal cells
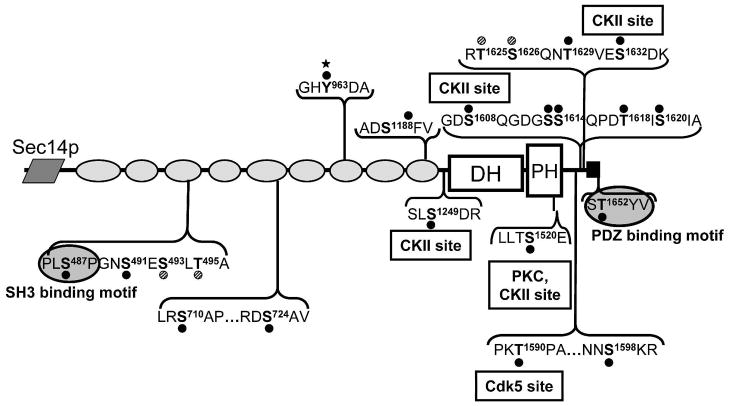
Since purified myc-Kal7 was phosphorylated by multiple protein kinases, we next sought to determine sites in myc-Kal7 that were phosphorylated in cells. For the initial studies, myc-Kal7 was expressed in pEAK Rapid cells, a derivative of HEK293 cells that adopt a compact, round morphology in response to expression of myc-Kal737. In an attempt to maximally phosphorylate myc-Kal7 and identify multiple phosphorylation sites, we stimulated transfected cells with agonists of the PKA (8Br-cAMP) and PKC (phorbol myristate acetate) pathways as well as a calcium ionophore (A23187) and included cell permeant phosphatase inhibitors (calyculin A and fenvalerate) in the incubation medium. Analysis of tryptic phosphopeptides of myc-Kal7 isolated from these cells revealed 20 unique phosphorylation sites (Figs. 2 and 3). These sites were identified from analysis of over 1700 individual peptides identified from 8 different cell culture preparations. Data for all of the tryptic phosphopeptides identified in myc-Kal7 purified from stimulated pEAK Rapid cells are summarized in Table 2, a representative spectrum is shown in Fig. 2 and the remaining spectra are shown in Supplementary Figs. S23 to S41.
Figure 2.
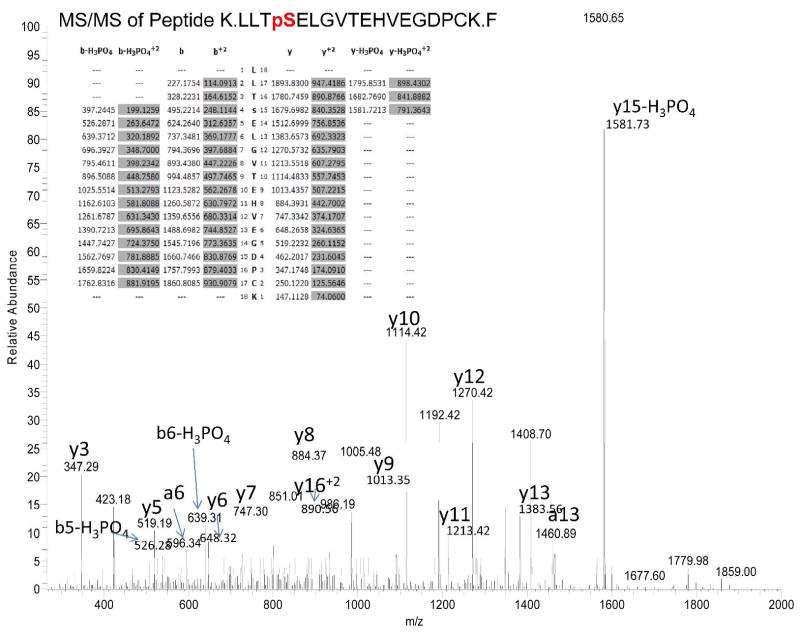
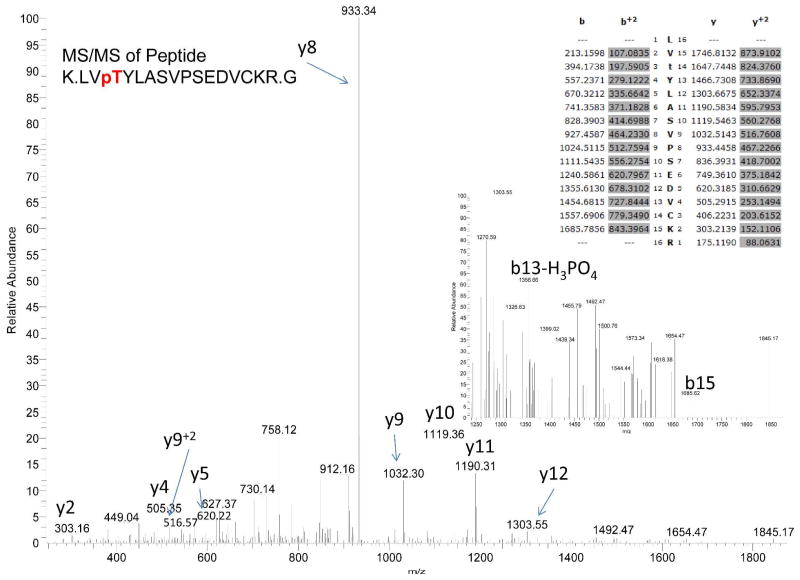
Identification of phosphorylated Ser1520 in myc-Kal7 expressed in pEAK Rapid cells. The spectrum from MS/MS of the doubly charged M+2 1004.46 ion of peptide K.LLTpSELGVTEHVEGDPCK.F is shown. Based on the y10 and y15-H3PO4 ion, residue 1520 is phosphoserine. Observed M=2005.8886; calculated M=2005.9068.
Figure 3.
