Abstract
To genetically transform plants, Agrobacterium transfers its T-DNA into the host cell and integrates it into the plant genome, resulting in neoplastic growths. Over the past two decades, a great deal has been learned about the molecular mechanism by which Agrobacterium produces T-DNA and transports it into the host nucleus. However, T-DNA integration, which is the limiting, hence, the most critical step of the transformation process, largely remains an enigma. Increasing evidence suggests that Agrobacterium utilizes the host DNA repair machinery to facilitate T-DNA integration. Meanwhile, it is well known that chromatin modifications, including the phosphorylation of histone H2AX, play an important role in DNA repair. Thus, by implication, such epigenetic codes in chromatin may also have a considerable impact on T-DNA integration, although the direct evidence to demonstrate this hypothesis is still lacking. In this review, we summarize the recent advances in our understanding of Agrobacterium T-DNA integration and discuss the potential link between this process and the epigenetic information in the host chromatin.
Keywords: Agrobacterium, T-DNA integration, DSB repair, chromatin modifications, histone codes
1. Introduction
Agrobacterium-mediated genetic transformation of plants is the only known natural example of trans-kingdom gene transfer. During transformation, Agrobacterium exports a single-stranded copy of the bacterial transferred DNA (T-DNA) into the host cell and ultimately integrates it into the host genome. In nature, Agrobacterium (A. tumefaciens) infects plant wounded tissues and causes neoplastic growths called crown gall tumors. In addition, under laboratory conditions, this phytopathogen has the ability to transform virtually any eukaryotic species, from fungal to human cells (reviewed in [1]). This unique feature distinguishes Agrobacterium as a versatile and powerful tool for molecular genetic studies as well as for plant biotechnology.
The Agrobacterium transformation process is coordinately regulated by the bacterial proteins and the host factors (for recent reviews, see [2–5]). Upon perception of plant phenolic compounds exuded from wound sites, Agrobacterium activates expression of several effectors, termed virulence (Vir) proteins, via the two-component (VirA-VirG) signal transduction system. Among the induced Vir proteins, VirD1 and VirD2 function together as an endonuclease complex and generate a single-stranded copy of T-DNA (T-strand) from a specific DNA segment that is defined by two border sequences of 25-bp direct repeats in the tumor-inducing (Ti) plasmid. Subsequently, the T-strand, with one VirD2 molecule covalently attached to its 5′ end (Fig. 1A), is exported into the host cell through a type IV secretion system (T4SS) composed of the VirB and VirD4 proteins. Moreover, with the help of their C-terminal export signals [6], at least four other bacterial effectors (VirD5, VirE2, VirE3, and VirF) are also translocated into the host cell through the T4SS channel [6, 7], facilitating the rest of the transformation process.
Figure 1.
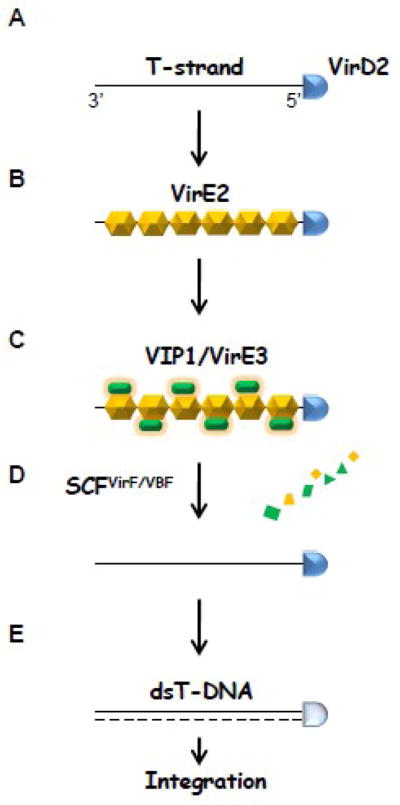
Schematic overview of the T-complex formation and uncoating
(A) The Agrobacterium protein VirD2 is covalently attached to the 5′ end of the single-stranded (ss) T-DNA (T-strand) within the bacterial cell. (B) Numerous VirE2 molecules, which are most likely to be exported into the host cell independently of the T-DNA, directly bind to the T-strand, forming the T-complex. (C) In addition, the plant factor VIP1 (VirE2-interacting protein 1) and/or the Agrobacterium effector VirE3 interact with VirE2, facilitating the nuclear import of the T-complex. (D) Once the T-complex reaches the host cell nucleus, VIP1 and VirE2 are presumably removed from the T-strand by the Agrobacterium effector VirF and/or the plant factor VBF (VIP1-binding F-box protein). Both VirF and VBF are F-box proteins that function in the SCF (Skp1-Cul1-F-box protein) ubiquitin E3 ligase complex (SCFVirF and SCFVBF, respectively) and target VIP1 as well as its associated protein VirE2 for proteasome-dependent degradation. It remains elusive whether and how VirE3 and VirD2 dissociate from the T-strand. (E) The T-strand is likely to be converted into a double-stranded form (dsT-DNA) before T-DNA expression and/or integration. Whether VirD2 is still attached to the T-strand during this conversion is also unknown.
