Abstract
Large-scale sequencing studies in vertebrates have thus far focused primarily on the genomes of a few model organisms. Birds are of interest to genomics because of their much smaller and highly streamlined genomes compared to mammals. However, large-scale genetic work has been confined almost exclusively to the chicken; we know little about general aspects of genomes in nongame birds. This study examines the organization of a genomic region containing an Mhc class II B gene in a representative of another important lineage of the avian tree, the songbirds (Passeriformes). We used a shotgun sequencing approach to determine the sequence of a 32-kb cosmid insert containing a strongly hybridizing Mhc fragment from house finches (Carpodacus mexicanus). There were a total of three genes found on the cosmid clone, about the gene density expected for the mammalian Mhc: a class II Mhc β-chain gene (Came–DAB1), a serine–threonine kinase, and a zinc finger motif. Frameshift mutations in both the second and third exons of Came–DAB1 and the unalignability of the gene after the third exon suggest that it is a nonfunctional pseudogene. In addition, the identifiable introns of Came–DAB1 are more than twice as large as those of chickens. Nucleotide diversity in the peptide-binding region of Came–DAB1 (Π = 0.03) was much lower than polymorphic chicken and other functional Mhc genes but higher than the expected diversity for a neutral locus in birds, perhaps because of hitchhiking on a selected Mhc locus close by. The serine–threonine kinase gene is likely functional, whereas the zinc finger motif is likely nonfunctional. A paucity of long simple-sequence repeats and retroelements is consistent with emerging rules of chicken genomics, and a pictorial analysis of the “genomic signature” of this sequence, the first of its kind for birds, bears strong similarity to mammalian signatures, suggesting common higher-order structures in these homeothermic genomes. The house finch sequence is among a very few of its kind from nonmodel vertebrates and provides insight into the evolution of the avian Mhc and of avian genomes generally.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF205032 and AF241546–AF241565.]
Long DNA sequences provide one source of the genomic information that will revolutionize biology, yet cosmid-scale (25–40 kb) or longer DNA sequences are still almost exclusively confined to model organisms and microbial pathogens. Whereas several nonmodel mammal species are the focus of large-scale mapping and genome projects (O'Brien et al. 1999), cosmid-scale sequences of nonmammalian organisms are available only from chickens, Japanese quail, zebrafish, and pufferfish. We expect the genomic features gleaned from such models to predict aspects of the genomes of related species in their respective clades. Nonetheless, the full diversity of genomic structures will not be appreciated until a much larger number of genomes and DNA sequences from nonmodel species are investigated. To this end we have been investigating cosmid-scale sequences of birds, with particular attention to the immunologically important major histocompatibility complex (Mhc) region (Edwards et al. 1999). Here we report on the first cosmid-scale sequence from a songbird, the house finch (Carpodacus mexicanus), a member of the large clade Passeriformes that includes over half of all avian species (Edwards 1998).
The Mhc is a multigene family found thus far only in jawed vertebrates. Mhc genes have yet to be found in jawless fish or any lineage more ancient (Kandil et al. 1996), although allorecognition genes potentially related to Mhc genes have been found in tunicates (Magor et al. 1999). The primary function of the Mhc is to present foreign peptides from pathogens to T cells during the adaptive immune response (Klein 1986). Mhc genes are the most polymorphic genes found in vertebrates, and much research has been directed toward understanding their evolutionary dynamics, with particular emphasis on possible relationships between Mhc diversity and parasite resistance (Klein et al. 1993; Parham and Ohta 1996; Edwards and Hedrick 1998). Molecular interactions of Mhc genes and pathogen peptides may lead to a “molecular arms race” with recurring bouts of coevolution between the host and the parasite (the Red Queen hypothesis; Van Valen 1973; Hamilton 1982), or Mhc diversity may be elevated because of dissassortative mating between Mhc-dissimilar individuals (Penn and Potts 1998, 1999). This latter view is not inconsistent with a role for Mhc genes in defending hosts against parasites. Chickens have provided particularly powerful models for implicating Mhc genes in resistance to infectious disease (Briles and McGibbon 1948; Schat et al. 1994; Kaufman and Salamonsen 1997). Structurally, the coding regions of avian Mhc genes have many similarities to those of other vertebrates with both class I genes responsible for immune responses to intracellular parasites and class II genes that bind extracellular parasites (Kaufman et al. 1990; Shiina et al. 1999b). The chicken Mhc is also known to possess class III Mhc genes such as factor B that are involved in the complement system of the cellular immune response (Nonaka et al. 1994). The complete sequence of the chicken Mhc (B complex) is an order of magnitude smaller and much more densely packed with genes than mammalian Mhcs (Kaufman et al. 1999a,b).
Avian genes and genomes are thought to be subject to a variety of selective pressures imposed by flight. For example, the small size of avian genomes and chicken introns compared with those of mammals and the low frequency of simple-sequence repeats are thought to be due to selection for small cell size to optimize the high metabolic demands for flight (Tiersch and Wachtel 1991; Hughes and Hughes 1995; Primmer et al. 1997). The high gene density of the chicken Mhc is thought to reflect similar flight-induced genomic streamlining (Parham 1999). Birds are also known to posses a higher frequency of GC-rich isochores than mammals (Bernardi et al. 1997). However, the global similarities and differences of avian and mammalian genomes are still poorly understood. The concept of a “genomic signature” has emerged in recent years as one way to describe the higher-order structure, mutational biases, and selection pressures underlying genomes as revealed in the frequencies of DNA words of different length observed in long DNA sequences (Karlin and Burge 1995). Novel quantitative and qualitative methods permit description of the genomic signature in ways that are virtually independent of global base composition and isochore structure, thereby providing a common metric by which to compare genomes of different species (Jeffrey 1990, 1992). Deschavanne et al. (1999) reported that, contrary to intuition, the signature of an entire genome or of several megabases of a species' DNA can be accurately captured in just a few dozen kilobases and that a species' genomic signature is surprisingly robust to the isochore or gene region from which the DNA sequence for the signature is sampled. The few genomic signatures from mammals that have been published reveal, among other things, the characteristic deficiency of CG dinucleotides that had been noted in earlier analyses of mammalian sequences (Deschavanne et al. 1999), and we were curious to see how an avian genomic signature compared with those of mice and humans.