Phosphorylation of myc-Kal7 in non-neuronal cells. Experiments on transfected myc-Kal7 were undertaken to determine the extent of phosphorylation of the protein. Cells were treated with multiple stimulants (8-Br-cAMP, PMA, A23187) as well as multiple phosphatase inhibitors (Calyculin A, Fenvalerate) in order to induce maximal phosphorylation. Sites are indicated on the Kal7 schematic by a black dot and a bold letter followed by the residue number. There were two instances in which the phosphorylation site could not be pinpointed to one of two closely spaced residues; these residues are marked by a hatched dot. Residues on either side of the phosphorylation site are listed to provide a context for the site and do not indicate the entire peptide identified by LC-MS/MS (peptides are listed in Table 2). Text boxes indicate sites that were phosphorylated by the purified kinases tested. Phosphorylation sites in the SH3 binding motif and within the PDZ binding motif are highlighted, as they may be critical for protein-protein interactions. Tyr963, which was not dephosphorylated by phosphatase treatment, is indicated by a filled star.
Table 2.
Sites in myc-Kal7 Phosphorylated in Non-Neuronal Cells
| Residue | Score | Peptide Sequence | EN/FT | M/Z | Ion Mass | Charge |
|---|---|---|---|---|---|---|
| 487 | 82.08 | K.ALLDVLQRPLpSPGNSESLTATANYSK.A | FT | 942.474 | 2824.399 | 3 |
| 491 | 59.87 | K.ALLDVLQRPLSPGNpSESLTATANYSK.A | EN | 942.475 | 2824.404 | 3 |
| 493/495 | 56.12 | K.ALLDVLQRPLSPGNSEpSLpTATANYSK.A | EN | 942.475 | 2824.404 | 3 |
| 710 | 28.31 | R.pSAPPSLGEPTEAR.D | EN | 696.314 | 1390.614 | 2 |
| 724 | 47.39 | R.SAPPSLGEPTEARDpSAVSNNK.T | EN | 1104.002 | 2205.989 | 2 |
| 963 | 27.45 | K.AEALLQAGHpYDADAIRECAEK.V | FT | 1177.522 | 2353.030 | 2 |
| 1188 | 22.95 | K.LLIQLADpSFVEK.G | EN | 728.381 | 1454.747 | 2 |
| 1249 | 22.84 | K.DLELDIIPASLpSDR.E | EN | 818.896 | 1635.777 | 2 |
| 1520 | 53.17 | K.LLTpSELGVTEHVEGDPCK.F | FT | 1003.957 | 2005.898 | 2 |
| 1590 | 61.94 | K.GALKEPIQLPKpTPAK.L | EN | 835.967 | 1669.919 | 2 |
| 1598 | 92.54 | R.NNpSKRDGVEDGDSQGDGSSQPDTISIASR.T | EN | 1024.776 | 3071.307 | 3 |
| 1608 | 70.43 | K.RDGVEDGDpSQGDGSSQPDTISIASR.T | EN | 877.039 | 2628.096 | 3 |
| 1613 | 71.06 | K.RDGVEDGDSQGDGpSSQPDTISIASR.T | EN | 877.039 | 2628.094 | 3 |
| 1614 | 57.92 | R.DGVEDGDSQGDGSpSQPDTISIASR.T | EN | 1334.662 | 2667.310 | 2 |
| 1618 | 59.31 | R.DGVEDGDSQGDGSSQPDpTISIASR.T | EN | 825.007 | 2471.998 | 3 |
| 1620 | 65.83 | R.DGVEDGDSQGDGSSQPDTIpSIASR.T | EN | 825.006 | 2471.995 | 3 |
| 1625/1626 | 63.93 | R.pTpSQNTVESDKDGNLVPR.W | EN | 970.442 | 1938.870 | 2 |
| 1629 | 54.98 | R.TSQNpTVESDKDGNLVPR.W | EN | 970.443 | 1938.871 | 2 |
| 1632 | 75.24 | R.TSQNTVEpSDKDGNLVPR.W | EN | 970.441 | 1938.867 | 2 |
| 1652 | 24.06 | R.WHLGPGDPFSpTYV.- | EN | 778.336 | 1554.658 | 2 |
| 95-Non | 70.54 | R.GFTVIIDMR.G | FT | 526.284 | 1050.553 | 2 |
The peptides used to identify phosphorylation sites in Myc-Kal7 expressed in stimulated pEAK Rapid cells are identified; formatting for Table 2 is the same as Table 1. Closely spaced Ser and Thr residues, where the phosphorylation site could not be unambiguously assigned, are indicated. Thr95 was not identified as a phosphorylation site; data for the non-phosphorylated Thr95 peptide (95-Non) are provided. Representative spectra are shown in Supplementary Figs. S23 to S41.
Phosphorylation sites appeared in clusters throughout the protein, with a concentration of sites near the C-terminus, after the GEF domain and proximal to the PDZ-binding motif (Fig. 3). The catalytic DH domain was notably lacking in phosphorylation sites. This is of interest since modification of the DH domain might affect the ability of Kal7 to activate Rac and affect its downstream targets at the PSD43,44. Although multiple sites in the Sec14p domain were targets for purified CaMKII, PKA, PKC and CKII in vitro, no phosphorylation sites were identified in the Sec14p domain when phosphorylation occurred in these non-neuronal cells. Although the non-phosphorylated Thr95 peptide was detected more than twenty times (Table 2), the corresponding phosphopeptide was never identified.
Many of the sites phosphorylated by purified CKII and the single site phosphorylated by purified Cdk5 in vitro were identified in myc-Kal7 phosphorylated in cells (Fig. 3). Phosphorylation at CKII sites was especially common, with four of the six sites phosphorylated by purified CKII also utilized in non-neuronal cells; two of these sites are located in the linker region that precedes the PDZ-binding motif, one in the linker that connects the ninth spectrin repeat to the GEF domain, and one in the PH domain. The Cdk5 site (Thr1590) utilized in vitro27 also occurs in the region immediately preceding the PDZ binding motif and was phosphorylated in cells. Seven additional sites in this linker region were phosphorylated in Kal7 isolated from pEAK Rapid cells. Given that this C-terminal portion of the protein is known to be critical for Kal7 to interact with PDZ scaffolding molecules in the PSD, it is likely that some of the phosphorylation events in this region affect these interactions12,22,45.
