Abstract
The progress of human and mouse genome sequencing programs presages the possibility of systematic cross-species comparison of the two genomes as a powerful tool for gene and regulatory element identification. As the opportunities to perform comparative sequence analysis emerge, it is important to develop parameters for such analyses and to examine the outcomes of cross-species comparison. Our analysis used gene prediction and a database search of 430 kb of genomic sequence covering the Bpa/Str region of the mouse X chromosome, and 745 kb of genomic sequence from the homologous human X chromosome region. We identified 11 genes in mouse and 13 genes and two pseudogenes in human. In addition, we compared the mouse and human sequences using pairwise alignment and searches for evolutionary conserved regions (ECRs) exceeding a defined threshold of sequence identity. This approach aided the identification of at least four further putative conserved genes in the region. Comparative sequencing revealed that this region is a mosaic in evolutionary terms, with considerably more rearrangement between the two species than realized previously from comparative mapping studies. Surprisingly, this region showed an extremely high LINE and low SINE content, low G+C content, and yet a relatively high gene density, in contrast to the low gene density usually associated with such regions.
[The sequence data described in this paper have been submitted to EMBL under the following accession nos.: Mouse Genomic Sequence: Mouse contig A (AL021127), Mouse contig B (AL049866), BAC41M10 (AL136328), PAC303O11(AL136329). Human Genomic Sequence: Human contig 1 (U82671, U82670), Human contig 2 (U82695).]
As significant amounts of the human genome and more recently the mouse genome are sequenced, the opportunity to use cross-species sequence comparison as an analytical tool becomes increasingly attractive. The premise for this analysis is that functionally important sequences will be strongly conserved, whereas other regions will differ as a result of mutations that have accumulated since the time when the species shared a common ancestor. The detailed analysis and comparison of sequence in conserved segments may aid our understanding of the genomic organization of complex genes and suggest candidate regulatory regions. It is also anticipated that it will provide new insights into chromosome and genome evolution, e.g., by defining the sequence content of chromosomal evolutionary breakpoints.
A number of comparative sequence studies have begun to demonstrate the value of this approach in gene annotation and regulatory element identification (Hardison et al. 1997). Comparative sequencing of a number of regions in mouse and human, including
the Btk locus on the mouse and human X chromosomes (Oeltjen et al. 1997),
a gene-rich cluster on human 12p13 and mouse 6 (Ansari-Lari et al. 1998),
the mnd2 region on human chromosome 2p13 (Jang et al. 1999),
the ADA gene region (Brickner et al. 1999),
the T-cell receptor locus (Koop and Hood 1994), and
the ERCC2 gene regions (Lamerdin et al. 1996),
has underlined the value of comparative sequencing for gene annotation.
With the completion of the sequence of human chromosome 22 (Dunham et al. 1999) and the rapid progress towards a working draft of the human genome, the opportunities for sequence comparison of human with mouse genome sequence will increase, emphasizing the need to develop parameters for cross-species sequence comparison and to document the outcomes over extensive regions of the genome. Ab initio gene prediction methods applied to the finished sequence of human chromosome 22 suggest that there are at least 100 genes in this chromosome for which there is no supporting evidence in the sequence databases (Dunham et al. 1999). Moreover, sequence analysis has highlighted a large surfeit of CpG islands, which are not associated with defined transcription units, and may represent uncharacterized genes. Comparative sequencing might be expected to make a major contribution to the detection and annotation of these undefined mammalian gene loci.
The comparative sequence approach represents a potential universal method for gene prediction that can be applied to any and every genome region. An evolutionary conserved region (ECR) that exceeds a defined threshold of sequence homology is likely to represent a functional element. We have applied such an approach, together with conventional gene prediction and homology searching methods, to identify potential genes in a region of the mouse and human X chromosomes. In so doing we have made use of extensive published studies of orthologous genes in man and mouse (Makalowski et al. 1996) as well as employing available data from annotated mouse and human genomic sequence regions to help us set meaningful parameters for the detection of ECRs.
The mouse region sequenced encompasses a pair of murine X-linked dominant disorders, bare-patches (Bpa), and striated (Str). The comparative analysis of this region has facilitated the identification of the Bpa and Str gene, Nsdhl, as described previously (Liu et al. 1999). We now report a full analysis of the sequence comparison of this mouse sequence region and its human counterpart. The analysis has revealed a number of conserved elements that may represent novel genes not revealed by database searching. Moreover, the region is considerably more rearranged between the two species than previously realized from comparative mapping studies, highlighting that some regions of mammalian genomes may be highly mosaic in evolutionary terms.
RESULTS AND DISCUSSION
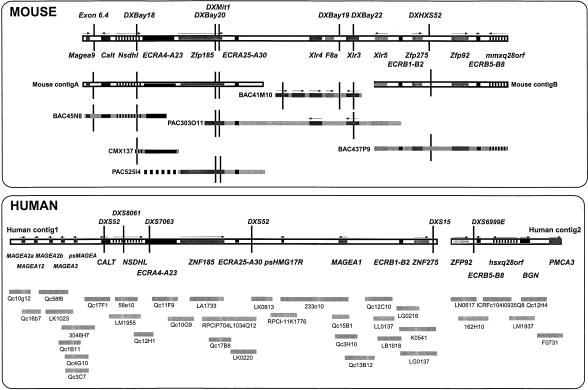
We have undertaken the sequencing of a 600-kb region that encompasses the Bpa and Str mutations on the mouse X chromosome. The critical region containing these two mutations is flanked by the loci DXHXS1104 and DXHXS52 (Levin et al. 1996). In addition, we have completed the sequencing of the homologous human region. In the mouse, cosmid contigs partially spanning the Bpa/Str critical region were already available (Chatterjee et al. 1994; Levin et al. 1996) and STS markers and previously identified genes were used to construct a BAC and PAC contig (see Methods). Thirteen markers were used to isolate clones and facilitate contig construction, resulting in 28 clones selected for further characterization. Fluorescent fingerprinting was used to assemble the minimal tiling path by comparison of overlaps between the clones. The mouse genomic sequence is enclosed in two contigs – A and B. Mouse contig A of 194 kb is assembled from three clones; BAC45N8, CMX137, and PAC525I4 and mouse contig B of 166 kb comprises BAC437P9 (Fig. 1). A central region separating mouse contigs A and B has also been sequenced. However, although a number of clones encompassing this interval were mapped, both STS content and fingerprint data suggested that this region demonstrated a high degree of instability in different clones. For example, whereas clone BAC41M10 from this region contained the marker F8a, this locus was absent from PAC303O11. This was confirmed by sequencing both these clones completely. PAC303O11 appears to encompass the whole region as its termini overlap mouse contig A and mouse contig B. Nevertheless, as expected the F8a gene was not contained within the finished sequence. The sequence from clone 41M10 contains as expected the F8a locus, but this clone is substantially rearranged with respect to PAC303O11.
Figure 1.
Clone- and STS-content map of the Bpa/Str critical region. Schematic representation of the gene and marker content of the mouse (top) and human (bottom) intervals. The available consensus gene and STS map assembled from genetic and physical mapping data is shown for the mouse and human regions, with assembled contigs (□) and sequenced clones (▪) displayed below. The dashed line part of clone PAC525I4 indicates the part of the clone not completed to a finishing stage as it had significant overlap with BAC45N8.
Originally, the human critical region was covered by a complete YAC map and cosmid map containing several gaps (Heiss et al. 1996). Two of these gaps were bridged by BAC/PAC clones. Despite all efforts, one gap remains between ZNF275 and ZFP92. Comparison of the available human sequence data to the mouse Zfp275–Zfp92 interval may provide a rough estimate of the gap size. If we assume no major human rearrangements, the gap may be about 20 kb composed of highly repetitive sequences. In total, the 32 cosmids, three BACS and one PAC span two genomic sequence contigs of 577 kb (Human contig 1) and 168 kb (Human contig 2) (Fig. 1).
