Abstract
The nematode Caenorhabditis elegans is the first animal whose genome is completely sequenced, providing a rich source of gene information relevant to metazoan biology and human disease. This abundant sequence information permits a broad-based gene inactivation approach in C. elegans, in which chemically mutagenized nematode populations are screened by PCR for deletion mutations in a specific targeted gene. By handling mutagenized worm growth, genomic DNA templates, PCR screens, and mutant recovery all in 96-well microtiter plates, we have scaled up this approach to isolate deletion mutations in >100 genes to date. Four chemical mutagens, including ethyl methane sulfonate, ethlynitrosourea, diepoxyoctane, and ultraviolet-activated trimethylpsoralen, induced detectable deletions at comparable frequencies. The deletions averaged ∼1400 bp in size when using a ∼3 kb screening window. The vast majority of detected deletions removed portions of one or more exons, likely resulting in loss of gene function. This approach requires only the knowledge of a target gene sequence and a suitable mutagen, and thus provides a scalable systematic approach to gene inactivation for any organism that can be handled in high density arrays.
The rapid progress in genome sequencing projects has propelled a shift in genome analysis from structural genomics to functional genomics, that is, the genome-driven systematic study of gene function (Hieter and Boguski 1997). In this postgenomic era, model organisms in which large-scale functional analyses and rapid genetic experiments are possible—chiefly the yeast Saccharomyces cerevesiae, the fruit fly Drosophila melanogaster, and the nematode Caenorhabditis elegans—are increasingly useful in understanding human disease pathways (Miklos and Rubin 1996; Oliver 1996; Ahringer 1997). With the recent completion of the C. elegans genomic sequence (The C. elegans Sequencing Consortium 1998), the first complete animal genome is now available. Computational analysis of partial C. elegans genome data has revealed that many positionally cloned human disease genes have C. elegans orthologs (genes encoding proteins with similar multidomain architecture and predicted function) (Mushegian et al. 1997, 1998). The C. elegans genome also contains a significant proportion of apparently nematode-specific protein families that may be relevant for nematode biology and parasitism (Sonnhammer and Durbin 1997; Blaxter 1998).
A rapid method to ascertain gene function by targeted gene inactivation in C. elegans would be highly desirable, but homologous recombination as in the mouse has not yet proven feasible (Plasterk 1995). Microinjection of target-specific RNA, for reasons not completely understood, is a remarkably effective means of transcriptional interruption in this small organism (Fire et al. 1998). However, the extent of RNA-mediated inactivation is difficult to assess in many cases, and such inactivation obviously does not produce a germ-line lesion necessary for genetic crosses, suppressor screens, and other longer-term genetic manipulations. One large-scale approach to germ-line inactivation is to induce random mutations in the animal population, followed by screening for mutations in a target gene of known sequence. The most well-developed method of this so-called target-selected gene inactivation in C. elegans has used random transposon Tc1 insertions to generate a collection of mutants, followed by PCR screening for the presence of Tc1 in a gene of choice (Zwaal et al. 1993). However, because Tc1 insertion alone does not usually result in gene inactivation, it is necessary to subsequently screen individual Tc1 alleles for animals in which the transposon and flanking DNA have been deleted through transposon excision, a relatively infrequent event (Plasterk 1995). An alternative approach is to use chemical mutagens to directly induce deletions in a population, and then to screen by PCR for deleted segments within a selected target region (Yandell et al. 1994). Jansen et al. (1997, 1999) have established the broader feasibility of this approach by isolating mutants of the C. elegans heterotrimeric G protein gene family.
In this study we describe our results using random chemical mutagenesis and PCR screening to rapidly isolate deletion mutations in a large number of genes encoding proteins with a broad range of functions. C. elegans is unique among model animal species in that it can be grown in liquid cultures and also can be stored frozen but viable at −80°C. We have taken advantage of these properties to devise a rapid and scalable procedure for gene disruption almost entirely on the basis of microtiter plate arrays of whole animals and genomic DNA. We used four different chemical agents to create mutagenized libraries, and found that all four mutagens induce detectable deletions. Almost all of the deletions were significant enough to result in loss of exons, frame shifts, and other molecular lesions likely to cause loss of gene function. We discuss the sensitivity, specificity, limitations, and broader utility of this approach to systematic gene inactivation in C. elegans.