We have been studying the Mhc region from house finches (Carpodacus mexicanus), both because house finches are well studied ecologically and because they represent an understudied avian lineage with respect to Mhc. House finches, a model species for studies of sexual selection and parasite resistance (Hill 1991; Luttrell et al. 1996; Dhondt et al. 1998), are socially monogamous songbirds found throughout the United States (Hill 1991, 1993). House finch Mhc class II B genes have been partially characterized via Southern blot analysis and by examining expressed genes through RT–PCR (Edwards et al. 1995a,b, 1999). The house finch Mhc contains fewer hybridizing elements when probed with a conspecific cDNA probe than do other songbird species, but we know nothing of house finch Mhc genes at the genomic level, nor anything about the noncoding genomic context of Mhc genes in any songbird (Edwards et al. 2000; Westerdahl et al. 1999). To add a phylogenetic perspective to chicken Mhc studies, and to characterize the genomic signature of house finches, we sequenced a 32-kb cosmid insert (HFcos10A) from a house finch that strongly hybridized with a house finch class II B clone.
RESULTS
Base Composition and Repeated Elements of Cosmid HFcos10A
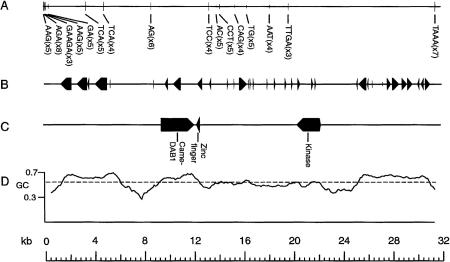
The sequence of the cosmid clone HFcos10A was 31,936-bp long (Fig. 1). The GC content averaged 56.9% over the entire cosmid and varied from 33.1% to 70.1% in moving windows of 500 bp. The highest GC content was found in coding regions and the lowest just proceeding these regions (Fig. 1). There were a total of 35 exons and 8 genes predicted by Genemark (Fig. 1B); these predictions also tended to occur in regions of relatively high (>60%) GC content. There were 16 simple sequence repeats (microsatellites) found. However, only a single microsattelite was longer than five repeat units (AGA8). In addition, we identified two LINE elements beginning at positions 802 and 22,221 using the program RepeatMasker (A. Smit and P. Green, unpubl.), but these LINEs were not verified by subsequent BLAST searches, suggesting that they may not be legitimate. However, their short lengths (32 and 261 bp) are within the range for truncated LINEs found in mammalian Mhcs and genomes (Yamazaki et al. 1999).
Figure 1.
Organization of cosmid HFcos10A containing simple sequence repeats (A), predicted exons found by the program GeneMark (B), three genes identified using various analysis packages (see text for details) (C), and the GC content plotted with a moving window size of 500 bp and offset length of 50 bp using the program PercentGC (D). Direction of arrows in B indicates the transcriptional orientation of each gene. Dashed line in C is the average GC content over the entire cosmid.
Structure and Diversity of Came–DAB1
SeqHelp identified an Mhc-like gene that had the highest GenBank alignment scores with other class II B Mhc genes from songbirds; we designate the gene Came–DAB1 as per Mhc nomenclature rules. Came–DAB1 contains the first three exons expected for typical Mhc class II B genes, but the final three exons are not identifiable (Fig. 2). Figure 3 shows an alignment of exons 2 and 3 of Came–DAB1 with homologous sequences from chicken and red-winged blackbird. There are at least three frameshift mutations located in the second and third exons of Came–DAB1. At 439 and 777-bp, respectively, the sizes of introns 1 and 2 are 2.11 and 8.93 times bigger than the corresponding chicken introns. We found only a single Mhc gene in >30 kb of cosmid sequence, suggesting a low density of Mhc-like sequences in this region of the house finch genome compared to the chicken Mhc.
Figure 2.
Mhc class II B structure in house finches (Came–DAB1), red-winged blackbirds (Agph–DAB1; Edwards et al. 1998), and chickens (B-LBII; Zoorob et al. 1990). The asterisks represent frameshift mutations indicating the nonfunctional nature of Came–DAB1. The shaded exon represents the polymorphic second exon encoding the peptide binding region. Intron and exon sizes are to scale. Dashes signify that the gene continues downstream.
Figure 3.
Alignment of Came–DAB1 exon 2 (A) and exon 3 (B) to a highly polymorphic chicken gene (B-LBI; Zoorob et al. 1993) and a house finch cDNA sequence (Edwards et al. 1995a). Deletions are indicated in the sequences by a − mark.
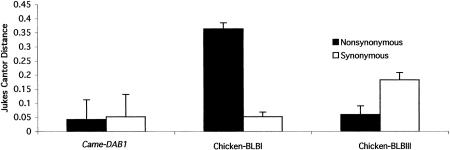
The nucleotide diversity of Came–DAB1 is consistent with the non-functional nature of the gene. Mhc peptide binding regions (PBRs, exon 2 in the case of class II B genes) tend to have a large number of nonsynonymous differences as compared with silent changes. We reconstructed the inferred haplotypes for exon 2 from direct sequencing diploid PCR products using the program HAPINFER (Clark 1990; Fig. 4). We used the reconstructed haplotypes to estimate the pattern of nucleotide substitution in the PBR of Came–DAB1 and for phylogenetic analysis of this exon. Figure 4 identifies the different haplotypes that occur in each individual and the specific base found at each segregating site. The program HAPINFER was unable to resolve the phase in two of the individuals, but all subsequent analysis on exon 2 used the inferred haplotypes and not the direct sequence data. The number of substitutions per nonsynonymous site (dn) is low compared with the number per silent site (ds) for Came–DAB1 and compared with typical functional genes from chickens (Fig. 5).
Figure 4.
List of variable sites as inferred from HAPINFER (Clark 1990) for exon 2 sequences. (Top) Unresolved sequences with reconstructed genotypes in parentheses; (bottom) the reconstructed alleles. HAPINFER was unable to resolve phase two of the individuals (see text for details). Column numbers refer to the site where the polymorphisms occur and are consistent with the alignment from Fig. 3.
Figure 5.
Comparison of the number of nonsynonymous (dn) and silent (ds) substitutions per site within house finches for exon 2 in Came–DAB1 (n=9), a functional chicken gene (n=12), and a “nonclassical” class II chicken gene (n=3). These comparisons were made with a number of chicken sequences downloaded from GenBank (Zoorob et al. 1993) and the nine inferred haplotypes from house finches obtained through PCR amplification, direct sequencing, and analysis using HAPINFER as described in text. The dn and ds values were calculated by the Jukes-Cantor method of Nei and Gojobori (1986) using the MEGA software package (Kumar et al. 1993).