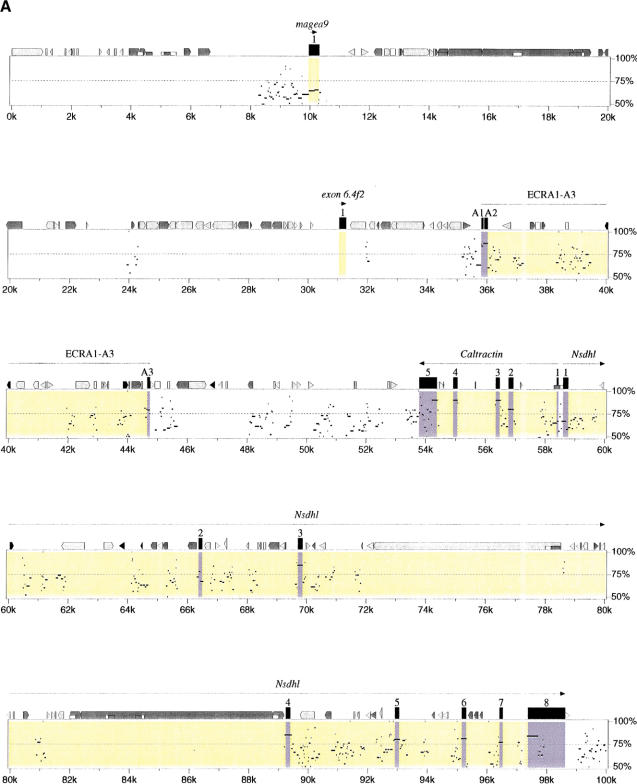
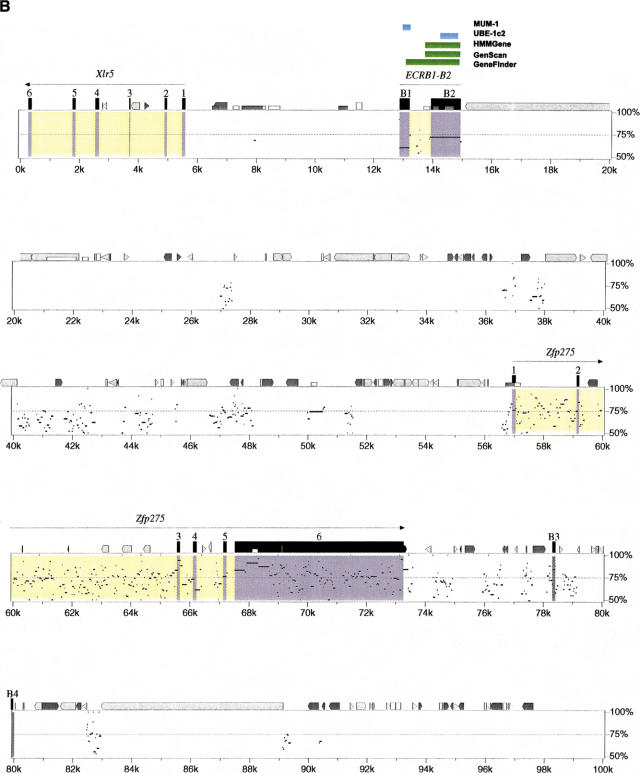
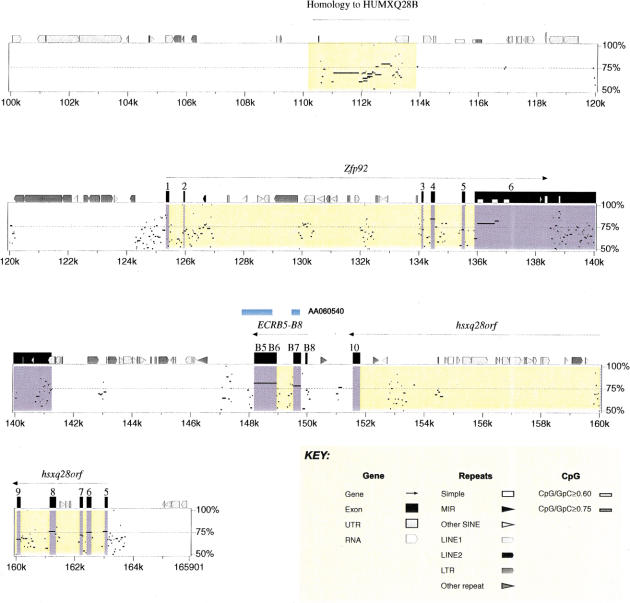
Extensive analysis using similarity searching and gene/exon prediction has enabled us to identify 11 genes in mouse, 13 genes and two pseudogenes in human, and to characterize their genomic structure (see Methods). The order, orientation, and conservation of these genes are displayed in the percent identity plot (PIP) (Fig. 2). In addition, we undertook extensive analysis of the sequence by pair-wise comparison to identify conserved gap-free alignments, which we have named ECRs that might represent additional unrecognized coding elements within the region. At least four potential novel genes were identified by this approach. We first describe those genes identified primarily on the basis of similarity searches and gene/exon predictions.
Figure 2.
(See pages 761–764.) PIP plot of mouse contig A (A) and mouse contig B (B). A PIP plot showing the mouse genomic sequence on the x-axis, and the percentage sequence identity (50%–100%) on the y-axis. Annotation is illustrated on the top of each main plot, with confirmed and putative exons depicted as numbered black boxes. The other icons along the top of the box depict repeats (grey pointed boxes are L1 repeats, light grey triangles are SINEs other than MIR, black triangles are MIRs, black pointed boxes are LINE2s, dark grey triangles are LTR elements, and dark grey pointed boxes are other kinds of interspersed repeats, such as DNA transposons, short dark grey boxes are CpG islands where the ratio CpG/GpC exceeds 0.75, short white boxes are CpG islands where the ratio CpG/GpC lies between 0.6 and 0.75). Specific annotation for each of the putative genes is illustrated above the computer-generated annotation, with gene predicitions shown as purple boxes, ORFs as green boxes, and sequence similarity as blue boxes.
Genes in the Bpa/Str Critical Region of Mouse and Human Identified through Similarity Searching and Gene/Exon Prediction
Melanoma Antigen Gene (MAGE) Family Cluster
MAGE genes encode tumor-specific proteins of unknown function, which are recognized by cytolytic T lymphocytes. A group of 12 genes, named MAGEA (1–12) have been previously located in the human Xq28 region and five other MAGE genes have been located elsewhere on the X chromosome (De Plaen et al. 1994; Rogner et al. 1995).
Five human MAGE genes and one mouse Mage gene have been identified from the genomic sequence generated in the critical region. A cluster of four MAGE genes are located in human contig 1 proximal to CALT (Fig. 1). Using the human contig 1 genomic sequence and published cDNAs, the gene structures of all four MAGE genes in the region have been characterized. The cluster contains two inverted copies of MAGEA2 (MAGEA2a and MAGEA2b) flanking the MAGEA12 gene. As with other MAGE genes, they contain a large 3′ coding exon and a variable number of 5′ UTR exons (MAGEA12:1, MAGEA2:3). The two MAGEA2 genes are part of an inverted repeat, which itself appears to consist of two parts, a constant segment (14 kb at 99.9% identity) and a comparatively variable segment (4.4 kb at 91.66% identity). Moreover, MAGEA3 has been detected in this cluster. Sequence comparison of overlapping bacterial clones containing this gene shows that MAGEA6 is an allelic variant of MAGEA3 (M. Platzer, unpubl.). In addition to these previously identified MAGE genes, a putative MAGEA pseudogene named psMAGEA exists in the last portion of the cluster and was identified through sequence similarity to MAGEA12.The open reading frame (ORF) of this pseudogene is truncated by a 2-bp deletion after codon 112. Another human MAGE gene family member, MAGEA1, lies >300-kb distal of this cluster (see Fig. 1). MAGEA1, consisting of two exons (5′UTR and coding) had been mapped previously proximal to GABRA3 (Rogner et al. 1995).
A putative murine Mage gene has been identified at the proximal end of mouse contig A. Similarity searching and exon prediction has enabled us to identify the putative 3′ coding exon which has an ORF of 729 bp encoding a putative peptide of 243 aa in length. FASTA alignment of this protein sequence with human proteins from the A, B, and C families shows highest similarity to the human MAGEA family and so we have named it Magea9 (Table 1). FASTA alignment of this protein sequence with the mouse proteins from the Magea family shows highest similarity to Magea8 and Magea3 (Table 1). To date, 11 murine Mage genes have been identified; three of which (Mageb3, Mageb1, and Mageb2) have been postulated to be the murine equivalents of the MAGEB genes (De Backer et al. 1995). The other eight Mage genes (Magea1–8) display a higher degree of similarity with the MAGEA ORFs than the MAGEB or MAGEC ORFs and, like MAGEA proteins, they are acidic (De Plaen et al. 1999). The putative Mage gene identified here is the first Magea murine gene to demonstrate local conservation of synteny with human. However, it shows most significant similarity to the human MAGEA8 gene and we cannot conclude that it is an ortholog of any of the human MAGE genes identified in the corresponding human region so far.
Table 1.