In addition to the phosphorylation hot spot detected in the C-terminal linker region of the protein, small clusters of phosphorylation sites were detected in the third and fifth spectrin repeat regions of Kal7 (Fig. 3). Each spectrin repeat is comprised of three anti-parallel a-helices connected by loop regions46. In both of these spectrin repeats, these clusters occur near the end of the second a-helix and in the linker region that joins it to the third a-helix. The cluster in the third spectrin repeat overlaps the SH3 binding motif (PLS487P) shown previously to be involved in an intramolecular interaction with the SH3 domain present in the longer isoforms of Kalirin47. One of the sites in spectrin repeat five immediately follows Arg722, which is exposed and susceptible to limited tryptic digestion of native Kal7.
Phosphorylation of cortical and striatal Kal7
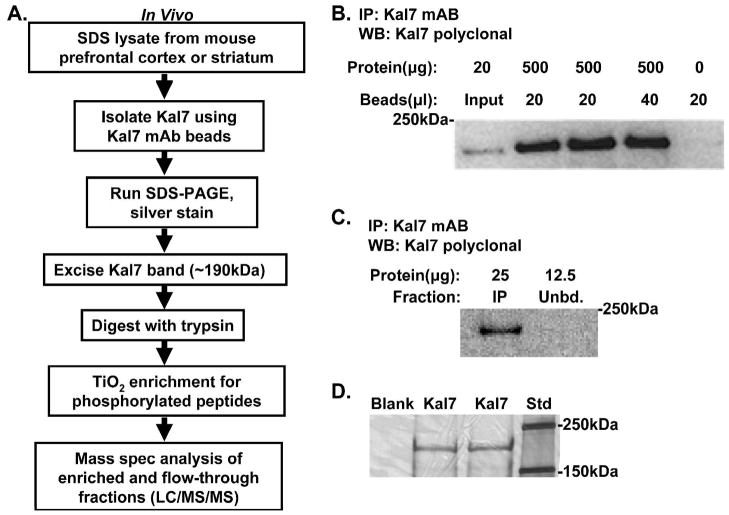
We next wanted to determine the phosphorylation status of endogenous Kal7 under baseline conditions so that we could evaluate factors that regulate its phosphorylation and dephosphorylation in vivo. We selected mouse prefrontal cortex and striatum for these experiments because these brain regions are involved in learning, emotion, decision making and drug addiction48–56, behaviors known to be altered in Kal7KO mice25,26. Given the higher levels of Kal7 in the cortex, we were able to obtain enough Kal7 from the prefrontal cortices of four mice; on average, six mice were used to prepare striatal Kal7. In order to solubilize Kal7, lysates were prepared using boiling SDS; Kal7 was not efficiently solubilized by milder detergents such as 1% TX-10025. Following the addition of NP-40 (to achieve an NP-40/SDS weight ratio of 7.5/1) and dilution into a buffer lacking detergent, endogenous Kal7 was immunoisolated using a monoclonal antibody specific for the C-terminus of Kal7 that was covalently attached to beads (Fig. 4A). All of the Kal7 in a lysate containing 0.5 mg of protein was bound to 20 μl of antibody resin (Fig. 4B,C). Kal7 eluted from the antibody resin was purified by SDS-PAGE; Kal7, which has a predicted mass of 190 kDa, was readily identified by silver staining (Fig. 4D). A gel slice containing Kal7 was excised, processed to eliminate detergent and digested with trypsin; following adsorption to TiO2 tips, LC-MS/MS analysis was carried out as described for myc-Kal7 isolated from transiently transfected cells. Coverage and data for all of the tryptic phosphopeptides identified in prefrontal cortex and striatal Kal7 are summarized in Table 3, a representative spectrum is shown in Fig. 5 and the remaining spectra are shown in Supplementary Figs. S42 to S62. The sites shown were identified from a total of eight Kal7 preparations and reflect analysis of almost 700 individual peptides.
Figure 4.
Purification of Kal7 from striatum and cortex. A. Flow chart summarizes the steps that were taken in purifying, digesting and analyzing Kal7 from adult mouse brain. B. The specificity and efficacy of Kal7 monoclonal antibody 20D8 was validated by varying the amount of antibody used to recover Kal7 from 0.5 mg of cortical protein and comparing the amount recovered to the input; the beads themselves created no background staining. C. After immunoprecipitation, an aliquot of the unbound fraction was analyzed (Unbd.); it contained no detectable Kal7, demonstrating effective recovery of Kal7 from the lysate. D. A representative silver stained gel of Kal7 immunoprecipitated from cortex revealed a single 190kDa band, which was excised from the gel and subjected to analysis.
Table 3.