Table 1 describes the overall diversity of Came–DAB1 using the statistics Π (average pairwise difference) and Θ = 4Neμ, where Ne is the effective population size and μ is the neutral mutation rate. Levels of genetic diversity of exon 2 are more similar to the levels for the nonclassical B-LBIII gene of chickens than to either of the classical chicken genes (B-LBI and B-LBII) or a polymorphic blackbird class II B gene (Table 1; Garrigan and Edwards 1999). In particular, we found much lower values of Θ and Π at the Came–DAB1 locus than found in similar surveys of polymorphic chicken B-LBI and B-LBII genes. Tajima's D statistic (Tajima 1989), which tests for neutrality in DNA sequence data, was negative for both the finch and the B-LBIII locus in chickens, a suggestion of a gene under directional selection although the values are not significantly different from the neutral expectation (P>>0.05). B-LBI, B-LBII, and the blackbird gene (Agph–DAB1) all had positive values for Tajima's D, consistent with balancing selection. HAPINFER was unable to resolve inferred haplotypes from exon 3 data from Came–DAB1, perhaps because of a number of alternate homozygotes found in the direct sequences. Nonetheless, we can still examine diversity (Θ) using the number of segregating (polymorphic) sites (Watterson 1975). As expected from a pseudogene, the values of Θ for exon 3 are similar to those of exon 2 (Table 1). Moreover, these values are somewhat higher than the neutral Θ found in other vertebrates such as humans. For example, Grimsley et al. (1998) found Θ values of 0.0262 for HLA-H, an Mhc pseudogene linked to the highly polymorphic HLA-A gene and reported that these Θ values were an order of magnitude higher than the background Θ for humans. A neutral Θ for house finches is not known and neutral intron diversity in some seabirds, which are known to have large population sizes, are about the same (H. Walsh, pers. comm.). Although it is not definitive we think that this value of Θ would be high for finches at a neutral locus.
Table 1.
Comparison of Nucleotide Diversity of Came–DAB1, Three Chicken Genes, and a Red-Winged Blackbird Gene Agph–DAB1
| Came–DAB1 | Chicken exon 2a | Agph–DAB1b | ||||
|---|---|---|---|---|---|---|
| Gene | exon 2 | exon 3 | B-LBI | B-LBII | B-LBIII | exon 2 |
| No. of sequences examined | 9 | 10 | 12 | 11 | 3 | 17 |
| Total no. of sites | 260 | 287 | 270 | 270 | 270 | 89 |
| No. of segregating sites(s) | 25 | 32 | 138 | 54 | 41 | 22 |
| Θ | 9.19 | 11.48 | 44.47 | 18.43 | 22.36 | 6.32 |
| Θ per site | 0.04 | 0.04 | 0.17 | 0.07 | 0.08 | 0.07 |
| Π | 8.89 | — | 47.99 | 22.64 | 22.33 | 8.90 |
| Nucleotide diversity (Π per site) | 0.03 | — | 0.18 | 0.08 | 0.08 | 0.10 |
| Tajima's D | −0.17 | — | 0.36 | 1.08c | −0.01 | 1.99c |
Data from both exons 2 and 3 from Came–DAB1 are shown. Θ is calculated using the finte sites method of Tajima (1996). All relevant statistics are discussed in Methods.
From Zoorob et al. (1993).
P < 0.03.
Origin of Came–DAB1
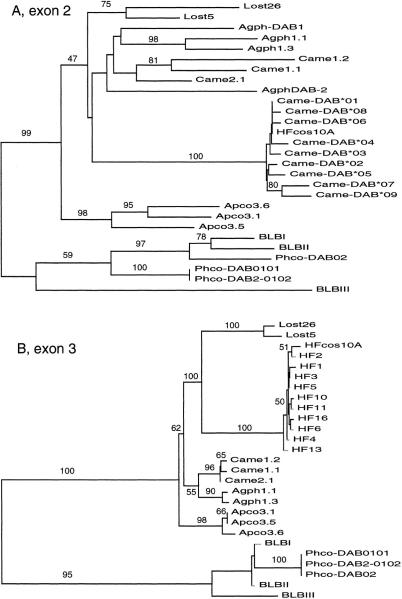
Came–DAB1 exon 2 and 3 sequences were easily aligned to those of other functional and nonfunctional Mhc genes. Phylogenetic trees of Came–DAB1 using the neighbor joining method (Saitou and Nei 1987), the inferred haplotypes for exon 2, and direct sequence data for exon 3 in general exhibit a strong trend toward clustering of sequences by species (Fig. 6a,b). The phylogenetic reconstructions of both exon 2 and exon 3 place the sequences from HFcos10A closest to the house finch sequences obtained for the polymorphism study. However, both the exon 2 and exon 3 trees suggest that the sequences from the Came–DAB1 locus are not the closest relatives of the expressed cDNA sequences of house finch class II B genes from Edwards et al. (1995a). For exon 3 the Came–DAB1 sequences are most closely related to sequences from the Bengalese finch (Lonchura striata, Lost; family Estrildidae; Vincek et al 1995). The branch lengths leading to the Bengalese finch in the exon 2 tree are deep compared to the branches leading to the Came–DAB1 sequences, and the bootstrap value supporting monophyly of finch and blackbird sequences is not high. However, the Bengalese finch and the Came–DAB1 sequences cluster strongly (100%) for exon 3 despite the fact that these two “finches” are in different taxonomic families.
Figure 6.
Phylogenetic analysis of exon 2 (A) and exon 3 (B) sequences from different avian species (Lost=Bengalese finch, Vincek et al. 1995; Apco=scrub jay, Came=house finch, Agph=red-winged blackbird, Edwards et al. 1995a; HFcos10A=house finch cosmid 10A, HF=unresolved exon 3 sequences; Phco=ring-necked pheasant, Witzell et al. 1999; BLB=chicken, Zoorob et al. 1993). The trees shown are neighbor-joining trees (Saitou and Nei 1987) using a Tamura-Nei (1993) distance. The numbers above the branches are bootstrap scores with 1000 replicates. The trees are rooted with the pheasant and chicken sequences used collectively as an outgroup.
Non-Mhc Genes
A serine–threonine kinase gene detected by SeqHelp ∼8340 bp from Came–DAB1 is predicted to be transcribed in the opposite direction as Came–DAB1. The sequence was aligned to two homologous sequences using a BLAST search. The highest similarity genes were members of the Ste20/PAK family from Xenopus, which is involved in the arrest of oocytes at Gs/prophase of the first meiotic cell cycle and prevention of apoptosis (Faure et al. 1997) and a Drosophila homolog of the serine–threonine kinase PAK gene that has a potential function in focal adhesion and colocalizes with dynamic actin structures (Harden et al. 1996). The house finch gene has three alignable exons, with the first and third exons slightly shorter than the genes mentioned above (not shown). Both a start and stop codon are found within a few amino acids of these features in the other sequences.