RESULTS
Library Construction and Screening
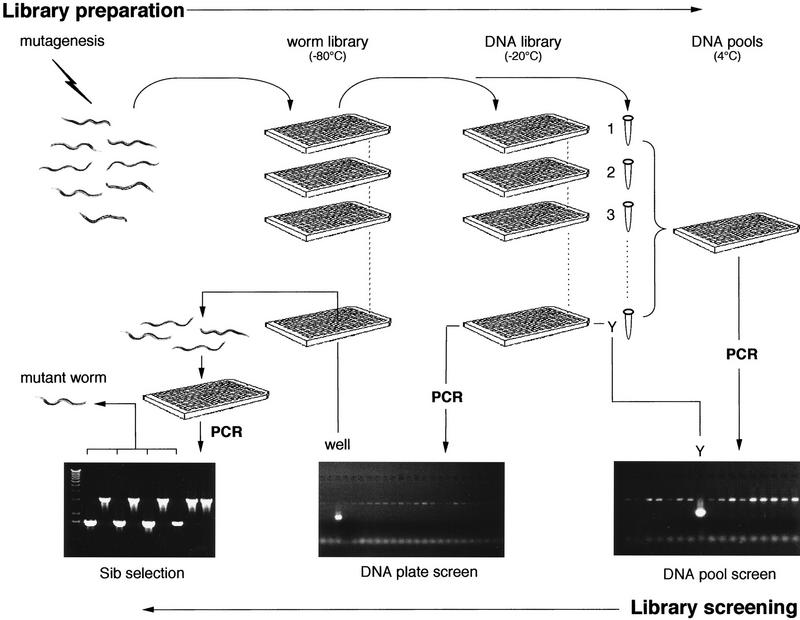
To enhance the scalability and speed of mutagenized library construction and screening, we relied on microtiter plate-based culture and analysis wherever possible (Fig. 1). F1 larvae of mutagenized P0 animals were distributed directly into microtiter plate wells (∼20 larvae representing 40 mutagenized genomes) and grown in liquid cultures to yield F2 larvae. Approximately one-half of the worms were processed into a corresponding genomic DNA microtiter plate array, with the remainder frozen at −80°C as viable animals. The initial round of nested PCR screening for deletion mutations in a targeted gene was performed on microtiter plate pools each containing ∼4000 mutagenized genomes. PCR primers were designed to flank ∼3 kb of exon-rich regions of the selected ORF, in many cases specifically targeting putative functional domains. Amplicons smaller than the wild-type amplicon were taken to represent candidate deletions. Such amplicons were expected to be preferentially amplified, compensating for the much lower representation of the mutant target template within the genomic DNA pool. Candidate-positive plate pools were resampled to eliminate false positives, which constituted about three-fourths of the first-round candidate deletions. After confirmation that a plate pool contained a deletion, the corresponding DNA array was then screened to identify a specific microtiter well. The corresponding well of the frozen worm library was thawed and individual animals were grown in microtiter wells; portions of these cultures were screened by PCR to identify heterozygous and homozygous mutant strains. A small number of thawed library wells failed to yield the expected deletion mutant, and the failure rate exceeded 50% when <100 viable animals were recovered from a well (data not shown).
Figure 1.
Schematic of mutant library construction and screening. F1 larvae of chemically mutagenized P0 animals are cultured in microtiter arrays to produce F2 larvae, constituting the worm library. An aliquot of worms from each well is removed and processed into a corresponding genomic DNA array; simultaneously, the 96 samples from each plate are pooled and processed to produce genomic DNA plate pools. The DNA plate pools are screened by nested PCR in 96-well plates for deletion amplicons, which identify a positive worm library plate Y. After confirmation by quadruplicate PCR (not illustrated), the genomic DNA array for that specific library plate is similarly screened to identify the specific well. The corresponding well is then retrieved from the frozen mutant worm library and thawed. Individual live worms are allowed to have self-progeny in microtiter wells, and aliquots of each well are tested by PCR to determine which of the thawed worms contain the deletion (see Methods for details).
To evaluate various chemicals that have been used to induce mutations in C. elegans, we created mutant libraries using different mutagens, including ethylmethane sulfonate (EMS), ethlynitrosourea (ENU), diepoxyoctane (DEO), and ultraviolet-activated trimethylpsoralen (UV-TMP). Each library consisted of forty-eight 96-well plate arrays and ∼200,000 mutagenized genomes. For any given target, usually 4–8 such libraries (comprising 0.8–1.6 million mutagenized genomes) were screened simultaneously for potential deletions. At this scale, deletion mutants were isolated from almost all loci that were targeted, and in many instances more than one deletion was identified; in the latter cases usually only the largest deletion was thawed from the worm library. We were unable to detect deletions in a small number (<5) of targeted genes, and these failed loci were too few to determine whether they shared any structural characteristics. The average number of animals recovered from frozen wells differed for each mutagen as follows: DEO 191 ± 159, EMS 359 ± 330, ENU 343 ± 249, and UV-TMP 199 ± 166.
Deletion Mutations
To date we have isolated deletion mutations in >100 independent gene targets using this method, and a representative listing is shown in Table 1. Among these are genes encoding a broad range of protein functional groups, including kinases, transcription factors, membrane receptors, metabolic enzymes, proteases, hormones, and nematode homologs of oncogenes and tumor suppressor genes. However, the largest target group comprises genes encoding proteins of unknown function, reflecting our strategic choice of using the C. elegans model to elucidate the function of novel predicted proteins.
Table 1.