Within the host cytoplasm, the T-DNA is believed to exist as a nucleoprotein complex (T-complex), in which it is coated with numerous VirE2 molecules (Fig. 1B; [8]). Furthermore, the plant factor VIP1 (VirE2-interacting protein 1), which contains a functional nuclear localization signal (NLS), interacts with VirE2 (Fig. 1C) and facilitates the nuclear import of T-DNA [9]. To augment this VIP1 function, Agrobacterium exports into the host cell another bacterial effector VirE3 [10]; like VIP1, the VirE3 protein also possesses functional NLSs and mediates the T-DNA nuclear import via its direct binding to VirE2 (Fig. 1C; [10]).
After the T-complex enters the cell nucleus, the coating proteins are most likely removed from the T-strand by the VirF-mediated protein degradation (Fig. 1D; [11]). VirF, the first F-box protein identified in prokaryotes [12], functions as a subunit of the SCF (Skp1-Cul1-F-box protein) ubiquitin E3 ligase complex in the host cell and targets VIP1 as well as its associated protein VirE2 for proteasome-dependent degradation [11]. In addition, the plant F-box factor VBF (VIP1-binding F-box protein) is involved in the T-complex uncoating in a manner similar to VirF (Fig. 1D; [13]). The finding that VirE3 and VirF bacterial effectors possess functional host analogs, VIP1 and VBF, respectively, indicates potential convergent evolution [14] and underscores the importance of the transformation steps mediated by these factors for the infection process. Furthermore, VIP1 and VBF are components of the plant defense system [13, 15, 16], indicating the ability of Agrobacterium to subvert the host defense machinery for active promotion of infection.
The T-complex proteasomal uncoating process is likely to be a prerequisite for conversion of the T-strand into the double-stranded DNA (dsT-DNA) and its subsequent expression and/or integration into the host genome (Fig. 1E). However, potentially in a defense response of the host plant, VirF is rapidly degraded via the host ubiquitin/proteasome pathway, and Agrobacterium has evolved another exported effector, VirD5, to interact directly with and stabilize the VirF protein (Magori S and Citovsky V, unpublished).
The entire process of Agrobacterium-mediated genetic transformation is reminiscent of the retrovirus-mediated gene transfer. However, unlike retroviruses, Agrobacterium does not export any proteins that function as an integrase. Moreover, none of the known exported bacterial effectors has been clearly demonstrated to play a direct role in T-DNA integration. Therefore, Agrobacterium most likely exploits the host factors to complete this process. In recent years, the host DNA double-strand break (DSB) repair has received increasing attention as a primary mechanism that facilitates T-DNA integration [17]. In this review, we focus on the potential role of the DSB repair machinery in Agrobacterium genetic transformation and also discuss how chromatin dynamics affects DSB repair and, by implication, T-DNA integration.
2. DSB represents the primary target site of T-DNA integration
As an indirect means to dissect the molecular mechanism underlying T-DNA integration, it is important to understand where in the host genome T-DNA is ultimately targeted. Large-scale analyses of T-DNA insertion distribution patterns in Arabidopsis suggest that the integration occurs preferentially in gene-rich euchromatic regions of the plant genome [18–20]. However, all these analyses were done using transgenic plants that had been positively selected based on the marker gene expression. Thus, the seemingly non-random integration pattern may be just a consequence of the variable transcription activity at the initial integration sites. To address this problem, a more recent work utilized Agrobacterium-transformed plant cells propagated under non-selective conditions and found a high frequency of T-DNA insertions even in the heterochromatic regions [21]. Furthermore, the integration pattern did not correlate with the genomic DNA methylation pattern [21]. Together, these observations suggest that T-DNA integration per se takes place randomly throughout the genome, regardless of the DNA sequences or the transcription activity at the pre-integration sites [21].
Given that T-DNA integration is truly random, what could be the limiting factor of this event? Several lines of evidence suggest that T-DNA integration may depend on the availability of naturally-occurring DNA double-strand breaks (DSBs) in the host genome. Indeed, exposure of plants to DSB-inducing agents, such as X-rays, is known to enhance integration of foreign genes [22]. In addition, it has been shown that induction of DSBs by transient expression of a rare-cutting restriction enzyme in plant genomes increases the T-DNA integration frequency [23–25]. Thus, Agrobacterium likely utilizes DSBs as the primary target sites of T-DNA integration. However, the possibility that other DNA lesions, such as single-strand breaks, may also serve as the potential integration sites cannot be excluded.
