Abstract
Affinity purification coupled to mass spectrometry provides a reliable method for identifying proteins and their binding partners. In this study we have used Drosophila melanogaster proteins triple tagged with Flag, Strep II, and Yellow fluorescent protein in vivo within affinity pull-down experiments and isolated these proteins in their native complexes from embryos. We describe a pipeline for determining interactomes by Parallel Affinity Capture (iPAC) and show its use by identifying partners of several protein baits with a range of sizes and subcellular locations. This purification protocol employs the different tags in parallel and involves detailed comparison of resulting mass spectrometry data sets, ensuring the interaction lists achieved are of high confidence. We show that this approach identifies known interactors of bait proteins as well as novel interaction partners by comparing data achieved with published interaction data sets. The high confidence in vivo protein data sets presented here add new data to the currently incomplete D. melanogaster interactome. Additionally we report contaminant proteins that are persistent with affinity purifications irrespective of the tagged bait.
Characterization of native protein interactions is essential to further our understanding of developmental processes and other complex pathways underpinning biological functions. For a comprehensive view of multicomponent protein complexes it will be necessary to develop high-throughput methods for identifying genuine interaction partners. To date the largest protein interaction data sets have come from yeast two-hybrid (Y2H)1 studies (1–3). Y2H data sets are useful initial protein interaction frameworks that provide a valuable resource for the community, but they are not without limitations. One of the major restrictions is that interactions are observed out of their in vivo context and may involve pairs of proteins that never meet in vivo because they are located in different cell types or subcellular compartments or are expressed at different times in the lifecycle. This is particularly problematic in multicellular organisms: without reliable temporal and tissue-specific protein expression data it is possible that interactions observed in yeast cells may be spurious. In addition, the Y2H methods do not allow the study of indirect interactions or interactions that involve post-translational modifications. As a result of these limitations, Y2H interaction data sets often contain a high level of false positives, estimated to be as high as 90% in some cases (4–6).
Complementary approaches to Y2H involve isolating multiprotein complexes from whole organisms via an in vivo tagged “bait” protein. Complexes can be captured using affinity purification and interacting proteins identified by high-throughput mass spectrometric methods. Commonly used tags include GFP, 6xHis, Myc, StrepII, and FLAG: each having advantages and disadvantages with respect to efficiency and purity (7). One of the most widely used versions of this approach is tandem affinity purification (TAP) tagging (8–10) that results in high stringency and low false positive rates. Although the tandem affinity purification approach has been very successful in unicellular organisms such as yeast (9), the serial protein purifications employed by this method result in reduced final protein yields that can be a problem when dealing with limited material from metazoans.
Homologous recombination is an efficient way to introduce affinity tags into endogenous proteins in yeast, but it is too laborious to use this approach on a large scale in multicellular organisms where the efficiency of homologous recombination is much lower. To circumvent this difficulty, we generated endogenously tagged proteins by mobilizing a transposable element containing an exon encoding a series of affinity tags flanked by splice donor and acceptor sites (14). When such an element integrates into the genome between protein coding exons of a gene in the correct orientation and reading frame, it can be spliced into the mRNA to generate a tagged protein. Several protein trap screens have been performed using a transposon carrying a GFP reporter to generate lines useful for the study of protein expression patterns and subcellular localization (14–16). We have adapted this approach by including affinity tags in the GFP exon so that the resulting lines can also be used for interactome mapping.
Here we present a robust pipeline for generating high quality and comprehensive interactomes suitable for use with multicellular organisms. We developed our method, interactomes by parallel affinity capture (iPAC), with the well-studied metazoan D. melanogaster model and demonstrate that a novel triple-tag system; yellow fluorescent protein (YFP), for screening and expression profiling, coupled with StrepII and FLAG tags for parallel affinity capture, allows the use of a parallel purification strategy to maintain protein complex yields suitable for MS analysis. The use of a tagless negative control facilitates the identification of proteins common to both FLAG and StrepII purifications and we have generated a list of common contaminants associated with affinity captured complexes. Finally, the use of MS exclusion lists derived from the contaminants is used to increase sensitivity and improve peptide coverage, thus allowing sampling of lower abundance proteins. We have optimized the methods using a selection of proteins of different sizes and subcellular localizations to test the robustness of the purification and identification procedures.