Comparison of Percentage Amino Acid Identity of Murine MAGEA9 Gene with Known Human and Mouse MAGE Genes Using FASTA
| Name | % Identity |
|---|---|
| Human genes | |
| MAGEA1 | 33.9% |
| MAGEA2 | 35.7% |
| MAGEA3 | 36.1% |
| MAGEA4 | 36.5% |
| MAGEA5 | 32.8% |
| MAGEA6 | 36.1% |
| MAGEA8 | 38.7% |
| MAGEA9 | 36.4% |
| MAGEA10 | 38.0% |
| MAGEA11 | 37.1% |
| MAGEA12 | 34.5% |
| MAGEB2 | 29.5% |
| MAGEC1 | 29.0% |
| Mouse genes | |
| MAGEA1 | 43.7% |
| MAGEA3 | 44.5% |
| MAGEA4 | 40.7% |
| MAGEA5 | 45.0% |
| MAGEA6 | 43.7% |
| MAGEA7 | 36.8% |
| MAGEA8 | 44.5% |
Caltractin and NAD(P)H Steroid Dehydrogenase-like Gene
Calt belongs to a family of calcium-binding proteins and is a structural component of the centrosome (Chatterjee et al. 1995). NAD(P)H steroid dehydrogenase-like gene (Nsdhl) encodes a novel 3β-hydroxysteroid dehydrogenase (3β-HSD) and was identified as the gene mutated in Bpa and Str mice (Liu et al. 1999). The gene structures of Nsdhl and Calt were determined from the comparative analysis of human and mouse genomic sequence, partial cDNAs, and ESTs. Calt and Nsdhl [originally designated H105e3 (Levin et al. 1996) or XAP104 (Heiss et al. 1996) in human] are arranged in a head-to-head orientation. Only 211 bp of mouse genomic sequence separates the 5′ UTRs. Both 5′ exons of each gene span a predicted CpG island that overlaps the 211-bp gap (Fig. 2).
Calt and Nsdhl are conserved in both orientation and exon organization between mouse and man, showing no gross differences in intergenic or intronic distances (Fig. 2). The coding (CDS) exons of Calt and Nsdhl are highly conserved with percentage identities for gap-free alignments extending from 74%–98%. With the exception of exon 2 of Nsdhl, the gap-free alignments extend the full length of the exon and splice junctions are conserved (see Fig. 2). A different pattern is observed in the UTRs of both genes, with the identified gap-free alignments being shorter and having lower percentage identity.
Zinc Finger Protein 185
The mouse Zfp185 and human ZNF185 are group 3 LIM domain proteins (Heiss et al. 1997). Group 3 LIM domain proteins contain between one and five LIM domains in the carboxyl terminus. Heiss et al. (1997) postulate that the composition of ZNF185 and Zfp185 indicates a function in regulating cellular proliferation and/or differentiation. Zfp185 has 18 exons, identified by alignment with the murine cDNA. Zfp185 has an ORF of 1056 bp encoding a protein of 352 aa and ZNF185 has 20 exons with an ORF of 1359 bp encoding a protein of 453 aa. The human gene structure is in agreement with that determined by Heiss et al. (1996, 1997) (Fig. 3a) except for an additional two 5′ UTR exons determined from mouse–human sequence comparison (exon 1 and 2).
Figure 3.
Gene structure and clustal alignment of ZNF185 and Zfp185. (a) Representation of the exon–intron organization of the human ZNF185 and mouse Zfp185 genes The first coding exon that contains the ATG codon is depicted. (b) FASTA alignment of ZFP185 and ZNF185, with the exons depicted for each gene.
The determination of the mouse gene structure allowed a complete and comprehensive comparison of the mouse and human gene structure. In addition to duplicated exons 13 and 14, which are only present in human, this comparison allowed us to demonstrate that the mouse lacks an exon corresponding to human exon 12 (Fig. 3b). Although gene structure is not completely conserved, high sequence similarity is observed between mouse and human in the exons (mouse exons 15 and 16) encoding the LIM domain (see Fig. 2). Other conserved gap-free alignments are present 5′ and 3′ of the Zfp185 and ZFP185 genes and may represent additional UTR sequences (see Fig. 2).
High Mobility Group Protein 17
The high-mobility group (HMG) proteins are the most abundant nonhistone chromosomal proteins in the nuclei of higher eukaryotes. HMG17, and the closely related protein HMG14, bind preferentially to the nucleosomal core particle and may modulate the chromatin configuration of transcriptionally active genes (Bustin et al. 1990). The mouse and human genome contain multiple copies of sequences homologous to the cDNA coding for HMG-17. Sequence analysis of the human genes by Srikanthat et al. (1987) showed that they were flanked by short repeats typical of processed retropseudogenes.
A predicted pseudogene for HMG protein HMG17 was identified in human contig 1 (psHMG17). No sequence similarity, however, is evident in the mouse genomic sequence generated across this region. This pseudogene is nearly a complete reverse transcribed copy of the HMG17 mRNA and is conserved at 86% identity over the entire region and at 90% identity over the translated region. The gene contains no introns, no initiation codon in the predicted position, and one in-frame stop codon.
X-linked Lymphocyte-regulated (Xlr) Related Genes
Xlr3a AND Xlr3b
The murine Xlr multi-gene family was originally identified by subtractive cDNA hybridization and cloning (Cohen et al. 1985). Various studies have shown that human genomic DNA does not cross-hybridize with murine Xlr probes, showing that at the DNA level, the Xlr family does not appear to be conserved between mouse and man. However, using anti-Xlr antibodies, Allenet et. al. (1995) characterized an Xlr-immunoreactive nuclear protein in human RAJI B-lymphoblastoid cells by flow cytofluorimetry, immunoblotting, and immuno-cytolabeling. Xlr3a and Xlr3b are two virtually identical genes, which represent a subfamily of X-linked lymphocyte-regulated genes (Bergsagel et al. 1994). Hybridization results had indicated previously that three copies of the Xlr3 subfamily lie within the critical region (Levin et al. 1996). However, because of the high sequence identity between family members, the relative locations of Xlr3a or Xlr3b were not determined. Xlr3a and Xlr3b differ by only 11 nucleotides in their coding sequence, representing eight amino acid differences. Genomic sequencing has identified Xlr3 subfamily genes in the central interval of the critical region in mouse. Clone 41M10 contains two Xlr3 genes, whereas 303O11 contains only one Xlr3 gene, emphasizing the apparent instability of clones from this region (see above).
The available sequence data from these two clones indicates, however, that the Xlr3 genes are split into nine exons, have an ORF of 678 bp encoding a protein of 226 aa and cover ∼10.7 kb of mouse genomic sequence. They are basic proteins and demonstrate significant homology to the other members of the Xlr family. Rigorous database searching identified one human EST (GenBank accession no. AI075991) with similarity to Xlr3a. However, this human EST may represent the human ortholog of hamster Cor-1 (meiotic chromosome core protein) rather than mouse Xlr3a, as it has a more significant similarity to Cor-1. No related sequences were detected in the corresponding human sequence contigs.
Xlr4
Sequence generated in this central interval in mouse also allowed the identification of an additional Xlr family member, subsequently named Xlr4. Two copies of Xlr4 were identified in 41M10, whereas only one copy of the gene was found in clone 303O11, again reinforcing the apparent clone instability in this region. Full-length cDNA sequence for Xlr4 was obtained through sequencing an EST (GenBank accession no. AA472809) initially identified by similarity searching. The cDNA is 1290 bp in length and alignment of this cDNA to genomic sequence identified that Xlr4 has nine exons, covering 8.5 kb of mouse genomic sequence. Xlr4 has a similar exon–intron structure to the Xlr3 genes in the region. Xlr4 has an ORF of 636 bp, encoding a putative protein of 212 aa. ProfileScan identified a potential bipartite nuclear localization signal (Prosite accession no. PS50079) in Xlr4. Xlr4 has a predicted pI of 9.51 and is therefore a basic protein like the Xlr3 subfamily. Over the full length of the protein, Xlr4 has 31% identity to Xlr3a and 25% identity to Xlr1 (Table 2). No significant similarities at the DNA or protein sequence level were identified in the human sequence databases. Furthermore, no related human sequences were identified in the corresponding human sequence. Xlr4 appears to define a new Xlr subfamily.
Table 2.