3. T-DNA integration largely relies on host factors
The VirD2 protein of Agrobacterium has long been proposed as a putative DNA ligase that functions during T-DNA integration [26]. After mobilization of the T-strand within the bacterial cell, VirD2 is conjugated to the 5′ end of the T-strand and escorts the T-DNA to the inside of the plant cell nucleus (see above). As a protein directly associated with T-DNA, VirD2 might possess additional functions in the cell nucleus. In fact, previous studies have shown that VirD2 has the ability not only to cleave the border sequence of T-DNA, but also to rejoin the cleavage products in vitro [27]. This ligation activity may be conferred by the conserved H-R-Y integrase motif found in the VirD2 amino acid sequence. However, an R-to-G mutation in this motif did not affect the T-DNA integration efficiency in vivo [26]. Furthermore, studies using an in vitro T-DNA ligation assay revealed that plant extracts, but not VirD2, are required for T-DNA ligation at the tested target sequence [28]. Together, these observations suggest that T-DNA integration largely relies on plant factors, but not any of the bacterial effector proteins.
4. The role of DSB repair machinery in T-DNA integration
As we discussed above, DSBs in the host genomes are thought to be the primary target sites of T-DNA integration, which is most likely mediated by the host factors. Thus, it makes biological sense that the host DSB repair proteins play a role in Agrobacterium T-DNA integration.
In eukaryotes, DSBs are known to be repaired by two conserved pathways: homologous recombination (HR) and non-homologous end joining (NHEJ). The HR pathway repairs DSBs by using sequence homology from an undamaged sister chromatid or homologous chromosome, whereas the NHEJ pathway directly rejoins damaged DNA ends (for recent reviews, see [29–32]). Possible involvement of both DSB repair pathways in T-DNA integration has been intensively studied in budding yeast, which can be transformed by Agrobacterium under laboratory conditions. For example, genetic studies using yeast mutants demonstrated that many of the NHEJ proteins, including Ku70, Rad50, Mre11, Xrs2 and Lig4, are required for integrating the T-DNA into the yeast genome (Table 1; [33]). Moreover, Rad51 and Rad52 have been shown to play an essential role in T-DNA integration by HR in yeast (Table 1; [34]). Although these studies were done in a non-natural host of Agrobacterium, the results clearly indicate that the host DSB repair machinery has a substantial involvement in T-DNA integration.
Table 1.
DNA double-strand break (DSB) repair machinerya and its possible involvement in T-DNA Integration
| Budding yeast | Arabidopsis | Proposed function | Requirement for T-DNA integration | References |
|---|---|---|---|---|
| Non-homologous end joining (NHEJ) | ||||
| Ku70 | AtKU70 | The Ku70-Ku80 heterodimer detects and juxtaposes the DSB ends | Yes (yeast) | [33] |
| Ku80 | AtKU80 | Yes/No (plants)b | [36–38] | |
| Mre11 | AtMRE11 | The Mre11-Rad50-Xrs2 (MRX) complex processes the DSB ends | Yes (yeast) | [33] |
| Rad50 | AtRad50 | Yes (yeast) | [33] | |
| Xrs2 | ND | Yes (yeast) | [33] | |
| Lig4 | AtLIG4 | The Lig4-Lif1 complex ligates the DSB ends | Yes (yeast), Yes/No (plants)b | [33,36,39] |
| Lif1 | AtXRCC4 | ND | ||
| Homologous recombination (HR) | ||||
| Rad51 | AtRAD51 | Facilitates strand invasion | Yes (yeast) | [34] |
| Rad52 | AtRAD52 | Helps to load Rad51 onto ssDNA | Yes (yeast) | [34] |
| Mre11 | AtMRE11 | The Mre11-Rad50-Xrs2 (MRX) complex processes the DSB ends. | No (yeast) | [34] |
| Rad50 | AtRAD50 | No (yeast) | [34] | |
| Xrs2 | ND | No (yeast) | [34] | |
| Rad54 | AtRAD54 | Facilitates strand invasion | Enhances gene targeting in plantsc | [77] |
ND, not determined; ss, single-stranded.
Only major DSB repair factors involved in NHEJ and HR are listed.
Not conclusive due to conflicting results between different studies.
Expression of the yeast Rad54 in Arabidopsis enhances gene targeting (i.e., HR-mediated T-DNA integration) by one to two orders of magnitude [77].
The role of the DSB repair proteins during Agrobacterium transformation has also been investigated in the model plant Arabidopsis thaliana. The homologs of most of the HR and NHEJ proteins have been identified in Arabidopsis (for a review, see [35]), and several of them have been tested for their effects on T-DNA integration mostly by genetic analyses with the corresponding mutants (Table 1). For example, it was reported that a mutant lacking the Arabidopsis homolog of Ku80 (AtKU80), a protein that recognizes the damaged dsDNA ends during NHEJ, exhibits a reduced T-DNA integration efficiency [36, 37]. On the other hand, overexpression of AtKU80 in Arabidopsis enhances T-DNA integration [37]. However, contrary to this result, another research group showed that AtKU80 is dispensable for the integration [38]. Such a discrepancy between different studies is also the case for Lig4 (AtLIG4), a DNA ligase essential for NHEJ. One study reported that AtLIG4 is required for T-DNA integration [36], while another study showed that it is not essential [39]. These contradictory results might simply reflect different assays used in different laboratories. For example, the floral-dip transformation method is thought to be a relatively imprecise means to analyze T-DNA integration, compared with the root tumor formation assay. It is also possible that, in multicellular organisms, the degree of the involvement of each DSB repair protein in T-DNA integration may vary among different cell types and/or developmental stages. Moreover, plant mutants defective in one NHEJ pathway may utilize other NHEJ pathways and/or HR as backup repair machinery, which could result in a relatively mild or undetectable phenotype of the mutants with regard to T-DNA integration.