Selected Deletion Mutations
| Cosmid name | GenBank acc. no. | Gene or protein | Allele name | Mutagen | Deletion size (bp) | Deletion effect on ORF | Preliminary phenotype | Referencea |
|---|---|---|---|---|---|---|---|---|
| B0478.1 | U57054 | Jun kinase homolog | nr2035 | DEO | 1825 | promoter–intron 7 | no obvious phenotype | |
| C02B8.4 | U40421 | Twist transcription factor homolog | nr2061 | EMS | 1267 | Intron 1–exon 3 | no obvious phenotype | S. Kostas and A. Fire |
| C03B1.12 | U40952 | Lysosomal-associated membrane protein | nr2045 | ENU | 1772 | promoter–intron 3 | no obvious phenotype | D. Fambrough |
| C04G6.3 | U55854 | phospholipase D homolog | nr2015 | EMS | 545 | exon 7–exon 8 | no obvious phenotype | |
| C05B5.7 | Z32679 | RGS GAP protein homolog | nr2017 | ENU | 637 | exon 1–intron 2 | mild egg-laying defective | M. Koelle |
| C18E3.8 | AF000265 | hop-1 presenilin | nr2003 | UV-TMP | 1196 | promoter–intron 3 | decreased brood size at 15° C | Westlund (1998) |
| C44H4.6 | Z79598 | GSK-3β kinase homolog | nr2056 | EMS | 1411 | intron 2–intron 6 | no obvious phenotype | |
| C47E8.5 | Z75530 | daf-21 HSP90 homolog | nr2081 | ENU | 863 | exon 1–exon 4 | larval arrest | |
| F02E8.6 | U53340 | Niemann–Pick type C protein homolog | nr2022 | EMS | 1120 | exon 3–exon 6 | no obvious phenotype | |
| F10B5.4 | Z48334 | Tubby homolog | nr2044 | DEO | 1816 | intron 1–beyond 3′ terminus | no obvious phenotype | |
| F13B12.a | Z70683 | insulin homolog | nr2091 | EMS | 1653 | deletes entire ORF | no obvious phenotype | S. Pierce; Gregoire et al. (1998) |
| F28H6.1 | AL031621 | akt-2 kinase | nr2021 | DEO | 2208 | promoter–exon 4 | no obvious phenotype | |
| F35H12.3 | U41540 | sel-12 presenilin | nr2011 | ENU | 1426 | intron 2–intron 6 | egg-laying defect | Westlund (1998) |
| F59G1.5 | U53332 | Frataxin homolog | nr2040 | DEO | 505 | promoter–intron 2 | no obvious phenotype | |
| K06A1.4 | U23449 | nuclear hormone receptor homolog | nr2034 | ENU | 1189 | exon 3–intron 5 | no obvious phenotype | P. Sengupta |
| K06A5.7 | AF039038 | cdc-25 homolog | nr2036 | UV-TMP | 2695 | deletes entire ORF | sterile with reduced germline | A. Golden and N. Ashcroft |
| K06C4.6 | U64843 | mod-1 serotonin-gated ion channel | nr2043 | EMS | 1209 | intron 2–exon 5 | resistant to serotonin-stimulated locomotion | R. Ranaganathan |
| K06E6.1 | U55375 | lim-6 homeodomain protein | nr2073 | ENU | 1703 | exon 4–beyond 3′ terminus | expulsion defective, nose-up foraging defective, synDaf-c, axonal defects | Hobert et al. (1999) |
| K10C3.6 | Z83235 | HNF4 transcription factor homolog | nr2041 | EMS | 893 | intron 2–exon 4 | no obvious phenotype | Sluder et al. (1999) |
| R11A5.4 | Z83122 | phosphoenolpyruvate kinase homolog | nr2043 | EMS | 1902 | intron 2–exon 5 | no obvious phenotype | |
| T01H8.1 | Z80219 | p90RSK kinase homolog | nr2031 | EMS | 758 | intron 2–exon 2 | no obvious phenotype | |
| T07A9.6 | AF036706 | daf-18 PTEN homolog | nr2037 | UV-TMP | 990 | intron 2–exon 3 | dauer defective | Gil et al. (1999) |
| T14B1.1 | Z46829 | tip-1 novel protein | nr2084 | ENU | 1206 | exon 1–beyond 3′ terminus | no obvious phenotype | D. Zarkower |
| T20H4.1 | U00037 | osm-10 novel protein | nr2076 | EMS | 1122 | intron 2–exon 5 | osmolarity response defective | Hart et al. (1999) |
| Y18D10 | AL008871 | GSK-3β kinase homolog | nr2047 | ENU | 1526 | intron 2–beyond 3′ terminus | maternal effect early embryonic lethal | |
| Y47D3 | AL031635 | p70S6 kinase homolog | nr2074 | ENU | 850 | intron 2–exon 3 | no obvious phenotype | |
| Y48E1B.13 | Z93393 | csp-1 caspase homolog | nr2018 | UV-TMP | 1220 | exon 4–intron 5 | no obvious phenotype | D. Hoeppner; Shaham (1998) |
| ZK154.7 | U70844 | TNF-converting enzyme homolog | nr2030 | EMS | 787 | exon 9–intron 14 | no obvious phenotype | |
| ZK520.4 | Z50796 | cul-2 cell cycle cullin | nr2020 | EMS | 1050 | exon 5–exon 5 | embryonic arrest | E. Kipreos |
| ZK632/6 | Z22181 | calnexin homolog | nr2010 | UV-TMP | 774 | exon 1–exon 3 | no obvious phenotype | |
| ZK675.1 | Z46812 | patched homolog | nr2029 | EMS | 960 | exon 6–exon 6 | sterile | P. Kuwabara |
Personal communications, unless otherwise noted.