FASTA Comparison of the XLR/SCP3 Gene Family
| XLR3A | XLR3B | XLR4 | XLR1 | SCP3 | XLR5 | |
|---|---|---|---|---|---|---|
| XLR3A | 100.0% | 100.0% | 30.5% | 24.5% | 29.3% | 23.6% |
| XLR3B | 100.0% | 30.5% | 24.5% | 29.3% | 23.6% | |
| XLR4 | 100.0% | 16.6% | 24.3% | 27.0% | ||
| XLR1 | 100.0% | 38.7% | 19.9% | |||
| SCP3 | 100.0% | 28.4% | ||||
| XLR5 | 100.0% |
Xlr5
Analysis of mouse contig B identified a further Xlr family member, based on EST sequence similarity and exon prediction. GENSCAN and HMMGene predicted a gene comprised of six exons. Several of these predicted exons overlapped exons predicted by Grail. An additional two exons of this gene were also detected in PAC303011. The assembled gene structure has an ORF of 708 bp, coding for a putative protein of 236 aa. A scan of the predicted protein sequence against PROSITE did not identify any functional domains. Database searches using BLASTP against Swiss–Prot identified homology to murine XLR3A, murine XLR3B, hamster SCP3 (Synaptonemal Complex Protein 3), rat SCP3, murine SYCP3, and murine XLR1. FASTA comparisons of this group of proteins showed that XLR5 demonstrated relatively low identity with other members of the XLR family; XLR5 showed highest identity with SCP3 (Table 2). Expression studies were performed to deduce the expression profile of this putative gene. RT–PCR assays between exon 1 and 2 on testis RNA detected the expected cDNA product (data not shown). Northern blot analysis of a probe amplified between exons 1 and 5 in testis cDNA detects a major transcript of 1.5 Kb in testis (data not shown). Finally, no related human sequences were identified in the corresponding human sequence contigs. Again Xlr5 appears to define a new Xlr subfamily.
Factor VIII-associated Gene
The human F8A gene was originally identified within intron 22 of the F8 gene (Levinson et al. 1992). It is a small (<2 kb), intronless gene and is GC-rich. There are two additional copies of F8A present in human, located telomeric of F8 in Xq28. However, pulsed-field mapping data has indicated that the mouse F8a gene is not located within F8 (previously called Cf8) (Faust et al. 1992). Further studies have demonstrated that F8a lies ∼200 kb proximal to the DXHXS52 locus (Chatterjee et al. 1994) (see Fig. 1). As expected from PCR analysis, F8a was identified within the sequence from the 41M10 clone, but not found in the sequence from clone 303O11. As would be expected from previous human mapping data, no F8A sequence was detectable in the corresponding human sequence contig.
Zinc Finger Protein 275
A new zinc finger protein gene, Zfp275, was identified distal of F8a in mouse contig B. A ZNF275 ortholog was also identified in human contig 1. Very few ESTs cover this coding region. However, the gene structure of exons 1–5 was determined utilizing a mouse partial cDNA clone. The terminal end of the 6th exon was defined by two mouse ESTs that included the polyA tail (GenBank accession nos. AA189691 and AA833132). The alignment of the available consensus mouse cDNA sequence with genomic DNA defined a gene of six exons, with the 6th exon being 6 kb in length. The gene structure in mouse and human is conserved with an ORF of 1392 bp encoding a protein of 464 aa. Analysis of this protein sequence showed that it contains 11 zinc finger motifs. The coding exons of Zfp275 are highly conserved between mouse and human. A different pattern of conservation can be observed in the 3′ UTR, with most of the conservation consisting of short gap-free alignments. It is apparent from the PIP plot that repeat density in the Zfp275 and ZNF275 gene regions is very low and overall sequence conservation is high. For example, conservation around exons 3 and 4 extends beyond the exons into intronic sequence. Expression studies using Northerns demonstrated a double band at about 7 kb in polyA+ mRNA from ES cells and embryos from E10.5–E18.5 (data not shown). RT–PCR within exon 6 detected signals in adult brain, kidney, heart, thymus, and spleen, all showing the same expected size in cDNA and genomic (data not shown).
Zinc Finger Protein 92
The Zfp92 gene was originally identified in the distal part of the DXHXS1104-DXHXS52 region in both human and mouse (Levin et al. 1996). Alignment of the Zfp92 cDNA with the mouse genomic sequence along with the identification of ESTs has facilitated the characterization of the genomic structure. This gene consists of six exons covering 15 kb of mouse genomic sequence, and has an ORF of 1464 bp encoding a putative protein of 488 aa. Exon 6 is 5.3 kb in length as defined from mouse–human homology and a rat EST (GenBank accession no. AI227988). It is a member of the large Kruppel gene subfamily of zinc finger proteins and contains the canonical KRAB A and KRAB B boxes at its amino-terminal end and eight consecutive zinc finger motifs. Analysis of the human sequence contig facilitated the prediction of the human gene, again comprising of six exons. Seven human ESTs confirm the terminal 2.6 kb of the predicted large final exon in human, which is therefore predicted to be 5.8 kb in length. The human ORF identified encodes a putative protein of 416 aa. The human ZFP92 protein is 66% identical to mouse ZFP92 over 374 aa. The human putative protein, like the mouse protein, appears to contain eight zinc finger motifs and the KRAB A and KRAB B boxes.
hsxq28orf /mmxq28orf
Significant sequence similarities were identified at the distal end of mouse contig B with a human cDNA (hsxq28orf or STS1769, GenBank accession no. X99270) in GenBank. It became apparent that this cDNA is chimeric due to the fact that its 5′ end (from 4–504 bp) has a 100% identity to GDP-D-mannose-4, 6-dehydratase mRNA (GenBank accession no. AF040260) and that this gene maps to 6p25. The 3′ end of this gene in human can be confirmed by many ESTs, however the 5′ end is presently defined by one EST (GenBank accession no. T66063). The human cDNA compiled from this data is 1.365 kb in length and identifies 10 exons. The homologous mouse cDNA as defined by ESTs, appears to be 1.363 kb in length. Exons 5–10 are in mouse contig B. The predicted peptide in mouse is 353 aa and 358 aa in human, showing 65% identity to each other and no significant similarity to anything in the protein database. Expression studies have shown that the expected 650 bp RT–PCR product from exon 4–9 was identified in all of the adult tissues tested. However, in addition a 300 bp-smaller product was identified and appears to be due to alternative splicing between exon 4 and 8 (data not shown). Northern blot analysis of this gene detects two bands in testis, with the larger being the expected 1.3 kb size (data not shown).
Novel Genes in the Bpa/Str Region Identified by Their Conservation with Human Sequence
With the aim of identifying additional putative genes, the mouse and human genomic sequences were analyzed extensively by pairwise comparison. At the outset of this analysis it was important to identify meaningful thresholds for the detection of ECRs that may represent undiscovered coding sequence. A previous study of 1196 orthologous mouse and human full-length mRNA sequences has described statistical distributions of sequence conservation in translated and untranslated regions (Makalowski et al. 1996). Our aim, therefore was to characterize these statistical distributions at a genomic level and to define the thresholds to be used to identify ECRs in novel genomic sequence. We examined the sequence conservation in coding (CDS) and noncoding exons (UTRs) of previously annotated genomic regions of the mouse and human (see Methods). Six mouse and human regions were chosen for this study:
mnd2 nonrecombinant region of human chromosome 2p13 and mouse chromosome 6 (Jang et al. 1999),
Gene-rich cluster on mouse chromosome 6 and its syntenic region on human chromosome 12p13 (Ansari-Lari et al. 1998),
BTK locus (Oeltjen et al. 1997),
ADA gene region (Brickner et al. 1999),
T-cell receptor locus (Koop and Hood 1994), and
ERCC2 gene regions (Lamerdin et al. 1996).
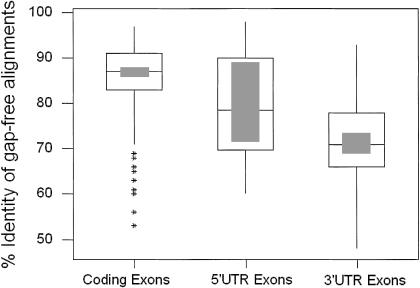
These regions comprised 581 kb of mouse genomic sequence and 595 kb of human genomic sequence. Overall, the regions chosen contained 32 annotated genes present in both the mouse and human. Comparison of mouse and human sequence from these six regions using BLASTZ with the standard parameter settings (Schwartz et al. 1999) and manipulation of its output resulted in the identification of 283 gap-free alignments overlapping known coding exons, 24 gap-free alignments overlapping known 5′ UTRs and 40 gap-free alignments overlapping known 3′ UTRs (http://www.mgc.har.mrc.ac.uk/comp_seq/reference.html). This reference set of gap-free alignments was determined using thresholds of 50-bp length and 50% identity. The percentage identity distributions for gap-free alignments in each category is illustrated in Figure 4. The analysis indicates that the distribution of percentage identity is broad for 5′ UTRs but more narrowly distributed for 3′ UTRs and coding exons. The average identity for 5′ UTRs is 79.08% (SD=11.65), 74.05% (SD=9.62) for 3′ UTRs, and 84.31% (SD=8.40) for CDS. In the study by Makalowski et al. (1996) the average percentage identity for 5′ UTRs is 67.47% (SD=13.2), 69.13% (SD=12.4) for 3′ UTRs , and 84.62% (SD=6.78) for CDS. Because each of the above studies was performed independently using different alignment algorithms, the results cannot be compared directly. Nevertheless, both studies indicated clear differences in the range of percentage identity observed between CDS and UTR regions and have aided us in setting parameters for the identification of ECRs.