Unlike yeast, plants predominantly utilize NHEJ rather than HR to repair DSBs [40, 41], suggesting that T-DNA is most likely integrated into the plant genome via NHEJ. This may be the reason why most studies thus far have been focusing on the plant NHEJ factors for their involvement in Agrobacterium transformation. However, one should be cautious about this notion; increasing evidence suggests that induction of DSBs by rare-cutting or site-specific endonucleases can enhance the HR machinery in plants [42–47]. Thus, under such conditions, the potential effect of the HR pathway on T-DNA integration should also be taken into account.
In addition to the highly-conserved HR and NHEJ machinery, plant-specific factors are also important in DSB sensing and/or repair. One such example is Arabidopsis SOG1 (SUPPRESSOR OF GAMMA RESPONSE 1), which encodes a putative transcription factor of the NAC domain family [48]. It has been shown that SOG1 is required for rapid induction of hundreds of genes in response to ionizing radiation in plants [48]. This suggests that SOG1, a protein unique to plants, represents a central transcriptional regulator that mediates DNA damage response [48]. It is tempting to speculate that such plant-specific DNA repair proteins might also play a role in Agrobacterium T-DNA integration.
Studies of DSB repair in the context of T-DNA integration have begun only recently and represent one of the most active fields of Agrobacterium research. Although the contradictions between different studies need to be resolved, it is now indisputable that Agrobacterium at least partly utilizes the host DSB repair machinery for its T-DNA integration. To understand the precise molecular mechanism of the integration process, further identification and characterization of DSB repair proteins in plants are necessary.
5. Chromatin modifications and T-DNA integration
Chromatin structure and modifications play an indispensible role in a wide range of cellular processes, including DSB repair. The basic unit of chromatin is a nucleosome, in which 147 bp of chromosomal DNA is wrapped around a protein octamer core comprising histones H2A, H2B, H3 and H4. Each histone molecule contains an N-terminal tail domain, which is susceptible to a variety of post-translational modifications, such as phosphorylation, methylation, acetylation and ubiquitylation. Recent studies have shown that several types of histone modifications are essential for the DSB repair response (for reviews, see [29–32]), and may also be important for Agrobacterium T-DNA integration (Table 2).
Table 2.
Chromatin-related proteins implicated in T-DNA integration
| Proteina | Description | Involvement in DSB repair | Effect on T-DNA integrationb | References |
|---|---|---|---|---|
| ScEaf7 | Subunit of the NuA4 HAT complex | Yes | Negative | [55] |
| ScYaf9 | Subunit of the NuA4 HAT complex and the SWR1 chromatin-remodeling complex | Yes | Negative | [55] |
| ScGcn5 | Catalytic subunit of the ADA, SAGA and SLIK HAT complexes | Yes | Negative | [55] |
| ScNgg1 | Subunit of the ADA, SAGA and SLIK HAT complexes | Unknown | Negative | [55] |
| ScHda2 | Subunit of a class II HDAC complex of the RPD3/HDA1 family | Unknown | Positive | [55] |
| ScHda3 | Subunit of a class II HDAC complex of the RPD3/HDA1 family | Unknown | Positive | [55] |
| ScHst4 | HDAC of the Sir2 family | Unknown | Positive | [55] |
| AtHAF1 | HAT of the TAFII250 family | Unknown | Positive | [62] |
| AtHAG3 | HAT of the GNAT-MYST family | Unknown | Positive | [62] |
| AtHDT1 | HDAC of the HD2 familyc | Unknown | Positive | [62] |
| AtHDT2 | HDAC of the HD2 familyc | Unknown | Positive | [62] |
| AtFAS1 | Subunit of the chromatin assembly factor 1 (CAF-1) complex | Yes | Negative | [64] |
| AtFAS2 | Subunit of the chromatin assembly factor 1 (CAF-1) complex | Yes | Negative | [64] |
| AtASF1Bd | Histone chaperone of the H3/H4 family | Unknown | Positive | [62] |
HAT, histone acetyltransferase; HDAC, histone deacetylase.
Sc and At indicate proteins from Saccharomyces cerevisiae (budding yeast) and Arabidopsis thaliana (plant), respectively.
