Abstract
The vertebrate vomeronasal system (VNS) detects intraspecific pheromones and environmental odorants. We sequenced segments of the gene encoding Trpc2, an ion channel crucial for vomeronasal signal transduction, in 11 species that represent all main basal lineages of Yinpterochiroptera, one of the two suborders of the order Chiroptera (bats). Our sequences show that Trpc2 is a pseudogene in each of the 11 bats, suggesting that all yinpterochiropterans lack vomeronasal sensitivity. The Trpc2 sequences from four species of Yangochiroptera, the other suborder of bats, suggest vomeronasal insensitivity in some but not all yangochiropterans. These results, together with the available morphological data from the bat VNS, strongly suggest multiple and widespread losses of vomeronasal signal transduction and sensitivity in bats. Future scrutiny of the specific functions of the VNS in the few bats that still retain the VNS may help explain why it is dispensable in most bats.
Keywords: bats, olfaction, pseudogenization, Trpc2, vomeronasal system
Olfaction plays multiple important roles in the life of a vertebrate organism. Tetrapod vertebrates use two olfactory systems, the main olfactory system (MOS) and the vomeronasal system (VNS), to detect environmental odorants and intraspecific pheromones (Dulac and Torello 2003; Grus and Zhang 2006). Although the MOS and VNS are both capable of detecting the two types of olfactory cues and are partially overlapping in function, they are anatomically and neurologically distinct, use different signal transduction pathways, and exhibit contrasting evolutionary patterns (Dulac and Torello 2003; Grus and Zhang 2008). Although all vertebrates except some cetaceans (Kishida et al. 2007) are known to possess the MOS, a number of lineages including birds and catarrhine primates (i.e., humans, apes, and Old World monkeys) have lost the morphological and genetic components of the VNS (Liman and Innan 2003; Zhang and Webb 2003; Shi and Zhang 2007). In the case of catarrhine primates, the origins of male trichromatic color vision and female sexual swelling may have rendered the VNS partially redundant and permitted its evolutionary deterioration (Liman and Innan 2003; Zhang and Webb 2003). This visual replacement hypothesis may also explain the loss of the bird VNS because birds have tetrachromatic vision and develop colorful plumages at sexual maturity (Zhang and Webb 2003). To test the above hypothesis in other vertebrates, we turn to bats, which are extremely variable in the morphology of their VNS, ranging from completely absent to fully developed (Wible and Bhatnagar 1996).
We use Trpc2 as a genetic marker to probe vomeronasal functionality because Trpc2 (transient receptor potential cation channel, subfamily C, member 2) is absolutely required for vomeronasal signal transduction but has no other function (Grus and Zhang 2006). For example, mice deficient for Trpc2 lack vomeronasal sensitivity and consequently show altered sexual and social behaviors (Leypold et al. 2002; Stowers et al. 2002). Catarrhine primates and birds, which lack a functional VNS, also lack functional Trpc2 (Liman and Innan 2003; Zhang and Webb 2003; Shi and Zhang 2007). The VNS-specific splicing variant of Trpc2 is encoded by 13 exons (Hofmann et al. 2000). We first sequenced the longest exon (exon 2) of Trpc2 from 13 bat species and subsequently sequenced several exons of Trpc2 from two additional bats with available draft genome sequences (supplementary table S1, Supplementary Material online). Based on molecular systematics, the order Chiroptera (bats) is now classified into two suborders: Yinpterochiroptera and Yangochiroptera (Teeling et al. 2005). Our sample includes 11 species representing all main basal lineages of Yinpterochiroptera and four species representing two of the three superfamilies of Yangochiroptera.