A zinc finger motif is found just downsteam of Came–DAB1 and also runs in the opposite transcriptional orientation. The total length of the motif is 79 amino acids, whereas the two sequences examined from GenBank with which it had the highest similarity were 574 and 1207 amino acids long. There was only a single alignable exon containing two C2H2 motifs [C2H2 motifs are tandemly repeated domains commonly found in zinc finger proteins and comprise one of the most common gene families in the human genome (Becker et al. 1995)], apparent from the BLAST search. A BLAST search of the 100 nucleotides upstream of this region did not yield any convincing hits.
Genomic Signature
We investigated the genomic signature implied by the 32-kb house finch sequence using the pictorial chaos–game representation (CGR) algorithm of Descavanne et al. (1999), in which the frequencies of different DNA words are depicted by varying shades, from black (most frequent) to white (least frequent). We investigated the frequency of DNA words of two (dinucleotides), five, and eight letters (Fig. 7) on both strands of the finch sequence. In the CGR method, a square image is divided into four quadrants signifying the four nucleotides. The pixel signifying the frequency of all DNA ‘words’ of any length ending in a given nucleotide occurs in that nucleotide's quadrant. Each quadrant is in turn divided into four quadrants that signify the nucleotide occurring in the second to last position of words. These secondary quadrants occur in the same positions relative to one another as do the original quadrants, and so on, until the appropriate number of pixels (4n, where n is the number of letters in the words being investigated) is achieved. In this way certain large-scale features of the sequence examined can emerge. For example, dark diagonals indicate stretches composed solely of purines or pyrimidines, and empty patches in upper left quadrant of the upper right quadrant indicate a deficiency of words containing CpG dinucleotides.
Figure 7.
Genomic signature of house finch DNA for two-, five-, and eight-letter words based on the 32-kb cosmid sequence. Dark pixels indicate high-frequency words and light pixels indicate low-frequency words. The 16 (42) words in the leftmost panel are indicated for convenience. The five-letter signature contains 45 words (pixels) and the eight-letter signature 48. The fact that the orientation of the four nucleotides is the same at all scales in the image, i.e., within any given quadrant of any size, contributes to the fractal nature of the image.
The genomic signature of the house finch exhibits strong signals on the diagonals for five- and eight-letter words, indicating a high frequency of purine and pyryimidine stretches (Fig. 7). It also exhibits a notable CpG depletion for all three word lengths, as indicated by the pale regions in the upper left quadrants of all three upper right quadrants, as well as a deficiency of TA dinucleotides, as indicated by the pale lower right quadrant of all three lower left quadrant (Fig. 7). This latter result occurs despite the presence of several TA-rich microsatellites (albeit short ones; Fig. 1). We conducted a quantitative analysis of the five-letter word frequencies. We found that two words are never met in the sequence or on its complementary strand—TACGC and GCGTA, both of which contain the two counter-selected dinucleotides CG and TA). Consistent with the G+C rich nature of sequence, the most frequent five-letter words are CCCTG, CAGGG, GGGGA, TCCCC, GGGGG, CCCCC, GGCCA, TGGCC, CCCAG, CTGGG, CCCCT, AGGGG, CCCCA, and TGGGG, all of which occurred between 255 and 292 times, about 5–6 times the median of the distribution of five-letter words in the entire sequence.
DISCUSSION
Status of Sequenced Genes on Cosmid HFcos10A
In conjunction with new sequences from red-winged blackbirds (Edwards et al. 2000), the cosmid we have sequenced is the first cosmid-scale sequence determined for any avian species other than chicken. It thus provides a glimpse into the genomic architecture of the most species-rich clade of birds, Passeriformes, as well as insight specifically into the structure of regions containing Mhc genes. At about one identified gene per 10 kb, the gene density of the region we have sequenced is more similar to that of the mammalian Mhc than to the chicken B complex (MHC Sequencing Consortium 1999; Kaufman et al. 1999b). Only one of the 35 exons predicted by Genemark corresponded accurately to the manually identified exons, the zinc finger, and neither Came–DAB1 nor the serine–threonine kinase genes were predicted accurately. We therefore suspect that many of these predictions are spurious. Although we cannot be sure that the house finch sequence occurs in the same genomic region as polymorphic and presumably functional Mhc genes, that is, in the canonical house finch Mhc (Edwards et al. 1995a, 1999), Mhc-containing regions of this gene density have not yet been reported for any avian species and our estimate of avian gene density is only the second (after the chicken B complex) for any bird based on cosmid sequences. Came–DAB1 is among a very few avian Mhc genes sequenced at the genomic level (Guillemot et al. 1988; Zoorob et al. 1990; Edwards et al. 1998; Kaufman et al. 1999b). It has none of the attributes of a classical Mhc gene. The gene does not have a high rate of nonsynonymous substitutions, an expected signature of a functional Mhc gene under balancing selection, nor high levels of total diversity (Π and Θ). All indications are that Came–DAB1 is a pseudogene. There are frameshift mutations in two of the three identifiable exons (including the PBR-encoding exon) and the sequence similarity to other Mhc genes declines after the third exon. We used the method of Miyata and Yasunaga (1981) to estimate the time since loss of function for this pseudogene. This method uses the difference in the dn/ds ratio in comparisons of the focal pseudogene, a functional homolog (ingroups) and an outgroup sequence to estimate time since loss of function in the pseudogene. We used the sequences from exon 2 of a functional blackbird gene Edwards et al. (1998) as the outgroup and Came–DAB1 and cDNA sequences from the house finch (Edwards et al. 1995a) as ingroups in this analysis. The estimated time since loss of function of Came–DAB1 is 0.9 T0, where T0 is the time since divergence of the blackbird and house finch. DNA–DNA hybridization studies (Sibley and Ahlquist 1990) place this split at about 50 million years ago (MYA), making the time since loss of function of Came–DAB1 at ∼45 MYA. This ancient date is consistent with the dn/ds ratios at Came–DAB1, which appear to have reached base substitutional equilibrium. The ratios were not significantly different than one, a pattern expected in a pseudogene after a long period of neutral evolution.