The effect of each chromatin-related protein on Agrobacterium T-DNA integration was predicted based on its mutant phenotype; “Positive” indicates that deletion or knockdown of the corresponding gene leads to a decreased T-DNA integration efficiency, whereas “Negative” indicates that the mutant shows an enhanced integration efficiency.
The HD2 family represents a plant-specific histone deacetylase group [78].
Also known as SGA1.
The first histone modification that becomes evident upon DSB induction is the rapid phosphorylation of the histone variant H2AX (or H2A in yeast). This modification, known as γ-H2AX, encompasses ~2 Mb of chromatin surrounding a DSB in mammalian cells (~50 kb in yeast) [49, 50]. The phosphorylated H2AX is believed to serve as a landing platform for DSB repair machinery and recruit a number of downstream factors, including histone modifying enzymes (NuA4) [51] and ATP-dependent chromatin remodeling complexes (INO80 and SWR1) [51–53]. NuA4 is a histone acetyltransferase (HAT) complex that acetylates the first four lysine residues in the N-terminal tail of histone H4. Mutations in these lysine residues of H4 or the NuA4 subunits cause hypersensitivity to DSB-inducing agents [51, 54], suggesting that the NuA4-mediated histone acetylation plays a critical role in DSB repair. Surprisingly, a genome-wide mutant screen revealed that yeast strains lacking Eaf7 or Yaf9, both of which are subunits of the NuA4 complex, exhibit a strongly enhanced Agrobacterium-mediated T-DNA integration efficiency (> 5-fold increase) [55]. It should be noted that Yaf9 is also a subunit of the SWR1 chromatin remodeling complex, which directly binds to the phosphorylated H2AX and replaces it with another histone H2A variant, H2AZ. Like NuA4, yeast strains lacking the functional SWR1 complex show hypersensitivity to DSB-inducing agents [56], suggesting the essential role of SWR1 in DSB repair. How the defects in the NuA4- and SWR1-regulated chromatin dynamics increase the T-DNA integration efficiency remains unclear, but one could speculate that disruption or delay of DSB repair at certain reaction steps might leave unrepaired DSBs in a form preferable for T-DNA integration. However, one should not forget that both NuA4 and SWR1 are also involved in transcriptional regulation of many genes [57, 58]. Thus, we cannot rule out the possibility that the increased T-DNA integration efficiency in mutants lacking NuA4 or SWR1 could be an indirect effect caused by misregulation of as yet unknown genes involved in the integration process.
In addition to the NuA4 HAT complex, other histone acetyltransferases, such as Gcn5 [59] and Hat1 [60], have been implicated in DSB repair, although their precise roles are relatively unclear. Interestingly, it has been reported that yeast strains lacking Gcn5 exhibit a highly increased T-DNA integration efficiency [55], suggesting that the Gcn5-mediated histone acetylation during DSB repair may negatively control T-DNA integration. However, again, this observation could be an indirect effect of misregulation of certain genes in the mutant because Gcn5 is known to be required for global transcriptional activation as a catalytic subunit of the SAGA, SLIK and ADA complexes (for a review, see [61]). Indeed, yeasts lacking Ngg1, another subunit of all these Gcn5-containing HAT complexes, also show an enhanced T-DNA integration frequency [55].
In contrast to these yeast HAT-related proteins (Eaf7, Yaf9, Gcn5 and Ngg1), two Arabidopsis HAT proteins, HAF1 and HAG3, seem to positively regulate T-DNA integration as RNAi-mediated knockdown of the corresponding genes leads to substantial reduction in T-DNA integration efficiency [62]. Thus, it is difficult to generalize the role of HATs in the integration process. Potentially, different HAT proteins/complexes may possess distinct functions via different pathways during Agrobacterium transformation.
Given that histone acetyltransferases affect Agrobacterium T-DNA integration, it is plausible that histone deacetylases (HDACs) may also play a role in the integration. Indeed, deletion of HDAC-encoding genes (HST4, HDA2 and HDA3) in yeast strongly decreases T-DNA integration efficiency [55]. Furthermore, in Arabidopsis, RNAi-mediated knockdown of at least two HDACs (HDT1 and HDT2) was found to attenuate the susceptibility to Agrobacterium-mediated root transformation [62]. The major function of HDACs is the repression of transcription by inducing chromatin condensation. Thus, it is plausible that mutations or knockdown of HDACs may lead to ectopic expression of as yet unknown negative regulators of Agrobacterium transformation. Alternatively, HDACs may positively regulate T-DNA integration via DSB repair. In fact, several HDACs, such as the Sin3/Rpd3 HDAC complex, have been suggested to play a critical role in DSB repair [59, 63]. Although the molecular basis for the effects of HATs and HDACs on T-DNA integration remains unclear, the observations in yeast and Arabidopsis suggest that histone acetylation balance controlled by the HAT/HDAC interplay is important to facilitate Agrobacterium transformation.