The exon 2 sequences we obtained from the 13 bats (10 yinpterochiropterans and 3 yangochiropterans) range in length from 436 to 491 nucleotides, accounting for about one sixth of the full coding sequence of Trpc2, which has 2673 nucleotides in mouse. We aligned the bat sequences with mouse Trpc2 (Hofmann et al. 2000) (fig. 1) and identified multiple insertions/deletions (indels) and premature stop codons in each of the 10 yinpterochiropterans examined, suggesting that Trpc2 is a pseudogene in all these species (fig. 2). Among the open reading frame (ORF)-disrupting mutations, five indels are shared by all examined species of Old World fruit bats (fig. 1), suggesting that these indels occurred in the common ancestor of Old World fruit bats. We also found 19 premature stop codons at different positions. Notably, no single premature stop codon or indel is shared by all 10 yinpterochiropterans (fig. 1). This lack of common disruptive substitutions suggests that the pseudogenization of Trpc2 may have occurred independently in Old World fruit bats and Rhinolophoidea, the two subgroups of Yinpterochiroptera (fig. 2). However, one cannot rule out the possibility that Trpc2 was completely functionally relaxed in the common ancestor of all yinpterochiropterans but no common ORF-disrupting substitutions are observed because of 1) a short evolutionary time between the start of the functional relaxation and the separation between Old World fruit bats and Rhinolophoidea, 2) the short sequence segment examined here, and/or 3) many subsequent indel and point substitutions that have blurred the signal of the initial ORF-disrupting mutation. We also examined the draft genome sequence of the Malayan flying fox Pteropus vampyrus, an Old World fruit bat. We were able to identify either complete or partial sequences belonging to 10 exons (no. 2–5 and 8–13) of a single Trpc2 gene and discovered several ORF disruptions. Because the draft genome sequence has a low coverage, we amplified and sequenced exons 3–5, 9–10, and 12–13 and part of exon 2 from another P. vampyrus individual and confirmed that the P. vampyrus Trpc2 is a pseudogene (supplementary fig. S1, Supplementary Material online). Specifically, within the sequenced region, the first premature stop codon occurs in exon 5 (supplementary fig. S1, Supplementary Material online), leading to the loss of all 6 transmembrane domains and the C-terminus of Trpc2. Hence, the truncated Trpc2 must be functionless. We cannot evaluate whether the five indels shared by all other Old World fruit bats examined are also present in P. vampyrus because the segment of exon 2 that harbors these indels is neither covered by its draft genome sequence nor by our sequence.
FIG. 1.

An alignment of the newly obtained bat Trpc2 exon 2 sequences with the mouse Trpc2 sequence. Dots indicate identical nucleotides to the first sequence, whereas dashes indicate alignment gaps. Codons in the correct reading frame are indicated by shading and premature stop codons are boxed. The numbers in parentheses indicate the order of premature stop codons, whereas the numbers after the alignment represent nucleotide positions. Full species names are presented in figure 2.
FIG. 2.
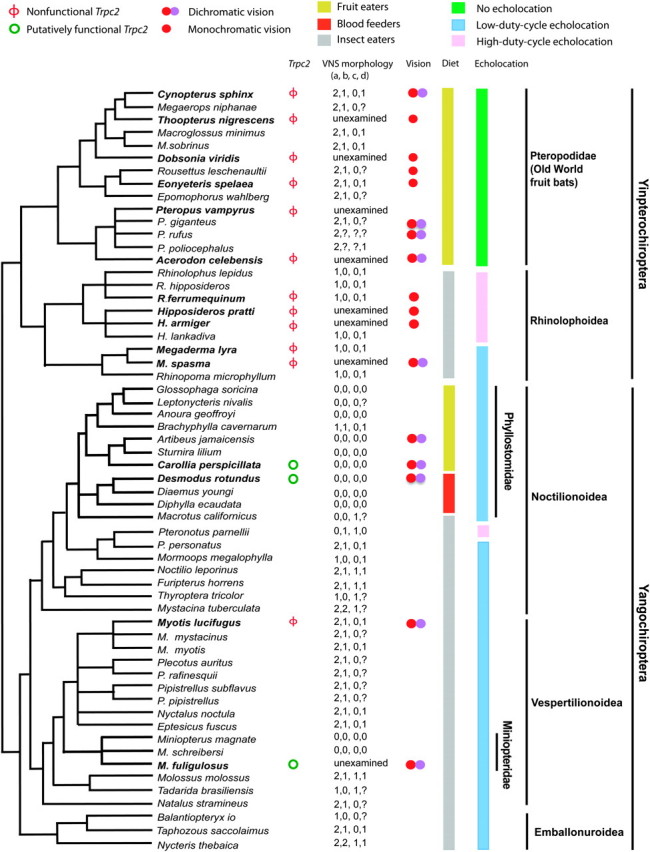
An established phylogeny of all bats with genetic or morphological data about theVNS. Information about Trpc2 functionality, VNS morphology, color vision, echolocation call type, and dietary preference is presented whenever available. Species in bold are those sequenced in this study. The VNS morphology is represented by four characters: “a” represents vomeronasal epithelial tube (0, well developed; 1, rudimentary; 2, absent), “b” indicates vomeronasal cartilage (0, “J”, “C”, “U” or “O” shaped; 1, bar shaped; 2, absent), “c” signifies nasopalatine duct (0, present; 1, absent), and “d” denotes accessory olfactory bulb (0, present; 1, absent). “?” represents missing data. Both the data and coding of the VNS morphology are from Wible and Bhatnagar (1996). Information about bat color vision is from Zhao, Rossiter, et al. (2009) and that about echolocation and diet is from Neuweiler (2000).