Sites in Kal7 Phosphorylated in Mouse Brain
| Residue | Score | Peptide Sequence | EN/FT | M/Z | Ion Mass | Charge |
|---|---|---|---|---|---|---|
| 79 | 53.91 | K.LVpTYLASVPSEDVCKR.G | FT | 930.4421 | 1858.87 | 2 |
| 189 | 90.2 | R.LpSLEEFFNSAVHLLSR.L | FT | 971.4714 | 1940.928 | 2 |
| 379 | 51.26 | R.LpSEAGHYASQQIK.Q | FT | 756.3428 | 1510.671 | 2 |
| 487 | 54.54 | K.ALLDVLQRPLpSPGNSESLTATANYSK.A | EN | 942.4711 | 2824.392 | 3 |
| 592 | 44.93 | K.RHDDFEEVAQNTYpTNADK.L | EN | 744.9724 | 2231.895 | 3 |
| 607 | 27.44 | K.LLEAAEQLAQpTGECDPEEIYK.A | FT | 1215.534 | 2429.054 | 2 |
| 672 | 37.24 | K.EVLEDVCADpSVDAVQELIK.Q | FT | 1077.983 | 2153.952 | 2 |
| 714 | 23.47 | R.SAPPpSLGEPTEAR.D | EN | 696.311 | 1390.607 | 2 |
| 943 | 36.62 | K.pTHQSALQVQQK.A | FT | 674.3194 | 1346.624 | 2 |
| 963 | 26.16 | K.AEALLQAGHpYDADAIRECAEK.V | FT | 1177.518 | 2353.022 | 2 |
| 1002/1003 | 45.91 | K.pTpSEQVCSVLESLEQEYR.R | FT | 1040.438 | 2078.862 | 2 |
| 1077 | 33.33 | R.NNVpSMPSVASHTR.G | FT | 748.3161 | 1494.618 | 2 |
| 1085 | 28.75 | R.NNVSMPSVASHpTR.G | FT | 748.314 | 1494.614 | 2 |
| 1111 | 33.29 | R.VLHFWpTLK.K | EN | 562.2866 | 1122.559 | 2 |
| 1198 | 24.52 | K.GHIHApTEIR.K | EN | 557.2586 | 1112.503 | 2 |
| 1215 | 21.59 | R.DFpSLR.M | EN | 717.2919 | 716.2846 | 1 |
| 1519 | 74.75 | K.LLpTSELGVTEHVEGDPCK.F | FT | 1003.954 | 2005.894 | 2 |
| 1520 | 63.71 | K.LLTpSELGVTEHVEGDPCK.F | EN | 1003.947 | 2005.88 | 2 |
| 1608 | 72.76 | K.RDGVEDGDpSQGDGSSQPDTISIASR.T | EN | 877.0382 | 2628.093 | 3 |
| 1613 | 46.57 | R.DGVEDGDSQGDGpSSQPDTISIASR.T | EN | 825.0034 | 2471.988 | 3 |
| 1614 | 54.29 | R.DGVEDGDSQGDGSpSQPDTISIASR.T | EN | 1236.996 | 2471.977 | 2 |
| 1632 | 32.83 | R.TSQNTVEpSDKDGNLVPR.W | EN | 647.2962 | 1938.867 | 3 |
| 95-Non | 49.3 | R.GFTVIIDMR.G | FT | 526.283 | 1050.552 | 2 |
The peptides used to identify phosphorylation sites in Kal7 immunoisolated from mouse brain (prefrontal cortex and striatum) are identified; formatting for Table 3 is the same as Table 1. Closely spaced Ser and Thr residues, where the phosphorylation site could not be unambiguously assigned, are indicated. Thr95 was not identified as a phosphorylation site; data for the non-phosphorylated Thr95 peptide (95-Non) are provided. Representative spectra are shown in Supplementary Figs. S42 to S62.
Figure 5.
Identification of phosphorylated Thr79 from mouse brain Kal7. The spectrum from MS/MS of the doubly charged M+2 930.44 ion of Peptide K.LVpTYLASVPSEDVCKR.G is shown. Based on the b and y ions, and the tentative b13-H3PO4, residue 79 is phosphorylated. Observed M=1858.8697; calculated M=1858.8900.
Kal7 isolated from mouse brain was extensively phosphorylated, with a total of 22 sites identified (Fig. 6A and Table 3). The only domains not phosphorylated were spectrin repeat six and the catalytic DH domain. Only nine of the 22 sites were also phosphorylated in myc-Kal7 expressed in non-neuronal cells (Fig. 6A; indicated by white stars); four of the nine common sites are located in the C-terminal linker region and two of these sites are CKII sites. Strikingly, only five of these 22 sites were phosphorylated in vitro by purified CaMKII, PKA, PKC or CKII, suggesting a major role for other protein kinases and for substrate and kinase subcellular localization and control. Of the 22 unique phosphorylation sites, only one was a tyrosine residue (Y963), which was not phosphorylated by recombinant Fyn in our in vitro experiments but was resistant to phosphatase treatment. To ensure that we were not missing tyrosine phosphorylations due to their relatively low stoichiometry, we immunoprecipitated Kal7 tryptic peptides using beads coated with a high concentration of anti-phosphotyrosine antibody. These samples were analyzed by LC-MS/MS to look for phosphorylated tyrosines; no additional tyrosine phosphorylation sites were identified (data not shown).
Figure 6.
Phosphorylation of cortical/striatal Kal7. A. Numerous phosphorylation sites were identified in Kal7 immunoisolated from mouse prefrontal cortex and striatum. Sites are again indicated by a black dot and the residue number of the phosphorylated site. Sites identified in myc-Kal7 from pEAK Rapid cells and Kal7 from mouse brain are marked with a white star, sites that were phosphatase resistant are marked with a filled star. Text boxes again indicate sites that were phosphorylated by purified kinases and hatched dots indicate where an exact identification of a phosphorylated residue was not possible. B. Top. Spectrin repeats are each composed of three antiparallel a-helices (A/B/C) connected by linker regions46. Bottom. Phosphorylation sites in the spectrin repeats of Kal7 are located with reference to the predicted secondary structure; filled circles, neuronal Kal7; open circles, non-neuronal myc-Kal7; gray circles, shared sites. Ambiguous assignments are shown by a single circle.
Unlike myc-Kal7 expressed in pEAK Rapid cells, the Sec14p domain of endogenous Kal7 was phosphorylated. The only phosphorylation site identified in the Sec14p domain was Thr79, a site phosphorylated by CaMKII, PKA and PKC in vitro. Once again, a tryptic peptide containing P-Thr95, the putative CaMKII site reported by Xie et al.30, was never detected; however, the non-phosphorylated peptide was detected repeatedly (Table 3). The other CaMKII site identified using purified kinase and purified substrate, Thr/Ser1002/1003, was also phosphorylated in Kal7 isolated from mouse brain. While our data support the conclusion that Kal7 can be phosphorylated by CaMKII, the phenomenon of kinase convergence means that additional experiments will be required to identify the protein kinase responsible for phosphorylation of these sites under different in vivo conditions.
In contrast to myc-Kal7 isolated from transiently transfected non-neuronal cells, the spectrin repeat regions in Kal7 purified from prefrontal cortex and striatum were riddled with phosphorylation sites. These phosphorylation sites were aligned with the previously predicted secondary structure39 of the spectrin repeats (Fig. 6B). As observed in non-neuronal cells, most of these phosphorylation sites occurred at the end of helix A or helix B or in the loop that follows; none occur in the linker regions that connect adjacent spectrin repeat regions. Although spectrin repeat regions 3 and 5 were multiply phosphorylated when Kal7 was expressed in non-neuronal cells, most of the sites identified in brain Kal7 differed from those identified in non-neuronal myc-Kal7 (Fig. 6B).