It is widely accepted that the highest fidelity protein interaction databases are likely to result from integrating data sets derived from a combination of different sources, including Y2H and high-throughput mass spectrometric methods (11–13).
EXPERIMENTAL PROCEDURES
Generation of Triple Tagged D. melanogaster Lines
The entire P element protein trap construct from the plasmid pPGA (14) was removed, using the ApaI and BmgBI restriction enzyme sites lying just outside the P element ends. The fragment was inserted into a unique HpaI site in the PiggyBac plasmid p3E1.2 (17) to give a hybrid PiggyBac/P-element. The synthetic exon sequences from pPGA were modified to replace the EGFP sequence with 3xFLAG-StrepTagII-VenusYFP-StrepTagII between the NcoI and SalI sites flanking EGFP. The 6xHis tag found in the original Morin vectors were retained in the construct for splice frame 1 but removed in the constructs for frames 0 and 2.
Transgenic lines containing this hybrid PiggyBac/P-element were generated by standard methods using a PiggyBac helper plasmid, pAct5C-orf (18). Mobilization of transgenic inserts to generate fluorescent protein-trap insertions was carried out by crossing to fly lines expressing transposase and selecting for fluorescent individuals, either manually as described by Morin et al. (14) or using a COPAS select sorter (Union Biometrica) as described by Buszczak et al. (15). Stocks established from YFP positive embryos were sequenced by inverse PCR to map the transposon insertions and identify the trapped protein.
Protein Extraction from D. melanogaster Embryos
For each transgenic D. melanogaster line, and the nontagged control w1118 progenitor line, three independent egg lay samples were prepared. Embryos were collected from apple juice agar plates supplemented with live yeast, from ∼200 females laying eggs over a 12 h period. Embryos were washed off the agar using tap water containing 0.1% Tween 20, collected in 100-μm sieves, rinsed in the same solution to remove any yeast, dechorinated in 50% bleach for 1 min, rinsed again and placed on ice. Where necessary, washed embryos were frozen at −80 °C until the required quantity for extraction and detection was achieved.
For each purification 200 μg wet-weight of embryos were manually homogenized with a 2-ml Dounce homogenizer in 1 ml of extraction buffer containing 50 mm Tris, pH 7.5, 125 mm NaCl, 1.5 mm MgCl, 1 mm EDTA, 5% Glycerol, 0.4% Nonidet P-40, and 0.1% Tween 20 (modified from Veraksa et al.) (21). For integral membrane proteins or unannotated protein lines where extract yields were low, 0.5% digitonin (Sigma) was included to efficiently extract proteins without disrupting complexes. To prevent degradation during the lengthy purification steps, protease mini EDTA inhibitor mixture 2× (Roche) was added at hourly intervals throughout the procedure. The homogenate was centrifuged at 10,000 × g for 15 min to isolate the soluble fraction. To determine the extraction efficiency, 1 μl protein pre- and post-extraction was quantified against an extraction buffer blank using the NanoDrop® (ND-1000 v3.3.0) instrument in protein A280 mode. Typical concentrations varied between 30 and 60 mg/ml. All samples were normalized to 30 mg/ml and divided into 1-ml aliquots for each parallel purification procedure.
Pull-down Procedure
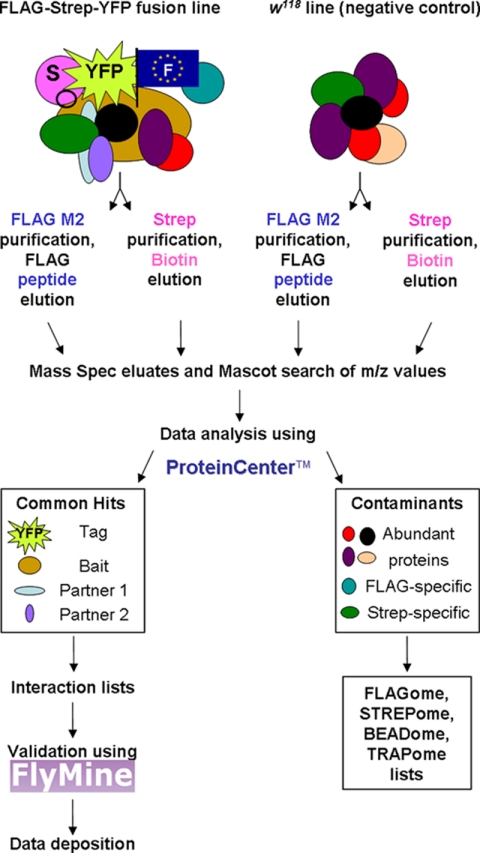
The iPAC protocol is outlined in Fig. 1. Each step is detailed below.