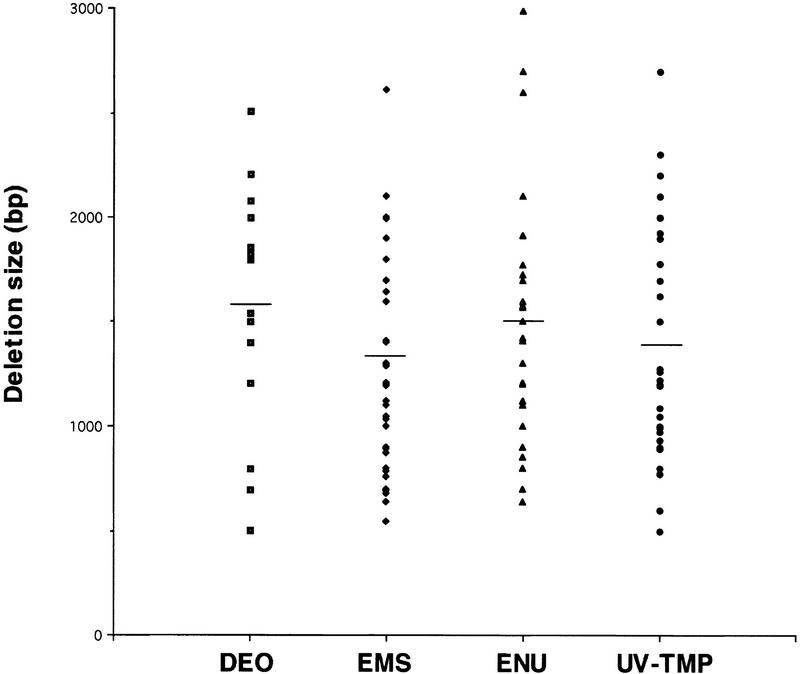
The average size of chemically induced deletions was ∼1400 bp (range 700–2900 bp), as detected by a PCR-screening window of ∼3 kb (Fig. 2). In several instances, smaller deletions (on the order of 400–500 bp) were fortuitously detected on screening of a specific library plate. The average size of deletions isolated from different mutagenized libraries ranged from ∼1320 bp for EMS up to 1580 bp for DEO, and a broad size range was observed for each agent (Fig. 2). DNA sequencing revealed that the majority were simple deletions, but about one-fifth of the sequenced deletions contained small (3–27 bp) insertions at the breakpoints. All of the latter deletions derived from libraries mutagenized by DEO, EMS, or ENU. Only one of our isolated deletions (xfl-1, nr2025), isolated from a UV-TMP library, contained a large inverted insertion, which is in contrast with the examples reported previously for gpa deletion mutants obtained by UV-TMP, in which two of three deletions contained large insertions (Jansen et al. 1997). In addition, in our series, both the proximal and distal deletion breakpoints in most cases fell within a region of two or more consecutive identical nucleotides, rendering the exact breakpoint ambiguous.
Figure 2.
Deletion size by mutagen, depicting those deletions selected with a PCR screening window of 2800–3400 bp. Not shown are a small number of larger (>3000 bp) deletions identified with PCR primers placed 4–5 kb apart in the target region, as well as smaller (<500 bp) deletions fortuitously detected on screening of specific library plates. Horizontal bar, average deletion size for each mutagen.
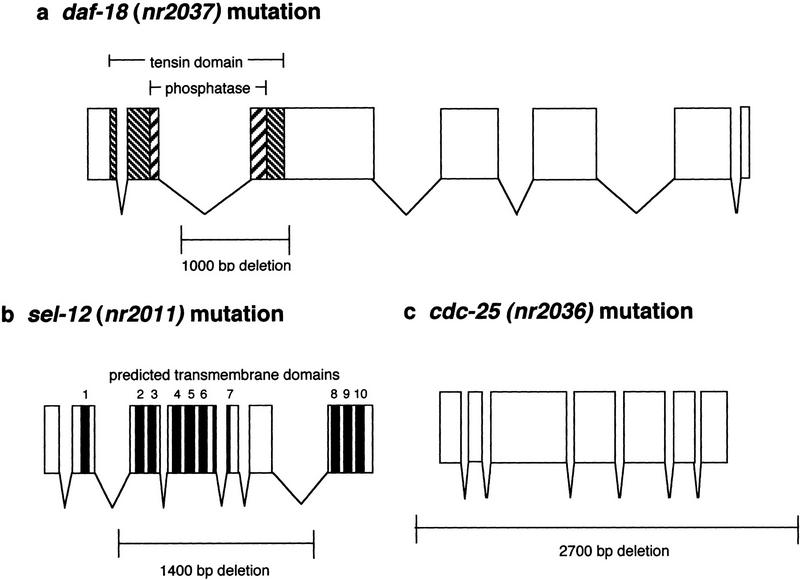
C. elegans genes are compact, with most introns ∼50 bp and exons 80–250 bp in length (Blumenthal and Steward 1997). Thus, deletions of >500 bp within a targeted C. elegans ORF are likely to significantly compromise gene function. Within an ORF, the position of such disabling deletions can be as follows: (1) the deletion begins or ends in an exon, removing portions of one or more exons, and usually leads to a premature stop codon (Fig. 3a); (2) the deletion begins and ends in introns, but removes one or more exons encoding domains essential for protein function (Fig. 3b); or (3) the deletion removes the entire ORF (Fig. 3c). Less than 2% of the deletion mutations that we isolated removed only noncoding DNA (data not shown), a figure that was optimized by selecting target regions that excluded long stretches of noncoding DNA. Given the characteristics of these molecular lesions, it is likely that most of these intragenic deletions represent severe or null mutations. The resulting phenotypes range from apparently wild-type to dramatic and lethal defects. Of note, less than one-half of the isolated deletion mutants (Table 1) displayed an obvious phenotype (such as lethal, uncoordinated, or sterile), but further analysis is necessary and ongoing in many cases.