Phosphorylation in the spectrin-like repeats could affect the alignment/orientation of the helices and alter protein function. The region between spectrin repeats 4 and 5 is of particular interest since it is responsible for the interaction of Kal7 with iNOS23. Additionally, spectrin repeat regions 4 and 5 are involved in the interaction of Kal7 with Arf6, DISC-1 and PAM20,39,40. The protein-protein interactions of spectrin repeats can be disrupted by small perturbations in their structure57. For example, phosphorylation of a single site in one of the 22 spectrin repeats of utrophin alters its ability to interact with dystroglycan58.
Additionally, the phosphorylation sites in spectrin repeats 4 and 5 fall at the junction of the two splice forms of Kalirin, Kal7 and ΔKal7. ΔKal7, which begins with the fifth spectrin repeat of full length Kal7, has the same unique 20 amino acid sequence as Kal7 at its C-terminus. The properties of ΔKal7 are very different from those of Kal7: unlike Kal7, ΔKal7 is soluble; in addition, when expressed in neurons, ΔKal7 increases dendritic spine size but does not increase dendritic spine number37. Phosphorylation sites in spectrin repeat 4 may be critical for some of the interactions that are unique to full length Kal7. Based on this, it seems likely that the six phosphorylation sites located in spectrin repeats 4 and 5 play an important role in regulating Kalirin function.
Two consecutive residues in the PH domain of endogenous Kal7 were phosphorylated (Thr1519, Ser1520). Interestingly, in preparing substrate for the kinase experiments presented in Fig. 1, we found P-Thr1519 resistant to dephosphorylation by either of the phosphatases utilized. Both recombinant PKC and recombinant CKII phosphorylated Ser1520 in purified Kal7, which may suggest a primed convergence mechanism wherein Thr1520 can be phosphorylated only if Thr1519 is also phosphorylated. Comparison of the Kal7 PH domain with the crystal structure of the highly homologous Trio PH domain predicts that Thr1519-Ser1520 are in the loop between the third and fourth β sheets of the PH domain59. This region is on the surface of the structure, should be accessible to kinases and might affect the ability of the PH domain to interact with phospholipids or other proteins60.
As with myc-Kal7 expressed in non-neuronal cells, the 60 amino acid linker region between the end of the PH domain and the beginning of the region unique to Kal7 was extensively phosphorylated in mouse brain under baseline conditions; four phosphorylation sites were identified in this region, all of which were also detected in myc-Kal7. Tryptic peptides containing Thr1590 or P-Thr1590, a known Cdk5 phosphorylation site, were not detected in digests of endogenous Kal7, perhaps reflecting the fact that complete tryptic cleavage produces a tetrapeptide (K.TPAK.L), which is too small for detection by the mass spectrometer. Detection of this phosphopeptide in myc-Kal7 relied upon missed cleavages by trypsin (Table 2).
Effect of phosphorylation on GEF activity
To determine if phosphorylation might have an effect on the GEF activity of Kal7, we made non-phosphorylatable Thr→Ala and phosphomimetic Thr→Glu/Ser→Asp point mutations at selected sites and assessed the effect on Rac activation using pEAK Rapid cells (Fig. 7). We used full length myc-Kal7 as a control, included Kal7 with its catalytic DH domain mutated (ND/AA: Asn1415-Asp1416/Ala-Ala) as a negative control and normalized Rac-GTP to expressed Kal7 using the Kal7-specific antibody. As expected, the ND/AA mutant was much less active than Kal7. Although, it was never identified as a phosphorylation site in any of our assays, we mutated Thr95 to Ala since this mutation was reported to eliminate the GEF activity of Kal730. In our hands, this mutation produced a nearly two-fold increase in Rac activation. To ensure that the construct encoded the desired mutant, it was re-sequenced, confirming that Thr95 had been mutated to Ala. As a further check on the effect of this region on GEF activity, we tested a mutant of Kal7 lacking the entire Sec14p domain (ΔSec14p); this mutant fails to mimic the effects of Kal7 on cytoskeletal organization in non-neuronal cells37. Kal7-ΔSec14p displayed Rac activation activity that was indistinguishable from that of Kal7. These results, coupled with our ability to identify Thr95, but not P-Thr95 in exogenous and endogenous Kal7 (Tables 1–3), call into question the conclusion that the ability of Kal7 to activate Rac requires phosphorylation of Thr95 by CaMKII30.
Figure 7.
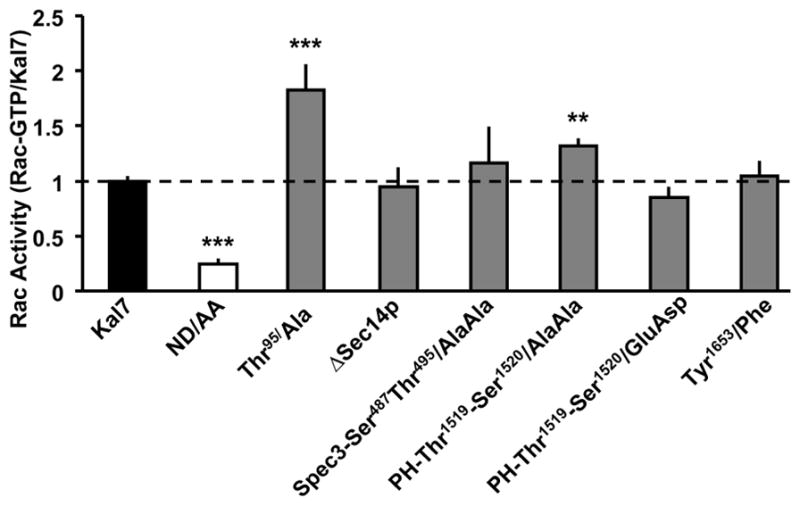
Rac activation activity of Kal7 point mutants. pEAK Rapid cells were transfected with vectors encoding the indicated proteins; Rac activation was assessed by adding GST-Pak-CRIB to the homogenization buffer and normalizing the amount of activated Rac (bound to glutathione agarose beads by GST-Pak-CRIB) to the amount of Kal7 protein in the lysate (assessed using a Kal7-specific antibody)25. Data are the mean of at least 4 separate transfections; error bars represent standard error. (**p<0.01; ***p<0.001; t-Test)
The increased activation of Rac by Kal7 bearing the Thr95/Ala mutation is hard to reconcile with the fact that deleting the entire Sec14p domain failed to alter Rac activation. Direct comparison of the catalytic activities of purified Kal7, ΔKal7 and Kal-GEF1 using GDP-Mant demonstrated a 4-fold decrease in the specific activity of both Kal7 and ΔKal7, revealing the presence of inhibitory intramolecular interactions7. The indirect assay used here to compare the GEF activities of Kal7-T95/A and Kal7-ΔSec14p assesses their ability to activate endogenous Rac1 in non-neuronal cells and could be influenced by differences in subcellular localization37 or by the presence of an HA-tag in Kal7-ΔSec14p. Further studies to assess GEF activity will require analysis of the purified proteins. Evaluation of the effect of phosphorylation site mutations in this essential region on the ability of Kal7 to stimulate spine formation will require expression of mutants in neurons.