Recent studies also suggest the role of histone chaperons in T-DNA integration. For example, Arabidopsis mutants lacking the chromatin assembly factor 1 (CAF-1) complex were found to exhibit an increased T-DNA integration efficiency [64]. CAF-1 is believed to mediate nucleosome assembly during DNA replication and nucleotide exchange repair (NER) (for recent reviews, see [65–67]). Interestingly, the loss of the CAF-1 activity in Arabidopsis leads to upregulation of several DSB repair proteins involved in HR but not NHEJ [64, 68–70]. Consistently, the CAF-1 mutants show an enhanced HR frequency [64, 69]. Thus, the enhanced T-DNA integration rate in the CAF-1 mutants may be the consequence of the hyperactivated HR process. However, we cannot exclude the possibility that mutations in CAF-1 result in formation of a relatively loose chromatin structure, which may be more accessible to a foreign DNA. In addition to CAF-1, the Arabidopsis homolog of Asf1, a member of the H3/H4 family of histone chaperons, has been implicated in T-DNA integration [62], but the molecular basis of its effect remains elusive.
6. Potential role of the histone code in T-DNA integration
Although some types of histone modifications are likely to be involved in Agrobacterium T-DNA integration, the molecular basis for this putative involvement is completely unknown. An attractive hypothesis is that epigenetic information at chromatins surrounding a DSB may serve as a “landmark” to be recognized by the T-complex (Fig. 2A). The resulting chromatin-T-complex interactions could bring T-DNA into close proximity to a DSB and facilitate its integration along the host DSB repair. In this model, the T-complex uncoating is not likely to occur until it reaches the host chromatin. It remains to be investigated whether the T-complex possesses a preferential affinity to any modified histones, but recent studies have shown that the plant factor VIP1, a component of the T-complex, directly binds to all of the core histones (H2A, H2B, H3 and H4) as well as purified plant nucleosomes [71–73]. Interestingly, the C-terminal truncated VIP1 (amino acids 1–164), which cannot interact with histone H2A, strongly decreases T-DNA integration efficiency [71]. Furthermore, the Arabidopsis mutant lacking histone H2A was shown to be defective in T-DNA integration [74]. These observations suggest that the association of the T-complex with the host chromatin via VIP1 is critical for T-DNA integration. This intrinsic interaction may be further stabilized by certain histone modification patterns in the host chromatin. However, it should be noted that non-plant species, which do not encode an apparent VIP1-like gene, are also susceptible to Agrobacterium transformation (reviewed in [1]). Thus, the VIP1-mediated T-complex targeting to the host chromatin is not the sole mechanism underlying T-DNA integration, but other unknown factors and pathways may be equally important for this process.
Figure 2.
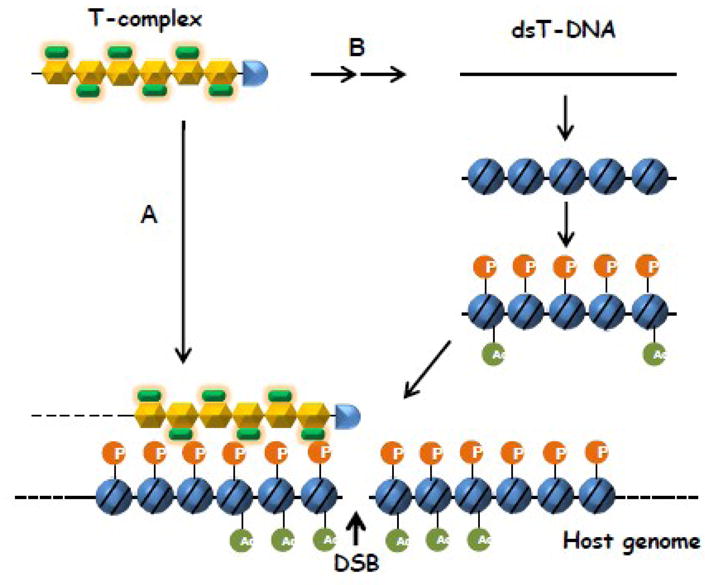
Potential roles of chromatin modifications in T-DNA integration
(A) A “T-complex-to-DSB targeting” model. The T-complex is preferably recruited to the host chromatin harboring certain histone modifications that occur nearby a DNA double-strand break (DSB). Such histone modifications may include the phosphorylation (“P”) of the histone H2A variant H2AX and the acetylation (“Ac”) of histone H4. In this model, the plant factor VIP1, a component of the T-complex, may serve as the molecular link between the DSB-containing chromatin and the T-DNA. Only after this association, the T-complex is uncoated and the single-stranded T-DNA is converted into a double-stranded (ds) T-DNA intermediate. Finally, the ends of the dsT-DNA are ligated with the DSB ends by the host DSB repair machinery. (B) A “T-complex-as-DSB disguise” model. First, the T-complex is uncoated and the T-strand is converted into dsT-DNA before its integration. This dsT-DNA is then assembled into a nucleoprotein complex composed of the host histones, which subsequently undergo certain modifications, such as phosphorylation (“P”) and acetylation (“Ac”). Finally, the resulting chromatin-like structure is mistakenly recognized by the host DSB repair machinery and incorporated into a naturally-occurring DSB in the host genome.