Figure 4.
Distribution of percent identities of ECRs in coding and noncoding regions of mouse and human genomes. A box-plot showing the distribution of percentage identity in gap-free alignments identified from three categories: 5′ UTRs, coding exons (CDS), and 3′ UTRs. The interquartile range is depicted as the open box with the median being shown as the line in the middle of this box. The shaded box indicates the 95% confidence interval. Outliers are shown as stars.
We have also assessed studies of exon size to assist us with setting parameters for identifying ECRs. A large study of human exon size has demonstrated that there is little constraint on exon length, the smallest identified being 15 bp (Zhang 1998). However in this study, which categorized exons into a number of different groups, the smallest average exon size (100 bp), was found in the iuexon (internal-untranslated) category. Nevertheless, to ensure that we identified the majority of exons, but also reduced noise from short conserved noncoding sequences, we set an arbitrary lower limit of 50 bp for the identification of ECRs. Taking together both percentage identity and exon size studies, we have defined two categories of ECR: category 1 [those with a percentage identity >80% and length >50 bp (http://www.mgc.har.mrc.ac.uk/comp_seq/category1.html)], and category 2 [those with a percentage identity >70% and a length >50 bp (http://www.mgc.har.mrc.ac.uk/comp_seq/category2.html)]. Clearly category 1 ECRs have a higher likelihood of representing true coding regions, as the 80% cutoff differentiates well between CDS and UTRs on the basis of both our analysis and that of Makalowski et al. (1996). Utilizing these filters and aided by visual interpretation from the PIP plot, we identified at least four further putative transcription units in the Bpa/Str region. It should be noted at this point that the visual interpretation of the plot identified additional ECRs not present in either category 1 or 2.
Identification of ECRs in Mouse Contig A
Analysis of mouse contig A identified 24 ECRs in category 1 and an additional 60 ECRs in category 2 not overlapping with previously annotated exons. From the 24 category 1 gapfree alignments, 20 (ECRA4–ECRA23) were identified in a 37-kb region between Nsdhl and Zfp185 (Fig. 2). The other four category 1 ECRs (ECRA1, A2, A3, and A24) are annotated on Figure 2 and may represent further transcription elements, but analysis did not uncover any other evidence to suggest they represent genes.
ECRA4–ECRA23
Following identification of these 20 ECRs, database searching identified a human EST (GenBank accession no. AI653754) with homology to ECRA20. None of the other ECRs showed matches to the nonredundant DNA database. The human ECRA20 sequence shows 100% identity to the EST whereas mouse ECRA20 shows 88% identity. An ORF was identified in seven of the ECRs (ECRA7, A8, A9, A17, A18, A20, and A23). Eight of the ECRs (ECR A9, A10, A17, A18, A19, A21, A22, A23) also have overlapping exon predictions, although no more than one package predicted each exon. ECRA18 and A19 also overlap a predicted CpG island (see Fig. 2).
Preliminary expression studies were carried out on six of the ECRs (ECR A4, A10, A11, A17, A20, and A23) using RT–PCR (see Methods). Of the seven regions identified with an ORF, three (ECRA17, 20, and 23) were tested by RT–PCR and two of these (ECRA20 and ECRA 23) gave positive results. ECRA20 was positive by RT–PCR in embryonic days 9.5, 11.5, 13.5, 15.5, and 16.5, neonate skin and in adult brain, heart, kidney, thymus, and testis (data not shown). ECRA23 is positive by RT–PCR in embryonic days 9.5, 11.5, 13.5, 15.5 and 16.5, neonate skin and in adult kidney, spleen, and testis (data not shown). Three other ECRs were tested (ECRA4, 10, and 11), one of which had an overlapping predicted exon (ECR10) and one of these (ECRA4) was shown by RT–PCR to be expressed. ECRA4 was positive by RT–PCR in 15.5d embryo, adult kidney, spleen, and thymus. We have not clarified if these putative exons can be connected into single or multiple transcription units. Nevertheless, several strands of data suggest the presence of at least one gene in this region: (1) the presence of multiple ECRs, (2) RT–PCR data from ECRs; and (3) the discovery of a single EST with matches to ECRA20.
ECRA25–ECRA30
PIP plots also identified a region of high overall conservation between Zfp185 and the end of mouse contig A. This island of conservation lying between 175 kb and 179 kb of mouse contig A is composed of six category 2 gapfree alignments (ECRA25–ERCA30) with percentage identities ranging from 70%–78% and lengths from 68–455 bp. GENSCAN, HMMGene, Grail, and Genemark all predicted a combination of exons over this island of conservation in both mouse and human. Both GENSCAN and HMMGene predicted a gene overlapping ECRA26, A27, and A28. ORFinder predicts a single ORF encoding a putative protein of 499 aa in mouse and 448 aa in human. BLAST analysis of the murine putative protein identified sequence similarity to Homo sapiens KIAA0883 (GenBank accession no. AB020690) at 38% identity over 328 aa and also to Homo sapiens paraneoplastic antigen MA1 (PNMA1) (GenBank accession no. NM_006029) at 34% identity over 309 aa. The first 102 aa of this putative protein also has 35% identity to part (311 bp) of HUMXQ28B (GenBank accession no. M89986), an anonymous X-linked STS. ECRA26 and ECRA27 also have similarity to a Rat EST (GenBank accession no. AI549430), at 91% identity and a predicted CpG island is identifiable over ECRA26. In summary, the comparative analysis indicates the presence of a putative gene with one coding exon encoding a protein of 499/448 aa in mouse and human.
Identification of ECRs in Mouse Contig B
Analysis of contig B identified 11 ECRs in category 1 and 56 ECRs in category 2 not overlapping with previously annotated exons. Eight of the category 1 ECRs were analyzed in this study; the others were excluded because they were identified within intronic sequence. ECRB3 and B4 are category 1 ECRs annotated on the PIP plot and may represent additional transcription units but, at present, we have not uncovered any other supporting evidence to indicate they represent genes.
ECRB1–ECRB2
Two ECRs, named ECRB1 and ECRB2, are localized between the Xlr5 and Zfp275 loci. ECRB1 was identified from the PIP as it was only 60% identical over 337 bp. However, an adjacent sequence, ECRB2, was identified as category 2, as it is 74% identical over 998 bp of mouse and human sequence. ECRB2 overlaps an exon predicted by GeneFinder, GENSCAN, and HMMGene. The predicted murine gene codes for a putative 528 aa protein that has 49% identity over 250 aa to mouse UBE-1c2 (GenBank accession no. AB030505). A search using ProfileScan also identified a bipartite nuclear localization signal (Prosite accession no. PDOC00015), between amino acids 385 and 402. ECRB1 is similar to melanoma ubiquitous mutated protein (MUM-1; GenBank accession no. U20896), at 44% identity over 127 aa. MUM-1 is a mutated intron sequence that codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. The gene is expressed in many normal tissues.
ECRB5–ECRB8
A second region of conservation was identified in mouse contig B between Zfp92 and hsxq28orf (Fig. 2). Four ECRs from category 1 and 2 were identified in this region (ECRB5, B6, B7, and B8), with percentage identities ranging from 70%–81% and lengths from 61–412 bp. Analysis of the nonredundant DNA database subsequent to the identification of ECRB5 and B4 identified matches to one mouse EST (GenBank accession no. AA060540). The EST is 1085 bp in length and has a polyA tail and polyadenylation signal at the 3′ end. The available EST sequence suggests a gene structure comprising at least two exons of 114 bp and 969 bp. Subsequent database searches have identified other mouse ESTs to support this organization (GenBank accession nos. AI595465, AI427513, AV021721) (see Fig. 2). The identified ECRs agree well with the proposed gene structure. The predicted ORF is 369 bp in length encoding a protein of 122 aa. This putative protein shows no matches to Swiss–Prot. RT–PCR of cDNA from adult tissue using primers from exon 1 and 2 detects the expected spliced product exclusively in skin. Northern blots of total RNA from adult tissues detected a 1.1-kb transcript only in skin (data not shown).