Figure 3.
Schematics of representative deletions. (a) The deletion of daf-18 PTEN removes a portion of exon 3, which encodes the distal portion of the tensin homology domain and the catalytic phosphatase domain known to be required for lipid phosphatase activity (Gil et al. 1999). (b) The sel-12 deletion removes exons 3–6, which encode predicted transmembrane domains necessary for proper protein topology (Westlund 1998). (c) The cdc-25 deletion removes the entire ORF.
Although generally 4–8 libraries were screened for each target, the total number of mutant libraries generated during the period of this study was greater. Additional libraries were produced as template DNA became depleted in the course of multiple target screens, and all of these libraries were included for analysis. In addition, the cumulative number of libraries prepared with each chemical mutagen was different for each agent. To estimate the likelihood of isolating a deletion with different mutagens, we compared the number of deletions relative to the total number of primary library screens for each mutagen (Table 2). By this analysis, UV-TMP appeared to generate a higher frequency of deletions compared with the other three chemical mutagens.
Table 2.
Deletion Yield by Mutagen
| Mutagen | No. of libraries | No. of 1° screens | No. of deletion mutants | Ratio of deletions/screens |
|---|---|---|---|---|
| DEO | 3 | 171 | 12 | 0.070 |
| EMS | 9 | 479 | 47 | 0.098 |
| ENU | 4 | 208 | 23 | 0.111 |
| UV-TMP | 4 | 135 | 29 | 0.215 |
The total number of pool screens initiated (using a target window of 2800–3400 bp) and the total number of isolated deletions were tabulated for each mutant library.
DISCUSSION
In this paper, we describe our experience with a rapid procedure for isolating deletion mutations in C. elegans that uses PCR to screen for deletions in a target gene within a randomly mutagenized population of animals. The growth of mutagenized animals, preparation of genomic DNA templates, PCR screens, and mutant worm recovery are all handled in scalable microtiter liquid plate well formats that maximize speed and utility. All of the mutagens we used resulted in significant deletion mutations in target genes almost certainly sufficient to produce loss of function.
In practice, this protocol yields a deletion in a selected target for roughly every 600,000 mutagenized genomes screened. However, many factors affect the apparent sensitivity of this procedure. Small deletions of <500 bp were not isolated in screening plate pools for two likely reasons. First, such amplicons would be difficult to distinguish from an abundant wild-type band on a gel, unless special and more time-consuming electrophoresis conditions were applied. Second, mutant templates with small deletions would be similar in size to the much more abundant (4000-fold in the primary screening pools) wild-type templates and less likely to be preferentially amplified. The fact that we discovered several smaller (<500 bp) deletions only on screening of plate arrays (100-fold less complex than the plate pools) suggests that smaller deletions may be more abundant than larger deletions. Reducing the complexity of the DNA pools may improve the sensitivity of detecting these smaller deletions.
Larger deletions of >3 kb would also be useful because of their increased likelihood of severely affecting coding regions; furthermore, in certain cases the simultaneous disruption of two adjacent genes (e.g., evolutionarily duplicated genes) by one large deletion is desirable. The upper range of detectable deletions is limited by the size of the PCR deletion screening window, which, in this series, was 2800–3400 bp, and the processivity of the thermostable polymerase and other components of the amplification reaction. Modification of PCR reagents and conditions would allow a larger screening window, and we have recently isolated larger deletions using a target window of up to 5 kb (data not shown). Increasing the sensitivity of deletion detection would also increase the likelihood of obtaining more than one allele for each target in a single screen, which would be desirable for comparative genetic and phenotypic analysis.
This PCR-based approach is highly specific because of the use of nested pairs of screening primers derived from the target gene.We sequenced a number of false-positive deletion amplicons from the primary round of plate pool screening; all were derived from the target gene rather than an extraneous locus (data not shown). Although the genesis of these truncated amplicons is incompletely understood, we observed a comparable number of false-positive amplicons using unmutagenized genomic DNA as a control template (data not shown), and suspect that they may arise from polymerase slippage across gaps formed by secondary loops in the DNA template.
A wide variety of chemicals and irradiation methods have been used to induce mutations in C. elegans (Anderson 1995), and we used four in making our libraries for reverse genetics: EMS, ENU, DEO, and UV-TMP. EMS is the most widely used mutagen for C. elegans because of its potency in generating loss-of-function or reduction-of-function alleles in classical phenotypic screens. Many EMS-induced mutations are point mutations (of which the vast majority are G/C–A/T transitions) (Anderson 1995), which, of course, would not be detected in a screen for deletions. However, ∼13% of EMS-induced mutations are reported to be deletions or other rearrangements, though most are small (Anderson 1995). ENU appears to generate a rate of mutations similar to EMS; studies of ENU-induced mutations in the unc-93 gene revealed an increased frequency of A/T–G/C transitions compared with EMS, with a frequency of intragenic deletions of 13% (De Stasio et al. 1997). DEO has not been as broadly used as a C. elegans mutagen, but a study of mutations of the unc-54 gene revealed that DEO generated mutations at a lower rate than EMS, and 27% of DEO-induced mutations were multilocus deletions and deficiencies, as defined by genetic criteria (Anderson and Brenner 1984). Treatment with trimethylpsoralen followed by ultraviolet activation, a procedure that leads to DNA cross-linking, appears to generate a lower frequency of mutations than EMS, as measured by a generation of unc-22 and pal-1 mutations, but a relatively high proportion of deletions (Yandell et al. 1994).