The two other genes found on the cosmid have not been found near Mhc genes of other birds (Kaufman et al. 1999a,b; Shiina et al. 1999b). However, genes found in the same broadly defined multigene families have been found inside the Mhcs of chickens and other vertebrates. For instance, the RING3 gene is a kinase that is found in the class II region of mammals, chickens, and frogs (Kaufman et al. 1999a). However, the house finch kinase is clearly not similar to RING3. Genes similar to the house finch serine–threonine kinase gene are involved in protein phosphorylation (Kruse et al. 1997) and the zinc-finger protein is a widespread transcription factor motif in eukaryotic genomes (Struhl 1989). There is no reason to expect that these genes are involved in the antigen presenting process.
Multigene Family Evolution
Because Came–DAB1 was easily aligned with other avian and vertebrate Mhc genes, and because phylogenetic analysis clearly showed that Came–DAB1 clustered with other expressed songbird Mhc genes, Came–DAB1 can justifiably be called an Mhc gene. In designating Came–DAB1 as an Mhc gene, we differ with Kaufman et al. (1999a), who suggest that only Mhc-like genes that reside in regions homologous to the chicken B-complex and are expressed and functionally important should be designated as such (Miller et al. 1994). We prefer a genealogical definition of Mhc genes, rather than a functional one. The fact that Came–DAB1 does not cluster specifically with functional chicken Mhc genes (Fig. 6a,b) does not bear on the question of whether it resides in the genomic region homologous to the chicken B-complex, given the possibility of functional Mhc-genes being dispersed in birds, as occurs in zebrafish (Bingulac-Popovic et al. 1997). Given what we know of multigene family evolution in avian Mhc genes (see below), we expect most songbird Mhc genes to form clusters separately from those of chickens, regardless of whether they are expressed or not.
Given that we can align Came–DAB1 to other Mhc sequences and perform a phylogenetic analysis, we are justified in discussing multigene family evolution in the avian Mhc, as we have in the past (Edwards et al. 1995a; 1999). That Came–DAB1 is a pseudogene supports the idea of a birth and death model of Mhc evolution (Ota and Nei 1994; Nei et al. 1997). The birth and death model predicts that there is frequent gene duplication and pseudogene formation in multigene families. Our phylogenetic analysis addressed some of the hypotheses about the mode of Mhc multigene family evolution in birds—whether a concerted evolution (Witzell et al. 1999) or a divergent evolution model is most prevalent. The data for both exons 2 and 3 largely support a concerted evolution model because the predominant pattern is clustering by species. However, exon 3 sequences from Came–DAB1 are more closely related to genes from other species (Bengalese finch, family Estrilididae) than they are to the presumably functional house finch genes obtained from cDNA. Either Came–DAB1 is orthologous to the genes from the Bengalese finch (exon 3) or the absence of stabilizing selection acting on Came–DAB1 has masked its phylogenetic signal through homoplasy (exon 2). Homoplasy is expected to be a problem more for the second exon of Mhc genes because of the increased likelihood of base substitutional saturation caused by balancing selection and, more importantly, the scrambling effects of recombination and gene conversion. Exon 3 is a better indicator of gene history because it is not under the diversifying selection pressures, and the strong clustering of the Bengalese finch with Came–DAB1 supports its orthology with the Lost sequences.
Genomic Signature
The HFcos10A sequence contained several microsatellites, mostly with five or fewer repeat units (Fig. 1). The low density of long microsatellites, with only two repeats with a total length >20 bp in 30 kb of sequence, is consistent with a low density of simple sequence repeats found in another survey of avian microsatellites (Primmer et al. 1997). These researchers found an average of one microsatellite of >20 bp total length per 39 kb, a low density compared to human DNA (1 microsatellite per 6 kb; Beckmann and Weber 1992). For the human Mhc class I region the number of microsatellites is ∼1 per every 2 kb (Shiina et al. 1999a).
The CGR genomic signature exhibited by the house finch sequence, the first reported for birds, displays a number of similarities to mammalian signatures and raises a number of predictions for signatures of other avian genomes. The house finch signature bears a number of similarities to other homeothermic vertebrates thus far examined (e.g., mouse and human). The deficiency of CpG dinucleotides, as well as the relatively high frequency of purine or pyrimidine runs (diagonals) suggests that these may be features of vertebrate genomes generally. The deficiency of CpG is intriguing given evidence for a much higher density of genes and CpG islands in the chicken than in the mammalian genome (McQueen et al. 1996) and the higher frequency of GC-rich isochores in birds compared with mammals (Bernardi et al. 1997). The deficiency of TA dinucleotides and the longer words embedding this and CG words is a novel feature that is not as pronounced in the mammalian signatures that have been investigated to date by the CGR method (Deschavanne et al. 1999). We note that the region we have sequenced is somewhat more GC-rich (and TA-depauperate) than mammalian sequences, as expected for birds (Bernardi et al. 1997). However, this TA deficiency of the genomic signature remains even after the effects of global base composition have been removed (data not shown). Some of these features could be the result of particular directional mutation pressures resulting from the high metabolic rates and high body temperatures of birds; such features are known to influence the mutational spectrum and base composition of animal mitochondrial DNA (Martin 1995). Pettigrew (1994) suggested that flying vertebrates should have elevated levels of A and T nucleotides because of higher metabolic demands. However, analysis by Van Den Bussche et al. (1998) showed that flying mammals such as bats do not have higher AT levels than other mammals. Our results suggest that birds also may not show the elevated AT levels predicted by Pettigrew (1994). Although the sequence we have analyzed is only 32 kb, we suspect it will capture many of the features of avian genomic signatures based on longer sequences, in part because of the patent similarities of the signature to those of non-Mhc regions in mammals; the signature for 8-mers may be less precise than those for shorter words because the frequency of each of the 48 8-mers may not be captured accurately even in 32kb. The house finch signature therefore can be tested for generality by analyzing sequences from avian species that are less well studied in this regard, such as the chicken.
METHODS
Samples
Cosmid 10A was isolated from a cosmid library as per Edwards et al. (2000). All birds used in the polymorphism screen are the same as those used in Edwards et al. (2000) as well as four more birds from the same Alabama population. All birds are unrelated. All sequences have been deposited in GenBank with accession numbers AF205032 and AF241546–AF241565.