We next explored the effect of a spectrin domain phosphorylation site mutation on GEF activity. We mutated two sites in the SH3 binding motif located in the third spectrin repeat (Ser487-Thr495/Ala-Ala) because this region is known to be involved in intramolecular interactions47. Rac activation activity was unaltered by this mutation (Fig. 7). Given the proximity of the PH and DH domains, we mutated the two adjacent phosphorylation sites identified in the PH domain, Thr1519-Ser1520/Ala-Ala and Thr1519-Ser1520/Glu-Asp. The non-phosphorylatable Ala-Ala mutant was significantly more active than myc-Kal7; the phosphomimetic Glu-Asp mutant was significantly less active than the Ala-Ala mutant, but was not significantly less active than the myc-Kal7. We also mutated Tyr1653, which is located in the 20 amino acid region unique to Kal7; mutation of this Fyn site, which lies in the PDZ binding motif, had no effect on Rac activation (Fig. 7).
The catalytic activity of a number of other RhoGEFs localized at the PSD is altered by phosphorylation; Tiam161, β-Pix62,63, Ephexin164, and Arhgef2 (a.k.a. LFC or GEF-H1) are regulated in this manner65,66. Indeed we have previously shown that the catalytic activity of Kal7 is increased by phosphorylation of Thr1590 in the unstructured C-terminal region of the protein27. The fact that the isolated GEF domain of Kal7 is more active than Kal7 or ΔKal737, demonstrates the common occurrence of N-terminal autoinhibition. Further support for this phenomenon comes from the ease with which Cdk5 phosphorylates Thr1590 in a protein that extends from the GEF domain to the end of Kal7 compared to the inaccessibility of this site in ΔKal727; the presence of spectrin repeats 5 through 9 clearly inhibits the ability of Cdk5 to access this site. Given the number of phosphorylation sites identified in the spectrin repeat regions, determining how their phosphorylation affects the status of the GEF domain will be a complex undertaking best carried out with purified Rac and purified Kalirin. Additionally, it is important to remember that the Sec14p, spectrin repeats, and PDZ binding motif play important functional roles in determining protein and lipid interactions of Kal7. A lack of change in GEF activity following phosphorylation of a particular site does not necessarily indicate a lack of functional importance.
Conservation of Phosphorylation Sites
In order to focus on phosphorylation sites of functional importance, we assessed their conservation in Kalirin and in Trio using Clustal analysis (Fig. 8). Interestingly, all of the phosphorylation sites identified were conserved evolutionarily from the human to the blowfish (fugu) genome; substitution of a serine for a threonine was counted as a conserved site. Nineteen of the sites are also conserved in the orthologous Trio gene. Finally, six of the phosphorylation sites that we identified are completely evolutionarily conserved from human through Drosophila and C. elegans, where Kalirin and Trio are encoded by a single gene. These findings suggest two interesting possibilities. The first is that sites that are fully conserved are of critical importance to controlling GEF activity. If the ability to phosphorylate these sites were critical to development or function of the nervous system, mutation could not be tolerated. Since the Kalrn and Trio genes fulfill non-redundant roles, the second suggestion is that phosphorylation sites unique to Kalirin are of importance to its unique roles during development, in specific tissues or at specific subcellular locations.
Figure 8.
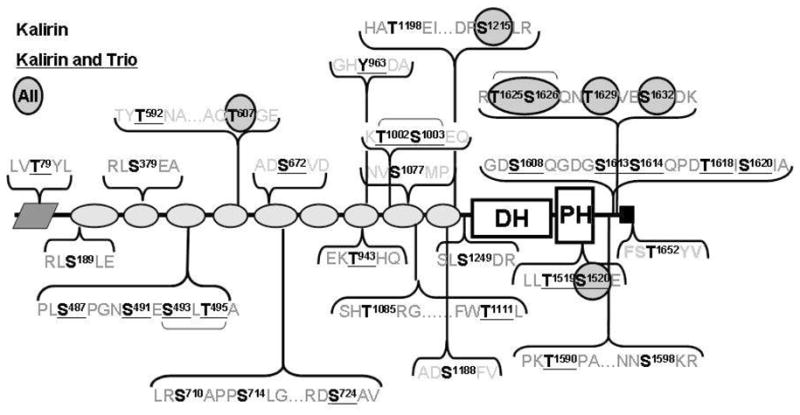
Evolutionary conservation of 35 identified phosphorylation sites. The sequences of the proteins encoded by Kalrn and Trio from human, mouse, chicken and fugu, D. melanogaster Trio and C. elegans Unc73 were aligned using Clustal. Identified Ser/Thr and Tyr phosphorylation sites conserved in both genes in all species are circled; sites conserved in species with separate Kalrn and Trio genes are underlined; the remaining sites (bold) are conserved in Kalrn but not in Trio. The only site unique to the Kal7 isoform is Thr1652.