In contrast to the yinpterochiropterans, each of the three yangochiropterans (Miniopterus fuliginosus, Carollia perspicillata, and Desmodus rotundus) whose exon 2 was sequenced has an intact ORF in the sequenced region (figs. 1,2). The probability for a sequence of 150 codons that has been functionless for a long time to contain no stop codon is approximately (61/64)150 = 7.4 × 10−4, suggesting that Trpc2 has been under functional constraint in these yangochiropterans. Consistent with this finding, the dN/dS ratio for the sequenced region is significantly lower than 1 in all pairwise comparisons of the three species (mean = 0.65, P < 0.01, Z-test). These results suggest that Trpc2 is functional and is under purifying selection in the three yangochiropterans studied. Nonetheless, a definitive proof of Trpc2’s functionality in these species requires sequencing its complete coding region and verifying its expression and function. We also examined the draft genome sequence of the little brown bat Myotis lucifugus, a yangochiropteran. We identified five exons (no. 9–13) of a single Trpc2 gene and found that its ORF is disrupted. We subsequently amplified and sequenced exons 9–13 from another M. lucifugus individual and confirmed that the ORF is disrupted. Within the sequenced region, the first premature stop codon occurs in exon 9 and causes the loss of the final transmembrane domain and the C-terminus of Trpc2. The truncated Trpc2 is most likely nonfunctional.
Our Trpc2 sequence data are fully consistent with the existing morphological data of the VNS. Specifically, morphological examinations of various components of the bat VNS showed that these components are either absent or poorly developed in all 16 yinpterochiropterans studied (Wible and Bhatnagar 1996), including 4 species that are sequenced here (fig. 2). The same study examined 11 of the 12 families of Yangochiroptera and found a morphologically intact VNS in only two (Phyllostomidae and Miniopteridae) of these (fig. 2). Furthermore, within Phyllostomidae, not all species possess a morphologically intact VNS (Wible and Bhatnagar 1996). Morphological data are available for three of the four yangochiropterans where the Trpc2 sequences are available. In all three cases, the two types of data are consistent with each other. In M. fuliginosus, our Trpc2 sequence suggests an intact vomeronasal transduction pathway. Although morphological data are lacking in this species, a morphologically intact VNS is present in other species of the same genus (M. magnate and M. schreibersi) (Wible and Bhatnagar 1996). Given the complete consistency between the previous morphological data and the present genetic data of the VNS, one can confidently infer that vomeronasal transduction is lost in those yangochiropterans that are not included in our study but are known to lack a morphologically intact VNS. These species are distributed in at least 10 of the 12 families of Yangochiroptera (Wible and Bhatnagar 1996). Together, the morphological and genetic data unambiguously demonstrate widespread losses of the VNS in bats.