Sequence Assembly
We sequenced the clone containing the most strongly hybridizing band as revealed by a Southern blot analysis probed with a partial (exon 1–4) RT–PCR product from a house finch class II B Mhc gene (see Edwards et al. 1998, 1999b for details on cosmid library construction and screening). We then sonicated the cosmid clone, subcloned the sonicated fragments into an M13 vector, and prepared multiple clones using Qiagen Prep Kits in a 96-well format. We sequenced at random 830 subclones on an ABI 373 cycle sequencer with Dye-terminator chemistry using a modified M13-T7 (5′-TGCCTGCAGGTCGACTCTAG) vector primer. These sequences were aligned and assembled into contigs using the program PhredPhrap (Ewing and Green 1998; Ewing et al. 1998). We visualized the assembled contigs using the program Consed (Gordon et al. 1998), designed primers for the end of each contig, and connected the contigs by walking. This method of primer design was also used to improve regions of low sequence quality after the first round of sequencing.
The final contig was analyzed for sequence similarity with GenBank sequences using the program SeqHelp (Lee et al. 1998). We also predicted open reading frames and exons using the program GeneMark (Lukashin and Borodovsky 1998). GeneMark uses a Markov model to statistically search for splice signals and start codons generated from a matrix derived from empirical observations. In our case a chicken matrix was used: simple sequence repeats using Sputnik (C. Abajian, unpubl.) and more complex repeats using RepeatMasker (A. Smit and P. Green, unpubl.). The genomic signature was analyzed for words of 1–8 letters using the methods outlined in Deschevanne et al. (1999). The frequencies of all words of varying lengths was determined after concatenating both strands into one sequence. This action was taken because the genomic signature is strand dependent (see Deschevanne et al. 1999 for details).
Polymorphism Analysis
We designed locus-specific PCR primers to amplify the sequences from exons 2 and 3 of Came–DAB1 (exon 2: 5′ HF10AEX2F–GCTGTGTCCTGCACTCACA, 3′ HF10AINT2R.1–GCAGGGTCCGAGGGGAC; exon 3: 5′ HF10AINT2F.1–CTGATTCCAGTGTGTCCCCA, 3′ HF10AINT3R.1–CCAGTGGCTCTCCCAGTG). This was accomplished by comparing the cosmid Mhc sequence to previously published cDNA sequences (Edwards et al. 1995a) and maximizing the areas of discrepancy. We directly sequenced 10 individuals (both strands) for all of exons 2 and 3 using the forward and reverse PCR primers as well as two internal sequencing primers both in the 5′ and 3′ directions (exon 2: 5′ HF10EX2F.2–GAGAGGTTCATCTACAACCG, 3′ HF10EX2R.2–AGCTCGTAGTTTCGCCAGC; exon 3: 5′ HF10AEX3F.2–CTCTCTCTCCCTCTCACAG, 3′ HF10AEX3R.2–CCGGGGGCTCCCCCATAT). These sequences were then aligned and a consensus sequence for each individual was generated using the alignment program Sequencher (Gene Codes). Sequences were checked manually and examined for the presence of heterozygous sites. We then used the program HAPINFER (Clark 1990) to generate haplotypes. These haplotypes were then used to generate estimates of the dn/ds ratios (the number of nonsynonymous to synonymous changes) using the Jukes–Cantor method (Nei and Gojobori 1986) and to infer Tajima's D (Tajima 1989), the number of segregating sites (s), and Θ (Watterson 1975) using a program DNAPOLY written by Dan Garrigan (unpubl.).
Phylogentic Analysis
We included the 9 inferred haplotypes for exon 2 and the 10 individuals yielded from direct sequencing for exon 3 in our analysis. We were unable to infer the haplotypes for exon 3 and therefore used diploid sequences in our phylogenetic analysis—technically a violation of standard phylogenetic analysis, but one with likely little effect given the low level of variability. Also included in these analyses were sequences downloaded from GenBank from the Bengalese finch (Lonchura striata, Vincek et al. 1995), chicken (Gallus gallus, Pharr et al. 1998), scrub jay, and red-winged blackbird (Edwards et al. 1995a). These sequences were aligned using the program Sequencher to lengths of 260 bp (exon 2) and 287 bp (exon 3). We used the neighbor-joining method (Saitou and Nei 1987) and a Tamura-Nei (1993) distance method for all phylogenetic analysis. The resulting trees were rooted using the chicken and pheasant sequences as outgroups. A total of 1000 bootstrap replicates (Felsenstein 1985) were completed to determine the robustness of phylogenetic groupings.
Acknowledgments
We are thankful for the helpful discussion provided by Dan Garrigan, Jim Kaufman, Takashi Shiina, Patrick Deschavanne, and Yoko Satta and for the technical assistance of Ming Lee, Patrick Deschavanne, Brent Ewing, Phil Green, and Mark Reider. Three anonymous reviewers provided helpful comments on the manuscript. This work was supported by a Graduate Recruitment Award to C.M.H., a Howard Hughes Predoctoral Fellowship to H.E.H., and NSF grants DEB-9419738 and DEB-9797548 to S.V.E .
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL edwards@zoology.washington.edu; FAX (206) 543-3041.