In an alternative model, histone modifications may help T-DNA “disguise” as a host chromatin that harbors a DSB (Fig. 2B). In this mechanism, after the T-complex uncoating, its T-strand needs to be converted to dsT-DNA, which is then packaged into a nucleoprotein complex composed of the host histones. Subsequently, the incorporated histones are subject to specific modifications, such as phosphorylation of H2AX. Finally, these histone codes are recognized by the host DSB repair machinery, leading to T-DNA integration at a nearby DSB (if available) in the host genome. The advantage of this model is that it does not require any plant-specific proteins and thus may explain how T-DNA is integrated into the genome of non-plant species. Although direct evidence to support this hypothesis is lacking, a recent study using an immunoprecipitation assay demonstrated that at least KU80, an essential protein that recognizes DSB ends during NHEJ, directly binds to dsT-DNA intermediates in vivo [37]. Moreover, it was shown that two or more dsT-DNA molecules can be ligated with each other in plant cells, most likely with the help of KU80 [37]. These observations imply that at least some fraction of T-DNA exists as free dsT-DNA in the cell nucleus and that this dsT-DNA can be recognized by the host DSB repair machinery, regardless of its targeting to the host genome. In this scenario, assembly of dsT-DNA into a chromatin-like structure with certain histone modifications may function as a “decoy” to misguide the repair proteins to the ends of the T-DNA molecule.
7. Future perspective
Despite intensive studies, little is known about the molecular mechanisms underlying T-DNA integration, the final and most critical step of Agrobacterium-mediated genetic transformation. Recent genetic studies have indicated the potential involvement of the host chromatin modifications in T-DNA integration. However, the host chromatin dynamics possesses a global impact on various cellular processes, including transcriptional regulation. Thus, it is still unclear whether the effect of the host chromatin modifications on the integration process is direct or indirect. This issue is important especially in higher eukaryotes, such as plants. Indeed, it is well known that mutations in many chromatin-modifying or chromatin-remodeling enzymes of Arabidopsis cause pleiotropic developmental defects, which could cofound the data interpretation in terms of the role of the corresponding gene in Agrobacterium-mediated transformation. To circumvent this problem, it may be beneficial to utilize simpler model organisms, such as yeast, and directly analyze the behavior of T-DNA and the host factors at DSBs. In budding yeast, physical monitoring of DSB repair can be performed using a well-established system which allows for induction of a single DSB in vivo by the HO endonuclease [75]. Alternatively, the zinc-finger nuclease (ZFN) technology, a recently developed strategy for gene targeting, can be exploited to induce a DSB at a specific genomic site even in plants (reviewed in [76]). Use of such molecular tools will help to understand how the host DSB repair and chromatin dynamics coordinately regulate Agrobacterium T-DNA integration.
Acknowledgments
The work in our laboratory is supported by grants from USDA/NIFS, NIH, NSF, BARD, OE, and BSF (to V.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet. 2006;22:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Gelvin SB. Agrobacterium and Plant Genes Involved in T-DNA Transfer and Integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 3.Tzfira T, Citovsky V. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 2002;12:121–129. doi: 10.1016/s0962-8924(01)02229-2. [DOI] [PubMed] [Google Scholar]
- 4.Gelvin SB. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol. 2010;48:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- 5.Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vergunst AC, van Lier MC, den Dulk-Ras A, Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 8.Citovsky V, Wong ML, Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc Natl Acad Sci U S A. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005;24:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 12.Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuink TJ, Crosby WL, Hooykaas PJ. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 13.Zaltsman A, Krichevsky A, Loyter A, Citovsky V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe. 2010;7:197–209. doi: 10.1016/j.chom.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–383. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 15.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 16.Pitzschke A, Djamei A, Teige M, Hirt H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci U S A. 2009;106:18414–18419. doi: 10.1073/pnas.0905599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Szabados L, Kovacs I, Oberschall A, Abraham E, Kerekes I, Zsigmond L, Nagy R, Alvarado M, Krasovskaja I, Gal M, Berente A, Redei GP, Haim AB, Koncz C. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 2002;32:233–242. doi: 10.1046/j.1365-313x.2002.01417.x. [DOI] [PubMed] [Google Scholar]
- 19.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 20.Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, Lepiniec L, Caboche M, Lecharny A. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 2002;3:1152–1157. doi: 10.1093/embo-reports/kvf237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SI, Veena Gelvin SB. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007;51:779–791. doi: 10.1111/j.1365-313X.2007.03183.x. [DOI] [PubMed] [Google Scholar]
- 22.Kohler F, Cardon G, Pohlman M, Gill R, Schieder O. Enhancement of transformation rates in higher-plants by low-dose irradiation - Are DNA-repair systems involved in the incorporation of exogenous DNA into the plant genome. Plant Mol Biol. 1989;12:189–199. doi: 10.1007/BF00020504. [DOI] [PubMed] [Google Scholar]