Why is the VNS dispensable in most bats? First, we observed no correlation between the VNS functionality and color vision in bats. Most bats are dichromatic, whereas bats with high-duty-cycle echolocation and several Old World fruit bats roosting in caves are monochromatic (Zhao, Rossiter, et al. 2009; Zhao, Xu, et al. 2009). However, all yinpterochiropterans have lost the VNS, regardless of dichromaticy or monochromaticy (fig. 2). Of the four yangochiropterans examined, three species with a functional VNS and one without are all dichromatic (fig. 2). Thus, the visual replacement hypothesis cannot explain the patterns of VNS evolution in bats. Second, in a modification of the visual replacement hypothesis, Webb et al. (2004) explained the coexistence of male trichromacy and vomeronasal sensitivity in howler monkeys by the factor that visual signals may not be preferred over olfactory signals in dense tropical rainforests where howler monkeys live. Although this modified visual replacement hypothesis explains why a group of trichromatic primates can still retain the VNS, it does not explain how monochromatic species could afford to lose the VNS, which is observed in bats (fig. 2). Third, in addition to color vision, other sensory systems such as echolocation may also have trade-offs with vomeronasal sensitivity. However, we found no correlation between vomeronsal sensitivity and echolocation abilities or types in bats (fig. 2). For example, the VNS is absent in all yinpterochiropterans, including species that echolocate and those that do not. All yangochropterans echolocate but some retain the VNS, whereas others do not. Fourth, pheromone-mediated behaviors have been reported in bats that lack the VNS (Bloss et al. 2002; Safi and Kerth 2003), suggesting that pheromones are detected by the MOS in these species. Is the absence of the VNS compensated by an exceptionally advanced MOS? The odorant receptors (ORs) of the MOS are encoded by the largest gene family in the vertebrate genome, and the fraction of functional genes in the OR gene family has been used as an indicator of the functionality and sophistication of the MOS (Nei et al. 2008). A recent study measured this fraction in 11 bat species, including two that retain the VNS (Hayden et al. 2010). But, we found no correlation between this fraction and the presence/absence of the VNS in bats. Fifth, it has been proposed that the absence of both the MOS and VNS in some cetaceans (Kishida et al. 2007; Yu et al. 2010) is compensated by their enhanced taste sensitivity (Kishida et al. 2007). In bats, it has also been suggested that the VNS is related to the examination of food in the mouth (Neuweiler 2000), but we fail to find correlation between the diet of a bat and the presence/absence of its VNS (fig. 2). For example, some fruit-eating bats retain the VNS, whereas others have lost it. It is interesting to note, however, that vampire bats, which have lost their sweet taste receptor gene Tas1r2, have all retained their VNS (Zhao et al. 2010). But, whether this relationship is causal or coincidental is unclear. Hence, no simple sensory trade-offs explain the widespread losses of the vomeronsal sensitivity in bats. Future in-depth studies of physiological functions of the VNS in the few lineages of bats that still maintain this system may provide a better understanding of why it is dispensable in most bats.
Methods
A pair of primers (5′-AYG TGG AGG AGG CCG AGG A-3′ and 5′-CTC BCG GCT GAG CTG GAA G-3′) were designed based on the alignment of horse, cow, dog, mouse, and rat Trpc2 sequences. Genomic DNAs were extracted from bat wing biopsies using DNeasy blood & tissue kits (Qiagen). Polymerase chain reactions (PCRs) were performed with Taq DNA polymerase (Takara). The PCR mixtures (10 μl) contained 1 μl (50 ng/μl) genomic DNA, 5 μl 2× buffer, 1.5 μl (50 mM) MgCl2, 1 μl (10 μM) of each primer, and 1 U Taq DNA polymerase (Takara). PCRs were conducted under the following condition: 5 min of initial denaturation, 30 cycles of denaturation at 94 °C for 30 s, annealing at 53–58 °C for 30 s, and extension at 72 °C for 60 s, and a final extension at 72 °C for 5 min. PCR products were purified and cloned into the pMD19-T vector (Takara). Multiple clones (>3) from a single PCR product were sequenced by the Sanger method in both directions. DNA sequences were aligned with CLUSTALX 1.81 (Thompson et al. 1997). Nonsynonymous (dN) and synonymous (dS) distances and their standard errors were estimated using the modified Nei-Gojobori method (Zhang et al. 1998) implemented in MEGA4 (Tamura et al. 2007). Mouse Trpc2 was used as the query to TblastN the draft genome sequences of P. vampyrus (2.6× coverage) and M. lucifugus (1.7×) available at www.ensemble.org.