REFERENCES
- Becker KG, Nagle JW, Cannin RD, Biddison WE, Ozato K, Drew PD. Rapid isolation and characterization of 118 novel C2H2-type zinc finger cDNAs expressed in human brain. Hum Mol Genet. 1995;4:685–691. doi: 10.1093/hmg/4.4.685. [DOI] [PubMed] [Google Scholar]
- Beckmann JS, Weber JL. Survey of human and rat microsatellites. Genomics. 1992;12:627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Hughes S, Mouchiroud D. The major compositional transitions in the vertebrate genome. J Mol Evol. 1997;44:S44–S51. doi: 10.1007/pl00000051. [DOI] [PubMed] [Google Scholar]
- Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, Klein J. Mapping of Mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics. 1997;46:129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- Briles WE, McGibbon WH. Heterozygosity of inbred lines of chickens at two loci affecting cellular antigens. Genetics. 1948;33:605. [PubMed] [Google Scholar]
- Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- Deschavanne PJ, Giron A, Vilain J, Fagot G, Fertil B. Genomic signature: Characterization and classification of species assessed by chaos game representation of sequences. Mol Biol Evol. 1999;16:1391–1399. doi: 10.1093/oxfordjournals.molbev.a026048. [DOI] [PubMed] [Google Scholar]
- Dhondt AA, Tessaglia DL, Slothower RL. Epidemic mycoplasmal conjunctivitis in house finches from eastern North America. J Wildlife Dis. 1998;34:265–280. doi: 10.7589/0090-3558-34.2.265. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Hedrick PW. Evolution and ecology of MHC molecules: From genomics to sexual selection. Trends Ecol Evol. 1998;13:305–311. doi: 10.1016/s0169-5347(98)01416-5. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Wakeland EK, Potts WK. Contrasting histories of avian and mammalian Mhc genes revealed by class II B sequences from songbirds. Proc Natl Acad Sci. 1995a;92:12200–12204. doi: 10.1073/pnas.92.26.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Grahn M, Potts WK. Dynamics of Mhc evolution in birds and crocodilians: amplification of class II genes with degenerate primers. Mol Ecol. 1995b;4:719–729. doi: 10.1111/j.1365-294x.1995.tb00272.x. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Gasper J, Stone M. Genomics and polymorphism of Agph-DAB1, an Mhc class II B gene in red-winged blackbirds (Agelaius phoenicus) Mol Biol Evol. 1998;15:236–250. doi: 10.1093/oxfordjournals.molbev.a025921. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Hess CM, Gasper J, Garrigan D. Toward an evolutionary genomics of the avian Mhc. Immunol Rev. 1999a;167:119–132. doi: 10.1111/j.1600-065x.1999.tb01386.x. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Nusser J, Gasper J. Characterization and evolution of Mhc genes from non-model organisms, with examples from birds. In: Baker AJ, editor. Molecular methods in ecology. Cambridge, UK: Blackwell Scientific; 2000. . (In press.) [Google Scholar]
- Edwards, S.V., J. Gasper, D. Garrigan, D. Martindale, and B. Koop. Submitted. Ghost of selection past and avian genome architecture revealed in a 39-kb sequence from the Mhc class II region of Red-winged Blackbirds (Agelaius phoenicus).
- Ewing B, Green P. Base-calling of automated sequencer using PHRED. II. error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier LD, Wendl MC, Green P. Base-calling of automated sequencer using PHRED. II. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Faure S, Vigneron S, Doree M, Morin N. A member of the Ste20/PAK family of protein kinases is involved in both arrest of Xenopus oocytes at G2/prophase of the first meiotic cell cycle and in prevention of apoptosis. EMBO J. 1997;16:5550–5561. doi: 10.1093/emboj/16.18.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Edwards SV. Polymorphism across an exon-intron boundary in an avian mhc class II B gene. Mol Biol Evol. 1999;16:1599–1606. doi: 10.1093/oxfordjournals.molbev.a026072. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grimsley C, Mather KA, Ober C. HLA-H: A pseudogene with increased variation due to balancing selection at neighboring loci. Mol Biol Evol. 1998;15:1581–1588. doi: 10.1093/oxfordjournals.molbev.a025886. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Billaulit A, Pourquie O, Behar G, Chausse AM, Zoorob R, Kreibech G, Auffray C. A molecular map of the chicken major histocompatability complex: The class II genes are closely linked to the class I genes and the nucleolar organizer. EMBO J. 1988;7:2775–2785. doi: 10.1002/j.1460-2075.1988.tb03132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD. Pathogens as causes of genetic diversity in their host populations. In: Anderson RM, May RM, editors. Population biology of infectious diseases. New York, NY: Springer-Verlag; 1982. pp. 269–296. [Google Scholar]
- Harden N, Lee J, Loh H, Ong Y, Tan I, Leung T, Manser E, Lim L. A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol Cell Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. Plumage color is a sexually selected indicator of male quality. Nature. 1991;350:337–339. [Google Scholar]
- ————— . House finch. In: Poole A, Gill F, editors. The birds of North America. PA: The Academy of Natural Sciences of Philadelphia (no. 46); 1993. [Google Scholar]
- Hughes AL, Hughes MK. Small genomes for better flyers. Nature. 1995;377:391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- Jeffrey HJ. Chaos game representation of gene structure. Nucleic Acids Res. 1990;18:2163–2170. doi: 10.1093/nar/18.8.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Chaos game visualization of sequences. Comput Graphics. 1992;16:25–33. [Google Scholar]
- Kandil E, Namikawa C, Nonaka M, Greenburg A, Flajnik M, Ishibashi T, Kasahara M. Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates: Implications for the origin of MHC class I-restricted antigen presentation. J Immunol. 1996;156:4245–4253. [PubMed] [Google Scholar]
- Karlin S, Burge C. Dinucleotide relative abundance extremes: A genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Salmonsen J. The “minimal essential MHC” revisited: Both peptide-binding and cell surface expression levels of MHC molecules are polymorphisms selected by pathogens in chickens. Hereditas. 1997;127:67–73. doi: 10.1111/j.1601-5223.1997.t01-1-00067.x. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Skoedt K, Salmonsen J. The MHC molecules of nonmammalian vertebrates. Immunol Rev. 1990;113:83–117. doi: 10.1111/j.1600-065x.1990.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Jacob J, Shaw I, Walker B, Milne S, Beck S, Salomonsen J. Gene organisation determines evolution of function in the chicken MHC. Immunol Rev. 1999a;167:101–117. doi: 10.1111/j.1600-065x.1999.tb01385.x. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Milne S, Göbel T, Walker BA, Jacob JP, Auffrey C, Zoorob R, Beck S. The chicken B locus is a minimal-essential major histocompatibility complex. Nature. 1999b;401:923–925. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- Klein J. Natural history of the major histocompatability complex. New York, NY: Wiley and Sons; 1986. [Google Scholar]