- 23.Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double- strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzfira T, Frankman LR, Vaidya M, Citovsky V. Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol. 2003;133:1011–1023. doi: 10.1104/pp.103.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chilton MD, Que Q. Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol. 2003;133:956–965. doi: 10.1104/pp.103.026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel AM, Hohn B. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci U S A. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziemienowicz A, Tinland B, Bryant J, Gloeckler V, Hohn B. Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. Mol Cell Biol. 2000;20:6317–6322. doi: 10.1128/mcb.20.17.6317-6322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 30.Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutat Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 31.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Sinha M, Peterson CL. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics. 2009;1:371–385. doi: 10.2217/epi.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Attikum H, Bundock P, Hooykaas PJ. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 2001;20:6550–6558. doi: 10.1093/emboj/20.22.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Attikum H, Hooykaas PJ. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:826–832. doi: 10.1093/nar/gkg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleuyard JY, Gallego ME, White CI. Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair. 2006;5:1–12. doi: 10.1016/j.dnarep.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003;34:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Vaidya M, White C, Vainstein A, Citovsky V, Tzfira T. Involvement of KU80 in T-DNA integration in plant cells. Proc Natl Acad Sci U S A. 2005;102:19231–19236. doi: 10.1073/pnas.0506437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallego ME, Bleuyard JY, Daoudal-Cotterell S, Jallut N, White CI. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 2003;35:557–565. doi: 10.1046/j.1365-313x.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- 39.van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, Hooykaas PJ. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003;31:4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray A, Langer M. Homologous recombination: ends as the means. Trends Plant Sci. 2002;7:435–440. doi: 10.1016/s1360-1385(02)02327-0. [DOI] [PubMed] [Google Scholar]
- 41.Britt AB, May GD. Re-engineering plant gene targeting. Trends Plant Sci. 2003;8:90–95. doi: 10.1016/S1360-1385(03)00002-5. [DOI] [PubMed] [Google Scholar]
- 42.Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiurazzi M, Ray A, Viret JF, Perera R, Wang XH, Lloyd AM, Signer ER. Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by the HO endonuclease. Plant Cell. 1996;8:2057–2066. doi: 10.1105/tpc.8.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orel N, Kyryk A, Puchta H. Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J. 2003;35:604–612. doi: 10.1046/j.1365-313x.2003.01832.x. [DOI] [PubMed] [Google Scholar]
- 46.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 48.Yoshiyama K, Conklin PA, Huefner ND, Britt AB. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci U S A. 2009;106:12843–12848. doi: 10.1073/pnas.0810304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 50.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 53.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 54.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 55.Soltani J, van Heusden GP, Hooykaas PJ. Deletion of host histone acetyltransferases and deacetylases strongly affects Agrobacterium-mediated transformation of Saccharomyces cerevisiae. FEMS Microbiol Lett. 2009;298:228–233. doi: 10.1111/j.1574-6968.2009.01723.x. [DOI] [PubMed] [Google Scholar]
- 56.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 57.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 58.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 59.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin S, Parthun MR. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol Cell Biol. 2006;26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 62.Crane YM, Gelvin SB. RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc Natl Acad Sci U S A. 2007;104:15156–15161. doi: 10.1073/pnas.0706986104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci U S A. 2004;101:1644–1649. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006;25:5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ridgway P, Almouzni G. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J Cell Sci. 2000;113:2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- 66.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 67.Ramirez-Parra E, Gutierrez C. The many faces of chromatin assembly factor 1. Trends Plant Sci. 2007;12:570–576. doi: 10.1016/j.tplants.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Schonrock N, Exner V, Probst A, Gruissem W, Hennig L. Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J Biol Chem. 2006;281:9560–9568. doi: 10.1074/jbc.M513426200. [DOI] [PubMed] [Google Scholar]
- 69.Kirik A, Pecinka A, Wendeler E, Reiss B. The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell. 2006;18:2431–2442. doi: 10.1105/tpc.106.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loyter A, Rosenbluh J, Zakai N, Li J, Kozlovsky SV, Tzfira T, Citovsky V. The plant VirE2 interacting protein 1. a molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol. 2005;138:1318–1321. doi: 10.1104/pp.105.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lacroix B, Loyter A, Citovsky V. Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc Natl Acad Sci U S A. 2008;105:15429–15434. doi: 10.1073/pnas.0805641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mysore KS, Nam J, Gelvin SB. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc Natl Acad Sci U S A. 2000;97:948–953. doi: 10.1073/pnas.97.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 76.Weinthal D, Tovkach A, Zeevi V, Tzfira T. Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci. 2010;15:308–321. doi: 10.1016/j.tplants.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 77.Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci U S A. 2005;102:12265–12269. doi: 10.1073/pnas.0502601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]