Fig. 1.
iPAC workflow outlining the parallel purification steps, data processing including the removal of negative controls and identification of contaminants, confirmation by validation, and deposition of interaction data sets. ProteinCenter™ url: http://www.proxeon.com/productrange/data_interpretation/introduction/index.html, FlyMine url: http://www.flymine.org/, FlAnnotator url: www.flyprot.org/.
Batches of soluble tagged protein were purified in parallel, alongside a negative control tagless line and, occasionally a no-protein (extraction buffer only) was included as a systematic control for both StrepII and FLAG.
The standard manufacturers' protocols were adjusted so that binding affinities, elution concentrations and volumes were the same for each pull-down experiment allowing direct comparison, quantitation, and a high-throughput system.
FLAG Procedure
EZview™ Red ANTI-FLAG® M2 affinity gel (Sigma) was used to capture FLAG tagged bait and its binding partners. 50 μl prewashed 50% slurry was added to 1 ml (30 mg) soluble protein and incubated at 4 °C for 2 h on a rotary mixer. Nonbinding material was removed by centrifugation (8000 × g for 1 min) and the resin washed three times in ice cold extraction buffer. FLAG tagged protein, with any associating proteins, was incubated and eluted twice each with 50 μl (100 μg/ml) FLAG peptide (Sigma) in extraction buffer for 30 min at 4 °C on a rotary mixer. The two eluates were combined and any residual resin was removed by centrifugation at 8000 × g for 2 min.
StrepTagII Procedure
50 μl prewashed Strep-Tactin Sepharose resin (IBA) was added to 1 ml (30 mg) soluble protein and incubated at 4 °C for 2 h on a rotary mixer. Non binding material was removed by centrifugation (5,000 × g for 2 min) and the resin washed three times in ice cold extraction buffer. StrepII tagged protein, with any associating proteins, was eluted twice with 50 μl of 10 mm Biotin in extraction buffer for 30 min at 4 °C on a rotary mixer. The two eluates were combined and any residual resin was removed by centrifugation at 5,000 × g for 2 min.
Tandem Procedure for Pilot Studies
Embryonic lysates from nine trap lines were split four ways therefore starting with the same total protein concentration and two FLAG and StrepII purifications performed as before. For tandem purifications the initial eluates were diluted in extraction buffer then incubated with the second resin for a further hour, washed, and eluted with the second competitor. Reciprocal tandems were also performed to compare yields qualitatively. Final yields were compared by immunoblot analysis and MS analysis. Each comparison was performed at least twice.
Immunoblot Confirmation of the Bait
Five micrograms each of initial soluble extract and eluate were sampled for immunoblot analysis using either mouse anti FLAG MAb (Sigma) at 1:1000 dilution and/or rabbit anti GFP PAb (Abcam, Cambridge, UK) at 1:2000 dilution followed by secondary antibodies anti-mouse IgG (1:5000) or anti-rabbit IgG (1:5000) (both BioRad) respectively to detect the tagged protein. The Amersham Biosciences ECL plus kit was used to visualize the detected proteins.
Preparation for Mass Spectrometric Analysis
Protein eluates were precipitated with ice cold 100% acetone overnight at −80 °C. The precipitate was vacuum dried then reduced in 2 mm dithiotreitol for 1 h at room temperature and alkylated in 10 mm iodoacetamide for 30 min at room temperature. Proteins were digested with 2 μg sequencing grade trypsin (Promega, Madison, WI) for 1 h at 37 °C, then a further 2 μg for overnight digestion to maximize complete digestion of complex mixtures.