Repeat and Gene Distribution
In the mouse genomic sequence generated to date, 34.85% is composed of repetitive elements as identified by RepeatMasker, compared to 36.55% of the human sequence. The number of repetitive elements identified in each class is summarized in Table 3 along with the percentage of sequence occupied by each class. Moreover, we aligned mouse and human genomic sequences without repeat masking to identify orthologous repetitive elements. There appeared to be fewer mammalian interspersed repeat (MIR) relics in the mouse sequence and eight out of nine of these were identified in aligned regions, suggesting their presence predated mouse–human divergence some 80 million years ago.
Table 3.
Distribution of Repetitive Elements
| Repeat | Number of elements (% of human sequence) | Number of elements (% of mouse sequence) |
|---|---|---|
| SINES | ||
| Alu | 130 (4.68%) | |
| B1s | 61 (1.93%) | |
| B2–B4 | 86 (3.78%) | |
| ID | 8 (0.16%) | |
| MIRs | 67 (1.29%) | 9 (0.27%) |
| LINEs | ||
| LINE1 | 142 (12.90%) | 83 (16.35%) |
| LINE2 | 78 (3.64%) | 17 (0.56%) |
| LTR elements | ||
| MaLRs | 81 (5.09%) | 62 (4.80%) |
| Retroviral | 30 (2.40%) | 19 (4.14%) |
| MER4_group | 29 (2.69%) | 1 (0.06%) |
| DNA elements | ||
| MER1_type | 32 (0.88%) | 9 (0.46%) |
| MER2_type | 12 (0.94%) | 1 (0.05%) |
| Mariners | 2 (0.04%) | 0.00 |
| Small RNA | 3 (0.03%) | 4 (0.07%) |
| Simple Repeats | 141 (1.58%) | 86 (1.42%) |
| Low complexity | 47 (0.50%) | 35 (0.60%) |
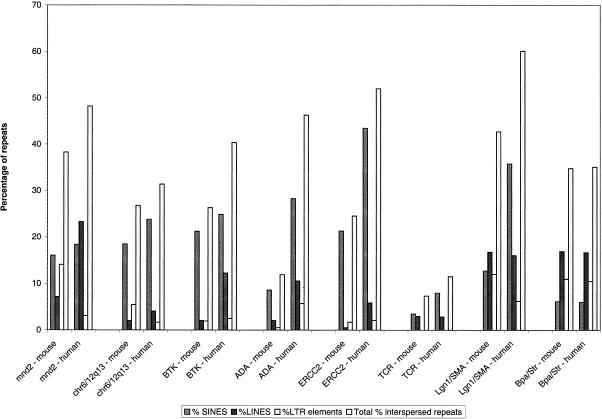
The frequency of LINE elements in the Bpa/Str region is significantly higher compared to most other regions of the mouse genome that have been sequenced (Fig. 5), the exception being the recently sequenced Lgn1/SMA region (Endrizzi et al. 1999). Perhaps most interestingly, the frequency of LINE elements is significantly greater than SINE elements. Only the T-cell receptor cluster (Koop and Hood 1994) shows a lower frequency of SINE elements. A similar pattern of LINE and SINE distribution was identified in the homologous human sequence. The high frequency of LINE elements compared to SINE elements is consistent with the relatively low G+C content of the mouse sequence from the Bpa/Str region (43.7%). This compares with an overall G+C content calculated from all available mouse genomic sequence of 45.6%. Most surprising, however, is that given the high LINE/low SINE content and low G+C content, the gene density across the Bpa/Str region is relatively high. Excluding those putative transcription units identified by mouse/human sequence comparison, a total of 11 genes have been definitively identified in the Bpa/Str region spanning ∼430 kb. This gives a gene density of 1 every 40 kb. In contrast, the human G+C content is more heterogeneous. Human contig 1 has a G+C content of 46.6% against a G+C content in human contig 2 of 57.3%. This difference is reflected in the gene densities of 1/64 kb and 1/42 kb, respectively. However, it is worth noting that the bulk of the mouse genomic sequence is spanned by human contig 1. A variety of evidence suggests that vertebrate genomes are a mosaic of isochores of differing G+C content, repeat and gene distribution (Bernardi 1995). G+C-rich isochores are rich in SINE elements and genes, whereas the G+C-poor isochores are gene poor and relatively depleted in SINE elements. G+C rich isochores predominantly localize to R-bands, whereas G+C-poor isochores are enriched in G-bands. In situ hybridization studies demonstrate that SINEs appear to be largely localized in R-bands and LINEs in G-bands (Boyle et al. 1990). However, this distribution was not evident on the mammalian X chromosome, which appeared to be abundantly rich in LINEs such that R-bands are virtually obscured. The Bpa/Str sequence region characterized here contrasts with a simplistic notion of a mosaic of two isochores in the mammalian genome. Given the high LINE/low SINE content and relatively low G+C content of the Bpa/Str region, it might be expected to show a lower gene density than was observed. The high LINE content of the region is consistent with the high frequency of LINE elements observed on the mammalian X chromosome. However, a similarly high frequency of LINE elements was not observed in genomic sequence from the Btk region of the mouse X chromosome (Oeltjen et al. 1997).
Figure 5.
Repeat distribution in various mouse and human genomic regions. Histogram showing the percentage of genomic sequence in various mouse and human regions occupied by SINEs, LINEs, LTR elements, and total interspersed repeats.
Conclusions
We have sequenced 430 kb from the mouse Bpa/Str critical region and 745 kb from the homologous region of the human X chromosome. Sequence from each species was subjected to gene prediction and homology searches to identify potential genes. These analyses had allowed us previously to undertake a comprehensive search for candidate genes for the bare-patches and striated mutants and ultimately lead to the identification of causative mutations (Liu et al. 1999). We also identified eight genes in mouse and human sequence not found previously by exon trapping or cDNA selection. These include a member of the melanoma antigen gene family (Magea9), two novel members of the X-linked lymphocyte-regulated family (Xlr4 and Xlr5), and a zinc-finger gene (Zfp275). However, additional analyses employing comparisons of mouse and human sequence allowed us to identify at least four potential additional genes based on their evolutionary conservation.
Using available genomic sequence from a variety of mouse and human regions, we developed an approach for the identification of ECRs that was likely to represent coding sequence. We searched for gapfree alignments of either 70% or 80% identity and with a minimum length of 50 bp. Our analyses of previously determined mouse and human genome sequence indicated that such gapfree alignments had a high probability of representing coding sequences. Using annotated PIP plots and these thresholds of sequence similarity, we identified four further potential transcribed regions in the 430 kb of mouse sequence analyzed: ECRA4–23, ECRA25–30, ECRB1–2, and ECRB3–6.
It appears that this approach provides a potentially significant enhancement to the process of identifying putative genes. For example, for the putative gene ECRA4–23, only one ECR homologous to an EST was identified and, though eight of the ECRs had overlapping exon predictions, no more than one package predicted each exon. It appears that the identification of ECRs has the potential to provide a much richer view of the putative gene sequences in this region and, indeed, this was confirmed by the demonstration that a number of the ECRs are transcribed. For example, ECRA4 is transcribed, yet neither similarity to ESTs or exon prediction would have highlighted these sequences as potential transcription units. Equally, ECRB5–B8 is another example of a candidate gene revealed by sequence comparison where evidence from transcription studies has subsequently emerged to substantiate the presence of a real transcription unit. It will remain to be seen if the untested ECRs in ECRA4–A23 are transcribed and if the two other ECR regions that have been identified do indeed represent true genes.
Although we employed relatively low thresholds (70% and 80%) to identify ECRs, a surprisingly low level of noise was present with only 35 category 1 and 116 category 2 ECRs being identified in the 430-kb region studied. Moreover, the cumulative data suggested the presence of only four additional transcription units that would not have been detected by homology searching and exon prediction methods. For two of these putative transcription units, we have provided evidence that they are transcribed. The results demonstrate that the approach we have employed to date is a productive means of identifying putative exons that may remain undetected by gene prediction and similarity searching. It would seem likely that with further enhancements to this method, the process of comparative sequence analysis could be made even more discriminating. This work and the comparative studies of others (Koop and Hood 1994; Lamerdin et al. 1996; Oeltjen et al. 1997; Ansari-Lari et al. 1998; Brickner et al. 1999; Jang et al. 1999) underlines the immense potential value of mouse genomic sequence as a means of annotating the human genome.
One cautionary note should be made, however, in that the Bpa/Str region appears to be a mosaic in evolutionary terms. It contains genes apparently specific to one species, members of the Xlr family, other genes that are conserved at the sequence level, but disrupt conservation of gene order, such as F8a, and others that show extremely high conservation of sequence and structure, such as Nsdhl. This may well be true of other parts of the genome and must be kept in mind when comparing the “working draft” mouse genomic sequence (to be available by 2003) with finished, high-quality human sequence. For these reasons, it would seem best to adopt a strategy of both clone-based and genome-wide approaches for the mouse genome sequencing project. Gaps in the draft sequence may well obscure gene relationships, unless workers make careful use of other positional information, such as genetic or physical maps.