For the purposes of a PCR-based reverse genetics screening approach, it is important to identify a mutagen that generates the highest frequency of small deletions (on the order of 100–5000 bp) rather than point mutations that cannot be detected. Furthermore, highly potent inducers of large numbers of point mutations would exacerbate the confounding background of extraneous mutations. In our experience, libraries prepared with each of the four mutagens yielded deletions at similar rates (Table 2). In addition, the average size of detected deletions induced by the different mutagens was not significantly different (Fig. 2). The frequency of deletions obtained from each of the four mutagens far exceeded the spontaneous rate of deletions of this size range, which is predicted to be <1 per 100 million genomes (P. Anderson, pers. comm.). About one-fifth of our chemically induced mutations contained short (≤7 bp) direct sequence repeats or longer (>2 bp) insertions at the deletion termini, in excess of what would be expected from spontaneous deletions (Pulak and Anderson 1988; data not shown). Such insertions may reflect mechanisms of break repair with nonhomologous endjoining, in response to chemically induced breaks in double-stranded DNA (Friedberg et al. 1995). To measure the efficiency of each mutagen in creating detectable deletions, we retrospectively determined the relative yield of deletions from each type of mutagen library (Table 2). By this indirect measure, UV-activated trimethylpsoralen appears to generate the highest frequency of detectable deletions, which confirms prior observations (Yandell et al. 1994). However, fewer animals, on average, were recovered from frozen UV-TMP-treated worm populations compared with EMS or ENU, suggesting that UV-TMP treatment may cause more generally deleterious effects. In our experience, γ irradiation or formaldehyde treatment, which reportedly causes more major rearrangements (Anderson 1995), did not produce significant numbers of deletion mutations in limited experiments (data not shown). Because deletions, rather than mutations per se (the majority of which are point mutations), are the goal of this reverse genetics screening approach, the identification of a specific deletogen chemical would be very helpful.
A disadvantage of random mutagenesis procedures is that substantial numbers of extraneous mutations are generated in the background of any selected target gene, and it can be difficult to titrate the mutagen dose so as to induce large numbers of mutations in the population while also minimizing the number of mutations in any one animal. A strong mutant phenotype may result from the deletion of interest or from mutation of a non-target gene. Similarly, a pleiotropic phenotype may be due to either multiple effects of a single mutation or to mutations in more than one gene. Thus, thorough outcrossing followed by transgenic rescue of the observed phenotype with a wild-type copy of the target gene (usually in the form of a cosmid or plasmid clone), is necessary to establish whether a putative loss-of-function phenotype is actually due to the deletion. In addition, reverse genetics may lead to no obvious phenotype at all. Earlier estimates suggested that up to one-half of C. elegans genes may cause no obvious phenotype when null (Park and Horvitz 1986). When no phenotype is seen on isolation of the mutant animal, it can be difficult to tell whether this lack of effect is because (1) the randomly generated molecular lesion does not produce loss or reduction of gene function, (2) the gene is functionally redundant, or (3) the phenotype requires specific environmental conditions for its expression or is otherwise too subtle to be distinguished by conventional methods. The complementary use of RNA-mediated interference (Fire et al. 1998) might be helpful in cases in which deletion mutants display a confusing pleiotropy or an apparent wild-type phenotype.
At the current state of development, this method requires ∼2 weeks for identification of a deletion and isolation of mutant worms. The rate of deletion identification is about one target per week, per person, without automation. This rapid approach to gene inactivation allows one to simultaneously approach a group of candidate genes, whether they belong to a single gene family or may be functionally related in a putative signaling pathway. Thus, deletion mutants of different, but potentially related, genes may be studied as a group for comparative phenotype analysis, genetic interactions, etc., rather than individually. For example, genetic and phenotypic analysis have revealed overlapping functions between the C. elegans presenilins sel-12 and hop-1 (Li and Greenwald 1997; Westlund et al. 1999) and in the human Niemann–Pick type-C disease gene homologs npc-1 and npc-2 (M. Sym, pers. comm.).
An obvious advantage of this target-selected approach is that specific regions of functional importance can be targeted to maximize the likelihood of obtaining null or severe loss-of-function mutations. On the other hand, this method is less well suited than traditional noncomplementation genetic screens for obtaining weaker (hypomorphic) alleles, which are often useful in genetic pathway analysis.
A procedure such as this one that uses random mutagenesis followed by deletion screening is ideally suited for organisms like C. elegans, in which pre-existing genomic sequence information is extensive but homologous recombination or transposon insertion methods are not practical. By improving the sensitivity of detection in more complex mutagenized genome pools, it may be possible to produce a clonal library of individually arrayed mutants, thereby eliminating the need to sort through thawed worms for the deletion. Furthermore, by increasing the density of C. elegans growth and DNA microarrays, and automating the PCR set up and analysis, it should be feasible to pursue the systematic disruption of all of the predicted genes in this model animal, as is currently being done, by different methods, for all of the genes in S. cerevisiae (Dujon 1998).