Supplementary Material
Supplementary figs. S1–S2 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Acknowledgments
We thank Meg Bakewell for valuable comments. This work was supported by a grant under the Key Construction Program of the National “985” Project and “211” Project of China to S.Z. and a research grant by US National Institutes of Health to J.Z. H.Z. was supported in part by the Fundamental Research Funds for the Central Universities.
References
- Bloss J, Acree TE, Bloss JM, Hood WR, Kunz TH. Potential use of chemical cues for colony-mate recognition in the big brown bat, Eptesicus fuscus. J Chem Ecol. 2002;28:819–834. doi: 10.1023/a:1015296928423. [DOI] [PubMed] [Google Scholar]
- Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Origin and evolution of the vertebrate vomeronasal system viewed through system-specific genes. BioEssays. 2006;28:709–718. doi: 10.1002/bies.20432. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Mol Biol Evol. 2008;25:1593–1601. doi: 10.1093/molbev/msn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20:1–9. doi: 10.1101/gr.099416.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann T, Schaefer M, Schultz G, Gudermann T. Cloning, expression and subcellular localization of two novel splice variants of mouse transient receptor potential channel 2. Biochem J. 2000;351:115–122. doi: 10.1042/0264-6021:3510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T, Kubota S, Shirayama Y, Fukami H. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol Lett. 2007;3:428–430. doi: 10.1098/rsbl.2007.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leypold BG, Yu CR, Leinders-Zufall T, Kim MM, Zufall F, Axel R. Altered sexual and social behaviors in trp2 mutant mice. Proc Natl Acad Sci U S A. 2002;99:6376–6381. doi: 10.1073/pnas.082127599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Innan H. Relaxed selective pressure on an essential component of pheromone transduction in primate evolution. Proc Natl Acad Sci U S A. 2003;100:3328–3332. doi: 10.1073/pnas.0636123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Neuweiler G. The biology of bats. Oxford (UK): Oxford University Press; 2000. [Google Scholar]
- Safi K, Kerth G. Secretions of the interaural gland contain information about individuality and colony membership in the Bechstein's bat. Anim Behav. 2003;65:363–369. [Google Scholar]
- Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male–male aggression in mice deficient for TRP2. Science. 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DM, Cortes-Ortiz L, Zhang J. Genetic evidence for the coexistence of pheromone perception and full trichromatic vision in howler monkeys. Mol Biol Evol. 2004;21:697–704. doi: 10.1093/molbev/msh068. [DOI] [PubMed] [Google Scholar]
- Wible JR, Bhatnagar KP. Chiropteran vomeronasal complex and the interfamilial relationships of bats. J Mammal Evol. 1996;3:285–314. [Google Scholar]
- Yu L, Jin W, Wang JX, Zhang X, Chen MM, Zhu ZH, Lee H, Lee M, Zhang YP. Characterization of TRPC2, an essential genetic component of VNS chemoreception provides insights into the evolution of pheromonal olfaction in secondary-adapted marine mammals. Mol Biol Evol. 2010;27:1467–1477. doi: 10.1093/molbev/msq027. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci U S A. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Rossiter SJ, Teeling EC, Li C, Cotton JA, Zhang S. The evolution of color vision in nocturnal mammals. Proc Natl Acad Sci U S A. 2009;106:8980–8985. doi: 10.1073/pnas.0813201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Xu D, Zhou Y, Flanders J, Zhang S. Evolution of opsin genes reveals a functional role of vision in the echolocating little brown bat (Myotis lucifugus) Biochem Syst Ecol. 2009;37:154–161. [Google Scholar]
- Zhao H, Zhou Y, Pinto CM, Charles-Dominique P, Galindo-González J, Zhang S, Zhang J. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq152. Advance Access published June 17, 2010, doi:10.1093/molbev/msq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