METHODS
Contig Construction
Murine Critical Region
BACs (129Sv library: Research Genetics) and PACs (RPCI21; 129/SvevTACfBr mouse spleen genomic DNA library: K. Osoegawa, P. de Jong, Roswell Park Cancer Institute, Buffalo; MRC HGMP-RC) were identified by radioactive hybridization and PCR screening. Resulting clones were tested subsequently for other markers from the region by PCR and Southern analysis. Pulsed field gel electrophoresis (PFGE) was used to estimate the size of the clones. DNA was digested with Not 1 to remove the vector, and run on a 1% low melting point (LMP) agarose gel. A 20 sec switch time was used at 170V over 20 hr. The size of the BAC clones varied between 85 and 165 kb, with the size of the PAC inserts being larger, between 140 and 200 kb. Fluorescent fingerprinting (Gregory et al. 1997) was used to aid the construction of a sequence-ready map. The purified clone DNA was digested with HindIII and Sau3AI, with the HindIII ends being labeled with a fluorescent dye-labeled dideoxynucleotide, followed by polyacrylamide gel electrophoresis on an ABI 377 automated DNA sequencer (PE-Applied Biosystems). The data was imported into Image3 (Sulston et al. 1989) for editing before transfer to fingerprinted contigs (FPC) (Soderlund et al. 1997) for analysis.
Human Critical Region
Cosmids indicated by ‘Qc’ (see Fig. 1) were isolated from a Xq28-specific cosmid library constructed from the hamster/human cell hybrid QIZ (Warren et al. 1990). Clones isolated from the Lawrence Livermore National Laboratory chromosome X cosmid library (LLOXNCO1) are indicated with ‘L’ (Fig. 1). A pool of cosmids binned to Xq28-specific YACs was kindly provided by D.L. Nelson (Baylor College of Medicine, Institute for Molecular Genetics, Houston TX, USA). BAC/PAC clones bridging gaps between cosmid contigs were identified by Genomic Library Screening Kits (Genome Systems) or BAC End Sequence Database searches (http://www.tigr.org/tigr_home/tdb/humgen/bac_search/bac_search.html).
Sequencing of the Mouse and Human Intervals
Mouse
The sequencing strategy adopted was the random shotgun approach with an 8- 10-fold redundancy. Typically, around 3500 subclones were sequenced for each BAC or PAC and 1000 for cosmids. Half of the sequencing reactions were carried out by the Dye Terminator cycle sequencing method (Rhodamine or dRhodamine) and half by Energy Transfer dye primer cycle sequencing. Automated editing of the reads was carried out using Pregap (Bonfield and Staden 1996) to mask vector and poor quality sequence. The assembly of the sequence reads was carried out using Pint (http://www.hgmp.mrc.ac.uk/Registered/Webapp/pint/), a Web-based interface to the sequence assembly process running the Phred base caller (Ewing et al. 1998) and Phrap assembly engine (P. Green, in prep.) as well as a number of postprocessing programs. The editing of the assembled sequence was carried out in Gap4 (Bonfield et al. 1995) according to the following criteria: each base is covered by a forward and a reverse read, or by two reads derived from different clones and different chemistries. A semiautomated strategy has been adopted for the contiguation and prefinishing processes based on reverse reads, primer panels, direct walks, and shatter clones (McMurray et al. 1998).
Human
Cosmid, BAC, and PAC DNA preparation and sequencing were performed as described previously (Kioschis et al. 1998).
Expression Studies
RT–PCR Assays and Northern Analysis
RT–PCR assays on Xlr5 were carried out on mouse Origene Multiple Choice™ cDNAs. PCR was carried out in a 20μl reaction volume, using 0.33 mM (final concentration) primers and HotstarTaq (Qiagen). The cDNA templates were denatured for 15 min at 95°C. This was followed by 35 cycles at 95°C, 5 sec; 58°C, 10 sec; 72°C, 1 min and a final extension of 5 min at 72°C. A probe for Northern blot hybridization was amplified from testis cDNA using primers spanning from exon 1–exon 5 of the Xlr5 gene (conditions above), resulting in a product of 373 bp. The PCR product was purified using Spin Columns (Quantum Prep® PCR Kleen, BIO-RAD) prior to labeling by random priming with 32P-dCTP using Megaprime DNA labeling system (Amersham Pharmacia Biotech). The probe was subjected to 2 hr of competition with mouse Cot-1 DNA (GIBCO BRL). Hybridization was carried out at 68°C for 1 hr in Expresshyb solution (Clontech) to a Multiple Tissue Northern (MTN™) from Clontech following the given protocol and washed in 0.1×SSC, 0.1% SDS for 45 min at 65°C.
RT–PCR analysis of ECRA4–A23, Zfp275, ECRB5–B8 and Northern analysis of Zfp275 and ECRB5–B8 were carried out as described previously (Levin et al. 1996; Liu et al. 1999).
Sequence Analysis
The sequence analysis of the genomic sequence was performed using Nix (http://www.hgmp.mrc.ac.uk/) and Rummage (Glockner et al. 1998). Nix is a WWW (World Wide Web) tool used to view the results of running multiple DNA analysis programs on DNA sequence. The analysis programs include GRAIL (Uberbacher and Mural 1991), Fex, Hexon (Solovyev et al. 1994), MZEF (Zhang 1997), Genemark (Borodovsky 1993), Genefinder (http://menu.hgmp.mrc.ac.uk/Nix/Help/genefind_washhelp.html), Fgene (Solovyev et al. 1994), GENSCAN (Burge and Karlin 1997), HMMGene (Krogh 1997), BLAST (Altschul et al. 1994) (against many databases), Polyah (Salamov and Solovyev 1997), RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker), and tRNAscan (Lowe and Eddy 1997).
ORFinder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to identify ORFs in query sequences, ClustalX (Thompson et al. 1994) to perform multiple sequence alignments, and ScanProsite (http://www.expasy.ch/tools/scnpsit1.html) and ProfileScan (http://www.isrec.isb-sib.ch/software/PFSCAN_form.html) to identify protein domains in PFAM (Bateman et al. 1999) and PROSITE (Hofmann et al. 1999). To determine the genomic structure of genes where cDNA sequence was available, sim4 was used to align the cDNA to genomic sequence (Florea et al. 1998). The mouse and human genomic sequences were compared using PipMaker (Schwartz et al. 2000; http://bio.cse.psu.edu/). PipMaker aligns two sequences using a program called BLASTZ, which is a new implementation of the gapped BLAST program (Altschul et al. 1997) that was designed specifically for determining local alignments of two long DNA sequences. Gap-free segments of these alignments are displayed in a PIP. The plot graphs gap-free segments according to their position in the query sequence on a percent identity scale from 50%–100% along the length of the chosen sequence. The light horizontal line through the middle of the plot indicates 75% nucleotide identity.
Analysis of Genomic Sequence Conservation in Annotated Regions of Mouse and Human Genome
Annotated regions of the mouse and human genome were aligned using BLASTZ (Schwartz et al. 1999). From the BLASTZ output, gap-free local pairwise alignments with a percentage identity ⋝50% were extracted and categorized into those that overlapped EMBL-annotated CDS (coding sequences), 5′ UTRs, and 3′ UTRs. The statistical analysis of the results was performed using Minitab (http://www.minitab.com).
Acknowledgments
This work was supported by the Medical Research Council (UK), German BMBF (BEO 0311108/0), and the European Commission (BMH4-CT96–0338). G.E.H. was supported in part by grant NIH R01 NS34953 and by funds from the Children's Research Institute. We thank Simon Gregory and Gareth Howell from the Sanger Center for their help with fingerprinting and Duncan Campbell for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL s.brown@har.mrc.ac.uk; FAX 44 1 235 824542.