Conclusions
Over the past decade, the importance of Kal7 as a regulator of dendritic spine morphology and synaptic plasticity has been established12,25,26. While previous studies have focused on single phosphorylation events27,30,67, it is now clear that Kal7 could serve as a PSD signaling node and hub for phosphorylation. Comparing the phosphorylation sites characterized in Kal7 to proteomic analyses of all phosphorylation events at the PSD identifies Kal7 as one of the most highly phosphorylated targets in the PSD36. Previous phosphoproteomics surveys identified only a few sites in Kalirin. One analysis of the PSD proteome identified no sites in Kalirin36 and the PHOSIDA project68 identified only six (Ser487, Ser491, Ser493, Thr495, Thr497 and Ser1376); four of these sites were identified in our analyses while two were not (S497 and S1376).
While many of the kinases that phosphorylate Kal7 remain to be identified, we have shown that it is phosphorylated by six kinases known to play critical roles in learning, memory and synaptic plasticity (PKA, PKC, CaMKII, CKII, Cdk5, Fyn). Future studies will focus on regulated phosphorylation of Kal7 at sites targeted by these protein kinases. Mice genetically lacking Kal7 have deficient fear conditioning memory formation and respond aberrantly to repeated administration of drugs of abuse25,26. We will now begin to look at how these phosphorylation events change in a quantitative fashion in animals that have been given a footshock or repeated injections of cocaine. Hopefully, learning about how phosphorylation events affect the catalytic activity or localization of Kal7 will help us clarify the specific role that this important protein plays in synaptic plasticity.
Supplementary Material
Acknowledgments
We thank Darlene D’Amato, Yanping Wang and Chris Mazzone for incredible technical assistance. This work was supported by grants from The National Institutes of Health (DA-15464, DA-18274, DA-23082, DA-26706, DA-18343, RR-24139, RR-24617, DA-26706), The Janice and Rodney Reynolds Fund, and The William Beecher Scoville Fund.
Footnotes
Supporting Information Available: Supplementary Figures S1 - S62 are available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 2.Yoshihara Y, De RM, Muller D. Dendritic spine formation and stabilization. Curr Opin Neurobiol. 2009;19(2):146–153. doi: 10.1016/j.conb.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10(9):647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Wang XB, Frerking M, Zhou Q. Spine expansion and stabilization associated with long-term potentiation. J Neurosci. 2008;28(22):5740–5751. doi: 10.1523/JNEUROSCI.3998-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57(5):719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Tashiro A, Yuste R. Role of Rho GTPases in the morphogenesis and motility of dendritic spines. Mtds Enzymol. 2008;439:285–302. doi: 10.1016/S0076-6879(07)00421-1. [DOI] [PubMed] [Google Scholar]
- 7.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 8.Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;5(Suppl 2):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 9.Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25(4):191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10(10):981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 11.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 12.Kiraly DD, Eipper-Mains JE, Mains RE, Eipper BA. Synaptic plasticity, a symphony in GEF. ACS Chem Neurosci. 2010;1(5):348–365. doi: 10.1021/cn100012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, Harris M, Nelson S, Hale AB, Granger CB, Haines JL, Jones CJ, Crossman D, Seo D, Gregory SG, Kraus WE, Goldschmidt-Clermont PJ, Vance JM. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80(4):650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Shiino T, Okochi T, Fukuo Y, Ujike H, Suzuki M, Inada T, Hashimoto R, Takeda M, Kaibuchi K, Iwata N, Ozaki N. Resequencing and Association Analysis of the KALRN and EPHB1 Genes And Their Contribution to Schizophrenia Susceptibility. Schizophr Bull. 2010 doi: 10.1093/schbul/sbq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11(6):557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 16.Youn H, Jeoung M, Koo Y, Ji H, Markesbery WR, Ji I, Ji TH. Kalirin is under-expressed in Alzheimer’s disease hippocampus. J Alzheimers Dis. 2007;11(3):385–397. doi: 10.3233/jad-2007-11314. [DOI] [PubMed] [Google Scholar]
- 17.Penzes P, Johnson RC, Alam MR, Kambampati V, Mains RE, Eipper BA. An Isoform of Kalirin, a Brain-Specific GDP/GTP Exchange Factor, Is Enriched in the Postsynaptic Density Fraction. J Biol Chem. 2000;275:6395–6403. doi: 10.1074/jbc.275.9.6395. [DOI] [PubMed] [Google Scholar]
- 18.Ma XM, Ferraro F, Mains RE, Eipper BA. Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. J Neurosci. 2008;28(3):711–724. doi: 10.1523/JNEUROSCI.5283-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma XM, Huang JP, Wang Y, Eipper BA, Mains RE. Kalirin, a multifunctional Rho GEF, is necessary for maintenance of hippocampal pyramidal neuron dendrites and dendritic spines. J Neurosci. 2003;23:10593–10603. doi: 10.1523/JNEUROSCI.23-33-10593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13(3):327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Photowala H, Cahill ME, Srivastava DP, Woolfrey KM, Shum CY, Huganir RL, Penzes P. Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J Neurosci. 2008;28(24):6079–6091. doi: 10.1523/JNEUROSCI.1170-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29(1):229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 23.Ratovitski EA, Alam MR, Quick RA, McMillan A, Bao C, Hand TA, Johnson RC, Mains RE, Eipper BA, Lowenstein CJ. Kalirin inhibition of inducible nitric oxide synthase. J Biol Chem. 1999;274:993–999. doi: 10.1074/jbc.274.2.993. [DOI] [PubMed] [Google Scholar]