METHODS
Bioinformatics
C. elegans homologs of non-nematode genes and proteins were identified by BLAST search (Altschul et al. 1990) of finished C. elegans sequences available at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and unannotated sequences available at the Sanger Centre (www.sanger.ac.uk/Projects/C_elegans/blast_server.html) and the Washington University Genome Sequencing Center (genome.wustl.edu/gsc/blast/blast_servers.html). Putative ORFs of unannotated sequences were determined with the GeneFinder algorithm available from the Baylor College of Medicine Human Genome Center (dot.imgen.bcm.tmc.edu:9331/gene-finder/gf.html). Nested PCR primer pairs were designed to flank a target window of 2.8–3.2 kb with MacVector 6.0. Whenever possible, the target window was chosen to include the most proximal exons, except in those cases in which the first intron was >1 kb in size.
Construction of Mutagenized Nematode and Corresponding DNA Libraries
Wild-type Bristol N2 nematodes were cultured by standard methods (Sulston and Hodgkin 1988). A synchronized population of L4 larvae was treated with one of the following four mutagens: 25 mm ethylmethane sulfonate for 4 hr, 0.4 mm ethylnitrosourea for 4 hr, 1 mm diepoxyoctane for 4 hr, or 30 μg/ml trimethylpsoralen (Yandell et al. 1994), followed by UV irradiation at 3.5 uW/cm2 for 15–30 sec (Anderson 1995). The efficiency of mutation was evaluated by quantitating the frequency of F2 resistance to ivermectin (Rand and Johnson 1995); libraries were discarded if the proportion of genomes carrying newly induced ivermectin resistance mutations was <0.5%. F1 eggs of mutagenized worms were collected for 4 hr, then allowed to hatch for 16 hr. Hatched larvae were distributed to 96-well microtiter plates at ∼20 worms per well in 50 μl of NGM culture medium containing 1% (by volume) Escherichia coli HB101 as food (Sulston and Hodgkin 1988). A library from each chemical mutagenesis comprised 48 microtiter plates and included the arrayed progeny of nearly 105 F1 animals representing 2 × 105 mutagenized genomes. Worms were grown in wells until the food was exhausted, generating, on the average, 100 F2 progeny per F1 animal. One-half of the worms by volume were removed from each well to make genomic DNA; of these, one-half were transferred to equivalent wells of V-bottom 96-well microtiter plates and one-half were pooled with other samples from the same 96-well plate (plate pools) into microfuge tubes. Worms in V-bottom wells were digested for up to 16 hr with 10 μl/well of worm lysis buffer containing 50 mm KCl, 10 mm Tris-Cl (pH 8.3), 2.5 mm MgCl2, 0.45% Tween 20, 0.45% NP-40, 0.01% (wt/vol) gelatin and 200 μg/ml proteinase K. These worm DNA plates were stored without further purification at −80°C. Plate pools were digested for 4 hr with 0.4 ml of lysis buffer containing 100 mm NaCl, 100 mm Tris-Cl (pH 8), 50 mm EDTA, 1% SDS, 1% β-mercaptoethanol and 100 μg/ml proteinase K (Birren et al. 1997). Plate pool genomic DNA was then isolated with the Purgene kit (Gentra Systems) according to the manufacturer’s protocol, followed by ethanol precipitation and transferral to sealed microtiter plates for storage at 4°C. The remaining half of the worms in the original plates were frozen in the wells by adding an equal volume of freezing solution, mixing, and graduated freezing to −80°C (Sulston and Hodgkin 1988). Survival following freezing was measured by counting motile animals thawed from 10 selected wells from each of 2 plates, and libraries were discarded if <100 viable animals per well on average were recovered.
PCR Screening for Deletions
Genomic DNA plate pools were screened by nested PCR with AmpliTaq Gold (Perkin-Elmer) and primer concentrations of 50 μm for 35 cycles of 94°C for 30 sec, 58°C for 1 min, and 72°C for 1.5–2.5 min. Aliquots (1 μl) of template DNA from two libraries (48 plate pools each) were added to a 96-well PCR plate, and up to 8 libraries were screened simultaneously for a given gene target in a Tetrad thermocycler (MJ Research, Watertown, MA). Potential deletions detected by gel electrophoresis of PCR products were confirmed by quadruplicate PCR of the original library plate pool. On identification of a positive plate, PCR was performed on the unpooled DNA plates to identify the positive well. Primary screen data and deletion sizes were available for 127 screened targets.
Isolation of Mutant Animals and Deletion Analysis
Deletions were mapped by restriction enzymes on the basis of the known genomic DNA cosmid sequence, and the mapping information was used to design three new PCR primers, two of which flanked the deletion and a third from within the deletion. Worms were thawed from the positive wells onto NGM plates and then picked individually into microtiter plate liquid cultures. Following growth and reproduction in liquid medium, aliquots of progeny were removed for PCR with flanking primers to identify clones bearing the deletion mutation. Positive lines were transferred to agar plates, and single worms were tested by multiplex PCR with both the flanking and internal primers to distinguish homozygous from heterozygous animals in subsequent generations. Mutant animals were subsequently outcrossed with wild-type N2 animals to remove other potential mutations.