REFERENCES
- Allenet B, Escalier D, Garchon HJ. A putative human equivalent of the murine Xlr (X-linked, lymphocyte- regulated) protein. Mamm Genome. 1995;6:640–644. doi: 10.1007/BF00352372. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Boguski MS, Gish W, Wootton JC. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont J, Miller W, Gibbs RA. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel PL, Timblin CR, Kozak CA, Kuehl WM. Sequence and expression of murine cDNAs encoding Xlr3a and Xlr3b, defining a new X-linked lymphocyte-regulated Xlr gene subfamily. Gene. 1994;150:345–350. doi: 10.1016/0378-1119(94)90450-2. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The Human Genome: Organisation and Evolutionary History. Annu Rev Genetics. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Staden R. Experiment files and their application during large-scale sequencing projects. DNA Seq. 1996;6:109–117. doi: 10.3109/10425179609010197. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith K, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky MM, McIninch JD. GeneMark: Parallel gene recognition for both DNA strands. Comp Chem. 1993;17:123–133. [Google Scholar]
- Boyle AL, Ballard SG, Ward DC. Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci. 1990;87:7757–7761. doi: 10.1073/pnas.87.19.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner AG, Koop BF, Aronow BJ, Wiginton DA. Genomic sequence comparison of the human and mouse adenosine deaminase gene regions. Mamm Genome. 1999;10:95–101. doi: 10.1007/s003359900951. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Bustin M, Lehn DA, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Faust CJ, Molinari-Storey L, Kiochis P, Poustka A, Herman GE. A 2.3-Mb yeast artificial chromosome contig spanning from Gabra3 to G6pd on the mouse X chromosome. Genomics. 1994;21:49–57. doi: 10.1006/geno.1994.1223. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Tanaka T, Parrish JE, Herman GE. Refined mapping of caltractin in human Xq28 and in the homologous region of the mouse X chromosome places the gene within the bare patches (Bpa) and striated (Str) critical regions. Mamm Genome. 1995;6:802–804. doi: 10.1007/BF00539008. [DOI] [PubMed] [Google Scholar]
- Cohen DI, Hedrick SM, Nielsen EA, D'Eustachio P, Ruddle F, Steinberg AD, Paul WE, Davis MM. Isolation of a cDNA clone corresponding to an X-linked gene family (XLR) closely linked to the murine immunodeficiency disorder xid. Nature. 1985;314:369–372. doi: 10.1038/314369a0. [DOI] [PubMed] [Google Scholar]
- De Backer O, Verheyden AM, Martin B, Godelaine D, De Plaen E, Brasseur R, Avner P, Boon T. Structure, chromosomal location, and expression pattern of three mouse genes homologous to the human MAGE genes. Genomics. 1995;28:74–83. doi: 10.1006/geno.1995.1108. [DOI] [PubMed] [Google Scholar]
- De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- De Plaen E, De Backer O, Arnaud D, Bonjean B, Chomez P, Martelange V, Avner P, Baldacci P, Babinet C, Hwang SY, Knowles B, Boon T. A new family of mouse genes homologous to the human MAGE genes. Genomics. 1999;55:176–184. doi: 10.1006/geno.1998.5638. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Endrizzi M, Huang S, Scharf JM, Kelter A-R, Wirth B, Kunkel LM, Miller W, Dietrich WF. Comparative sequence analysis of the mouse and human Lgn1/SMA interval. Genomics. 1999;60:137–151. doi: 10.1006/geno.1999.5910. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Faust CJ, Levinson B, Gitschier J, Herman GE. Extension of the physical map in the region of the mouse X chromosome homologous to human Xq28 and identification of an exception to conserved linkage. Genomics. 1992;13:1289–1295. doi: 10.1016/0888-7543(92)90048-w. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner G, Scherer S, Schattevoy R, Boright A, Weber J, Tsui LC, Rosenthal A. Large-scale sequencing of two regions in human chromosome 7q22: Analysis of 650 kb of genomic sequence around the EPO and CUTL1 loci reveals 17 genes. Genome Res. 1998;8:1060–1073. doi: 10.1101/gr.8.10.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Howell GR, Bentley DR. Genome mapping by fluorescent fingerprinting. Genome Res. 1997;7:1162–1168. doi: 10.1101/gr.7.12.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Rogner UC, Kioschis P, Korn B, Poustka A. Transcription mapping in a 700-kb region around the DXS52 locus in Xq28: isolation of six novel transcripts and a novel ATPase isoform (hPMCA5) Genome Res. 1996;6:478–491. doi: 10.1101/gr.6.6.478. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Gloeckner G, Bachner D, Kioschis P, Klauck SM, Hinzmann B, Rosenthal A, Herman GE, Poustka A. Genomic structure of a novel LIM domain gene (ZNF185) in Xq28 and comparisons with the orthologous murine transcript. Genomics. 1997;43:329–338. doi: 10.1006/geno.1997.4810. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Hua A, Spilson SV, Miller W, Roe BA, Meisler MH. Comparative sequence of human and mouse BAC clones from the mnd2 region of chromosome 2p13. Genome Res. 1999;9:53–61. [PMC free article] [PubMed] [Google Scholar]
- Kioschis P, Wiemann S, Heiss NS, Francis F, Gotz C, Poustka A, Taudien S, Platzer M, Wiehe T, Beckmann G, et al. Genomic organization of a 225-kb region in Xq28 containing the gene for X-linked myotubular myopathy (MTM1) and a related gene (MTMR1) Genomics. 1998;54:256–266. doi: 10.1006/geno.1998.5560. [DOI] [PubMed] [Google Scholar]
- Koop BF, Hood L. Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nat Genet. 1994;7:48–53. doi: 10.1038/ng0594-48. [DOI] [PubMed] [Google Scholar]
- Krogh A. Two methods for improving performance of an HMM and their application for gene finding. ISMB. 1997;5:179–186. [PubMed] [Google Scholar]
- Lamerdin JE, Stilwagen SA, Ramirez MH, Stubbs L, Carrano AV. Sequence analysis of the ERCC2 gene regions in human, mouse, and hamster reveals three linked genes. Genomics. 1996;34:399–409. doi: 10.1006/geno.1996.0303. [DOI] [PubMed] [Google Scholar]
- Levin ML, Chatterjee A, Pragliola A, Worley KC, Wehnert M, Zhuchenko O, Smith RF, Lee CC, Herman GE. A comparative transcription map of the murine bare patches (Bpa) and striated (Str) critical regions and human Xq28. Genome Res. 1996;6:465–477. doi: 10.1101/gr.6.6.465. [DOI] [PubMed] [Google Scholar]
- Levinson B, Bermingham JR, Jr, Metzenberg A, Kenwrick S, Chapman V, Gitschier J. Sequence of the human factor VIII-associated gene is conserved in mouse. Genomics. 1992;13:862–865. doi: 10.1016/0888-7543(92)90170-w. [DOI] [PubMed] [Google Scholar]
- Liu XY, Dangel AW, Kelley RI, Zhao W, Denny P, Botcherby M, Cattanach B, Peters J, Hunsicker PR, Mallon AM, et al. The gene mutated in bare patches and striated mice encodes a novel 3beta-hydroxysteroid dehydrogenase. Nat Genet. 1999;22:182–187. doi: 10.1038/9700. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W, Zhang J, Boguski MS. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 1996;6:846–857. doi: 10.1101/gr.6.9.846. [DOI] [PubMed] [Google Scholar]
- McMurray AA, Sulston JE, Quail MA. Short-insert libraries as a method of problem solving in genome sequencing. Genome Res. 1998;8:562–566. doi: 10.1101/gr.8.5.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Rogner UC, Wilke K, Steck E, Korn B, Poustka A. The melanoma antigen gene (MAGE) family is clustered in the chromosomal band Xq28. Genomics. 1995;29:725–731. doi: 10.1006/geno.1995.9945. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Recognition of 3′-processing sites of human mRNA precursors. Comput Appl Biosci. 1997;13:23–28. doi: 10.1093/bioinformatics/13.1.23. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer A, Smit C, Riemer J, Bouck R, Gibbs R, Hardison R, Miller W. PipMaker – A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R. FPC: A system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- Solovyev VV, Salamov AA, Lawrence CB. Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res. 1994;22:5156–5163. doi: 10.1093/nar/22.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Landsman D, Bustin M. Retropseudogenes for human chromosomal protein HMG-17. J Mol Biol. 1987;197:405–413. doi: 10.1016/0022-2836(87)90554-7. [DOI] [PubMed] [Google Scholar]
- Sulston J, Mallett F, Durbin R, Horsnell T. Image analysis of restriction enzyme fingerprint autoradiograms. Comput Appl Biosci. 1989;5:101–106. doi: 10.1093/bioinformatics/5.2.101. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberbacher EC, Mural RJ. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren ST, Knight SJ, Peters JF, Stayton CL, Consalez GG, Zhang FP. Isolation of the human chromosomal band Xq28 within somatic cell hybrids by fragile X site breakage. Proc Natl Acad Sci. 1990;87:3856–3860. doi: 10.1073/pnas.87.10.3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ. Identification of protein coding regions in the human genome by quadratic discriminant analysis Proc. Natl Acad Sci. 1997;94:565–568. doi: 10.1073/pnas.94.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]