- Klein J, Satta Y, O'Huigin C, Takahata N. The molecular descent of the major histocompatability complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- Kruse M, Miller IM, Muller WEG. Early evolution of metazoan serine/threonine and tyrosine kinases: Identification of selected kinases in marine sponges. Mol Biol Evol. 1997;14:1326–1334. doi: 10.1093/oxfordjournals.molbev.a025742. [DOI] [PubMed] [Google Scholar]
- Lee M, Lynch ED, King M-C. SeqHelp: A program to analyze molecular sequences utilizing common computational resources. Genome Res. 1998;8:306–312. doi: 10.1101/gr.8.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell MP, Fischer JR, Stallknecht DE, Kleven SH. Field investigation of Mycoplasma gallisepticum infections in house finches (Carpodacus mexicanus) from Maryland and Georgia. Avian Dis. 1996;40:335–341. [PubMed] [Google Scholar]
- Magor BG, DeTomaso A, Rinkevich B, Weissman IL. Allorecognition in colonial tunicates: Protection against predatory cell lineages? Immunol Rev. 1999;167:69–80. doi: 10.1111/j.1600-065x.1999.tb01383.x. [DOI] [PubMed] [Google Scholar]
- Martin AP. Metabolic rate and directional nucleotide substitution in animal mitochondrial DNA. Mol Biol Evol. 1995;12:1124–1131. doi: 10.1093/oxfordjournals.molbev.a040286. [DOI] [PubMed] [Google Scholar]
- McQueen H, Fantes AJ, Cross SH, Clark VH, Archibald AL, Bird AP. CpG islands of chicken are concentrated on microchromosomes. Nat Genet. 1996;12:321–324. doi: 10.1038/ng0396-321. [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium. Complete sequence and gene map of the human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Miller MM, Goto R, Bernot A, Zoorob R, Auffray C, Bunstead N, Briles WE. Two Mhc class I and two Mhc class II genes map to the chicken Rfp-Y system outside the B complex. Proc Natl Acad Sci. 1994;91:4397–4401. doi: 10.1073/pnas.91.10.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Yasunaga T. Rapidly evolving mouse α-globin-related pseudo gene and its evolutionary history. Proc Natl Acad Sci. 1981;78:450–453. doi: 10.1073/pnas.78.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:269–295. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka M, Takahashi M, Sasaki M. Molecular cloning of a lamprey homologue of the mammalian class III gene, complement factor B. J Immunol. 1994;152:2263. [PubMed] [Google Scholar]
- O'Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womak JE, Marshall Graves JA. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- Ohta T. On the pattern of polymorphism at major histocompatability complex loci. J Mol Evol. 1998;46:633–638. doi: 10.1007/pl00006343. [DOI] [PubMed] [Google Scholar]
- Ota T, Nei M. Divergent evolution and evolution by the birth-and-death process in the immunoglobin VH pseudogenes in chickens. Mol Biol Evol. 1994;12:94–102. [Google Scholar]
- Parham P. Soaring costs in defence. Nature. 1999;410:870–871. doi: 10.1038/44745. [DOI] [PubMed] [Google Scholar]
- Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- Penn D, Potts WK. Chemical signals and parasite-mediated sexual selection. Trends Ecol Evol. 1998;13:391–396. doi: 10.1016/s0169-5347(98)01473-6. [DOI] [PubMed] [Google Scholar]
- ————— The evolution of mating preferences and major histocompatibility complex genes. Am Nat. 1999;153:145–164. doi: 10.1086/303166. [DOI] [PubMed] [Google Scholar]
- Pettigrew JD. Flying DNA. Curr Biol. 1994;4:277–280. doi: 10.1016/s0960-9822(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Pharr GT, Dodgson JB, Hunt HD, Bacon LD. Class II MHC cDNAs in 15I5 B-congenic chickens. Immunogenetics. 1998;5:350–354. doi: 10.1007/s002510050369. [DOI] [PubMed] [Google Scholar]
- Primmer CR, Raudsepp T, Chowdhary BP, Møller AP, Ellegren H. Low frequency of microsatellites in the avian genome. Genome Res. 1997;7:471–482. doi: 10.1101/gr.7.5.471. [DOI] [PubMed] [Google Scholar]
- Reynolds PS, Lee RM. Phylogenetic analysis of avian energetics: passerines and nonpasserines do not differ. Am Nat. 1996;147:735–759. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schat KA, Taylor RL, Jr, Briles WE. Resistance to Marek's Disease in chickens with recombinant haplotypes of the Major Histocompatability (B) Complex. Poultry Sci. 1994;73:502–508. doi: 10.3382/ps.0730502. [DOI] [PubMed] [Google Scholar]
- Shiina T, Tamiya G, Oka A, Takishima N, Inoko H. Genome sequencing analysis of the 1.8 Mb entire human MHC class I region. Immunol Rev. 1999a;167:193–199. doi: 10.1111/j.1600-065x.1999.tb01392.x. [DOI] [PubMed] [Google Scholar]
- Shiina T, Shimizu C, Oka A, Teraoka Y, Imanishi T, Gojobori T, Hanzawa K, Watanabe S, Inoko H. Gene organization of the quail major histocompatibility complex (MhcCoja) class I gene region. Immunogenetics. 1999b;49:384–394. doi: 10.1007/s002510050511. [DOI] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE. The phylogeny and classification of birds: A study in molecular evolution. New Haven, NY: Yale University Press; 1990. [Google Scholar]
- Struhl K. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem Sci. 1989;14:137–140. doi: 10.1016/0968-0004(89)90145-X. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics. 1996;143:1457–1465. doi: 10.1093/genetics/143.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tiersch TR, Wachtel SS. On the evolution of genome size in birds. J Hered. 1991;82:363–368. doi: 10.1093/oxfordjournals.jhered.a111105. [DOI] [PubMed] [Google Scholar]
- Van Den Bussche RA, Baker RJ, Huelsenbeck JP, Hillis DM. Base compositional bias and phylogenetic analyses: A test of the “Flying DNA” hypothesis. Mol Phylogenet Evol. 1998;13:408–416. doi: 10.1006/mpev.1998.0531. [DOI] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- Vincek V, Klein D, Graser RT, Figueroa F, O'hUigin C, Klein J. Molecular cloning of a major histocompatability complex class II B gene cDNA from the Bengalese finch, Lonchura striata. Immunogenetics. 1995;42:262–267. doi: 10.1007/BF00176443. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Pop Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Westerdahl H, Witzell H, von Schantz T. Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics. 1999;49:149–157. doi: 10.1007/s002510050477. [DOI] [PubMed] [Google Scholar]
- Witzell H, Bernot A, Auffrey C, Zoorob R. Concerted evolution of two Mhc class II B loci in pheasants and domestic chickens. Mol Biol Evol. 1999;16:479–490. doi: 10.1093/oxfordjournals.molbev.a026130. [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Tateno Y, Inoko H. Genomic organization around the centromeric end of the HLA class I region: Large-scale sequence analysis. J Mol Evol. 1999;48:317–327. doi: 10.1007/pl00006475. [DOI] [PubMed] [Google Scholar]
- Zoorob R, Behar G, Kroemer G, Auffrey C. Organization of a functional chicken class II B gene. Immunogenetics. 1990;31:179–187. doi: 10.1007/BF00211553. [DOI] [PubMed] [Google Scholar]
- Zoorob R, Bernot A, Renoir D, M, Choukri F, Auffray C. Chicken major histocompatability complex class II B genes: Analysis of interallelic and interlocus sequence variance. Eur J Immunol. 1993;23:1139–1145. doi: 10.1002/eji.1830230524. [DOI] [PubMed] [Google Scholar]