Acknowledgments
We thank Bob Barstead and Ron Plasterk for sharing their technical results, Elizabeth Link and Marie Sutherlin for reviewing the manuscript, the C. elegans Genome Sequencing Consortium for primary sequence data and on-line search engines, and our many collaborators in the C. elegans community who provided biological and genetic expertise for the selection of targets.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lliu@cambriabio.com; FAX (781) 736-3107.
REFERENCES
- Ahringer J. Turn to the worm. Curr Opin Genet Dev. 1997;7:410–415. doi: 10.1016/s0959-437x(97)80157-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson P. Mutagenesis. in Caenorhabditis elegans: In: Epstein HF, Shakes DC, editors. Modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. pp. 31–58. [Google Scholar]
- Anderson P, Brenner S. A selection for myosin heavy chain mutants in the nematode Caenorhabditis elegans. Proc Natl Acad Sci. 1984;81:4470–4474. doi: 10.1073/pnas.81.14.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birren B, Green ED, Klapholz S, Myers RM, Roskams J.Genome analysis: A laboratory manual. 1997. Volume 1: Analyzing DNA1Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 20–22. [Google Scholar]
- Blaxter ML. Caenorhabditis elegans is a nematode. Science. 1998;282:2041–2046. doi: 10.1126/science.282.5396.2041. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Steward K. RNA processing and gene structure. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- De Stasio E, Lephoto C, Azuma L, Holst C, Stanislaus D, Uttam J. Characterization of revertants of unc-93 (e1500) in Caenorhabditis elegans induced by N-ethyl-N-nitrosourea. Genetics. 1997;147:597–608. doi: 10.1093/genetics/147.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. European Functional Analysis Network (EUROFAN) and the functional analysis of the Saccharomyces cerevisiae genome. Electrophoresis. 1998;19:617–624. doi: 10.1002/elps.1150190427. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- Gil E, Link EM, Liu LX, Johnson CD, Lees JA. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc Natl Acad Sci. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire FM, Chomiki N, Kachinskas D, Warden CH. Cloning and developmental regulation of a novel member of the insulin-like gene family in Caenorhabditis elegans. Biochem Biophys Res Comm. 1998;249:385–390. doi: 10.1006/bbrc.1998.9164. [DOI] [PubMed] [Google Scholar]
- Hart AC, Kass J, Shapiro JE, Kaplan J. Distinct signalling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci. 1999;19:1952–1958. doi: 10.1523/JNEUROSCI.19-06-01952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P, Boguski M. Functional genomics: It’s all how you read it. Science. 1997;278:601–602. doi: 10.1126/science.278.5338.601. [DOI] [PubMed] [Google Scholar]
- Hobert O, Tessmar K, Ruvkun G. The C. elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development. 1999;126:1547–1562. doi: 10.1242/dev.126.7.1547. [DOI] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RHA. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RHA. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Li X, Greenwald I. HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc Natl Acad Sci. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos GLG, Rubin GM. The role of the genome project in determining gene function: Insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Bassett DE, Jr, Boguski MS, Bork P, Koonin EV. Positionally cloned human disease genes: Patterns of evolutionary conservation and functional motifs. Proc Natl Acad Sci. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Garey JG, Martin J, Liu LX. Large-scale taxonomic profiling of eukaryotic model organisms: A comparison of homologous and orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 1998;8:590–598. doi: 10.1101/gr.8.6.590. [DOI] [PubMed] [Google Scholar]
- Oliver SG. From DNA sequence to biological function. Nature. 1996;379:597–600. doi: 10.1038/379597a0. [DOI] [PubMed] [Google Scholar]
- Park EC, Horvitz HR. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 1986;113:821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RHA. Reverse genetics: From gene sequence to mutant worm. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. pp. 59–80. [DOI] [PubMed] [Google Scholar]
- Pulak RA, Anderson P. Structure of spontaneous deletions in Caenorhabditis elegans. Mol Cell Biol. 1988;8:3748–3754. doi: 10.1128/mcb.8.9.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB, Johnson CD. Genetic pharmacology: Interactions between drugs and gene products in Caenorhabditis elegans. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. pp. 187–204. [DOI] [PubMed] [Google Scholar]
- Shaham S. Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J Biol Chem. 1998;273:35109–35117. doi: 10.1074/jbc.273.52.35109. [DOI] [PubMed] [Google Scholar]
- Sluder AE, Mathews SW, Hough D, Yin VP, Maina CV. The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res. 1999;9:103–120. [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. Analysis of protein domain families in Caenorhabditis elegans. Genomics. 1997;46:200–216. doi: 10.1006/geno.1997.4989. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Westlund B, Parry D, Clover R, Basson M, Johnson CD. Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signalling. Proc Natl Acad Sci. 1999;96:2497–2502. doi: 10.1073/pnas.96.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell MD, Edgar LG, Wood WB. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc Natl Acad Sci. 1994;91:1381–1385. doi: 10.1073/pnas.91.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal RR, Broeks A, van. Meurs J, Groenen JTM, Plasterk RHA. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc Natl Acad Sci. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]