- 24.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 25.Ma XM, Kiraly DD, Gaier ED, Wang Y, Kim EJ, Levine ES, Eipper BA, Mains RE. Kalirin-7 is required for synaptic structure and function. J Neurosci. 2008;28(47):12368–12382. doi: 10.1523/JNEUROSCI.4269-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiraly DD, Ma XM, Mazzone CM, Xin X, Mains RE, Eipper BA. Behavioral and morphological responses to cocaine require kalirin7. Biol Psychiatry. 2010;68(3):249–255. doi: 10.1016/j.biopsych.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xin X, Wang Y, Ma XM, Rompolas P, Keutmann HT, Mains RE, Eipper BA. Regulation of Kalirin by Cdk5. J Cell Sci. 2008;121(Pt 15):2601–2611. doi: 10.1242/jcs.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayer KU, De KP, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411(6839):801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- 29.Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3(3):175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 30.Xie Z, Srivastava DP, Photowala H, Kai L, Cahill ME, Woolfrey KM, Shum CY, Surmeier DJ, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56(4):640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104(17):7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45(4):525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 33.Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR. Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron. 2008;57(1):94–107. doi: 10.1016/j.neuron.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25(13):3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11(6):427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 36.Coba MP, Pocklington AJ, Collins MO, Kopanitsa MV, Uren RT, Swamy S, Croning MD, Choudhary JS, Grant SG. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal. 2009;2(68):ra19. doi: 10.1126/scisignal.2000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiller MR, Ferraro F, Wang Y, Ma XM, McPherson CE, Sobota JA, Schiller NI, Mains RE, Eipper BA. Autonomous functions for the Sec14p/spectrin-repeat region of Kalirin. Exp Cell Res. 2008;314(14):2674–2691. doi: 10.1016/j.yexcr.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin X, Ferraro F, Back N, Eipper BA, Mains RE. Cdk5 and Trio modulate endocrine cell exocytosis. J Cell Sci. 2004;117:4739–4748. doi: 10.1242/jcs.01333. [DOI] [PubMed] [Google Scholar]
- 39.Alam MR, Johnson RC, Darlington DN, Hand TA, Mains RE, Eipper BA. Kalirin, a Cytosolic Protein with Spectrin-like and GDP/GTP Exchange Factor-like Domains That Interacts with Peptidylglycine α-Amidating Monooxygenase, an Integral Membrane Peptide-processing Enzyme. J Biol Chem. 1997;272:12667–12675. doi: 10.1074/jbc.272.19.12667. [DOI] [PubMed] [Google Scholar]
- 40.Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama Y, Kawabata I, Sobue K, Okabe S. Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat Methods. 2005;2(9):677–684. doi: 10.1038/nmeth783. [DOI] [PubMed] [Google Scholar]
- 42.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5(6):1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1(3):173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 44.Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290(5492):754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Mata R, Burridge K. Catching a GEF by its tail. Trends Cell Biol. 2007;17(1):36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513(1):119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 47.Schiller MR, Chakrabarti K, King GF, Schiller NI, Eipper BA, Maciejewski MW. Regulation of RhoGEF activity by intramolecular and intermolecular SH3 domain interactions. J Biol Chem. 2006;281(27):18774–18786. doi: 10.1074/jbc.M512482200. [DOI] [PubMed] [Google Scholar]
- 48.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10(8):561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- 50.Sotres-Bayon F, Quirk GJ. Prefrontal control of fear: more than just extinction. Curr Opin Neurobiol. 2010;20(2):231–235. doi: 10.1016/j.conb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters J, Kalivas PW, Quirk GJ. Extinction circuits for fear and addiction overlap in prefrontal cortex. Learn Mem. 2009;16(5):279–288. doi: 10.1101/lm.1041309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price JL, Drevets WC. Neurocircuitry of mood disorders. Neuropsychopharmacology. 2010;35 (1):192–216. doi: 10.1038/npp.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robbins TW, Arnsten AF. The neuropsychopharmacology of fronto-executive function: monoaminergic modulation. Annu Rev Neurosci. 2009;32:267–287. doi: 10.1146/annurev.neuro.051508.135535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8(11):1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- 55.Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry. 2005;162(8):1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- 56.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 57.Stabach PR, Simonovic I, Ranieri MA, Aboodi MS, Steitz TA, Simonovic M, Morrow JS. The structure of the ankyrin-binding site of beta-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties. Blood. 2009;113(22):5377–5384. doi: 10.1182/blood-2008-10-184291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamashita K, Suzuki A, Satoh Y, Ide M, Amano Y, Masuda-Hirata M, Hayashi YK, Hamada K, Ogata K, Ohno S. The 8th and 9th tandem spectrin-like repeats of utrophin cooperatively form a functional unit to interact with polarity-regulating kinase PAR-1b. Biochem Biophys Res Commun. 2010;391(1):812–817. doi: 10.1016/j.bbrc.2009.11.144. [DOI] [PubMed] [Google Scholar]
- 59.Skowronek KR, Guo F, Zheng Y, Nassar N. The C-terminal basic tail of RhoG assists the guanine nucleotide exchange factor trio in binding to phospholipids. J Biol Chem. 2004;279(36):37895–37907. doi: 10.1074/jbc.M312677200. [DOI] [PubMed] [Google Scholar]
- 60.Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J. Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem. 2007;282(40):29201–29210. doi: 10.1074/jbc.M703458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyamoto Y, Yamauchi J, Tanoue A, Wu C, Mobley WC. TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc Natl Acad Sci U S A. 2006;103(27):10444–10449. doi: 10.1073/pnas.0603914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59(6):914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin EY, Shin KS, Lee CS, Woo KN, Quan SH, Soung NK, Kim YG, Cha CI, Kim SR, Park D, Bokoch GM, Kim EG. Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. J Biol Chem. 2002;277(46):44417–44430. doi: 10.1074/jbc.M203754200. [DOI] [PubMed] [Google Scholar]
- 64.Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci. 2007;10(1):67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- 65.Birkenfeld J, Nalbant P, Bohl BP, Pertz O, Hahn KM, Bokoch GM. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell. 2010;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujishiro SH, Tanimura S, Mure S, Kashimoto Y, Watanabe K, Kohno M. ERK1/2 phosphorylate GEF-H1 to enhance its guanine nucleotide exchange activity toward RhoA. Biochem Biophys Res Commun. 2008;368:162–167. doi: 10.1016/j.bbrc.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 67.Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL. Rapid Induction of Dendritic Spine Morphogenesis by trans-Synaptic EphrinB-EphB Receptor Activation of the Rho-GEF Kalirin. Neuron. 2003;37(2):263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 68.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.