Mass Spectrometry
5 or 10 μl peptide were loaded onto a 5 cm C18 precolumn, 300 μm i.d., (LC packings) then concentrated peptides were subsequently loaded onto a PepMap C18 reverse phase, 75 mm i.d., 15 cm analytical column (LC Packings) and eluted using an Eksigent nano LC system at a flow rate of 300 nl/min attached to a LTQ Orbitrap (Thermo Electron). The gradient described in supplemental Table S1 online was applied to resolve and elute the peptides into the LTQ ion trap. The two 30-min washes with 85 and 65% acetonitrile were adopted to reduce carryover of residual abundant peptides, such as Actin, that bind nonspecifically. The Orbitrap was operated in data-dependent mode, MS then 2× tandem MS (MS/MS) with data dependent settings set to excluded contaminant masses of peptides from negative controls (supplemental Fig. S1 online) with a dynamic exclusion of 0.3 Da. m/z values were selected based on the protein abundance across multiple samples, including controls, from the same purification batch and from previous assays.
Protein Identification
Peak lists were generated using Bioworks Browser version 3.3.1 (2007). Resulting fragment masses (MS/MS) were searched using the MASCOT version 2.2 (Matrix Science) search engine against an in house database comprising the FlyBase D. melanogaster genome (version 5.9) totaling 21,064 proteins, plus the FASTA sequence for YFP as a secondary confirmation of the presence of the tagged protein. Parameters included a precursor mass tolerance of 1.0 Da and fragment ion mass tolerance of 0.8 Da, two missed cleavages and methionine oxidation variable- and carboxymethyl cysteine fixed-modifications. The decoy database option, comprising a scrambled D. melanogaster database in silico digested that generates a similar number of the same-sized peptides, was selected to automatically calculate the protein false discovery rate. Stringent parameters were used to ensure accuracy in the data sets. For example, proteins with single peptide hits were eliminated.
Interaction List Generation
All protein lists were imported into ProteinCenter™, the negative control proteins subtracted, proteins with single peptide hits removed and the resulting interaction lists exported as .csv files into Excel (see supplementary methods and supplemental Fig. S2 online for details). Experimental confidence scores were assigned to each interactor based on the number of times they were observed in multiple experiments with higher weighting toward proteins seen in different affinity pull-down lists than those seen multiple times in only one affinity method. A confidence scoring sigmoidal curve was constructed to assign scores for different sized data sets (see supplemental Fig. S3 online); for example, a protein seen four times in four experiments would be assigned 1.0, a protein seen three times (2× in FLAG, 1× in StrepII or 1× in FLAG, 2× Strep) in four experiments (2× FLAG, 2× StrepII) would be assigned a score of 0.8. Proteins seen twice, once each in a FLAG and Strep pulldown from four experiments (2× FLAG, 2× StrepII) would be assigned 0.5, however proteins seen twice but in both FLAG or both Strep would only be assigned 0.2. Finally, proteins seen only once in four experiments were assigned a score of 0.03 and were not included in interaction lists unless stated otherwise. Proteins were ranked according to their confidence score and then the Mascot protein probability score.
Interaction Validation Using FlyMine
To see if the interaction list members identified in this study agree with other published interaction sets, lists of UniProtKB identifiers for each identified protein in the interactions lists were imported into FlyMine v15 (www.flymine.org) (22) and searched with known genetic and protein interactor lists including IntAct (EBI), Biogrid, and FlyBase manual curations. Six templates (T1–6) were created to search our identified proteins against the D. melanogaster data in public repositories for both direct (binary, typically from Y2H screens) interactions between bait and prey (T1) and interacting pairs among all members of the list (prey-prey) (T2). We also searched for evidence of indirect interactions (nonbinary, from phenotypic enhancement and suppression genetic screens and protein affinity purification screens), again for both bait and prey (T3) and prey-prey (T4). In addition, we also searched for direct interactions in orthologous proteins in other species including C. elegans and S. cerevisiae, again for bait and prey interactions (T5) and prey-prey interactions (T6), because no one interaction repository is comprehensive and direct partners in other species would increase our confidence in our observed results. To determine the proportion of validations in random protein lists, a random number generator was used to select 100 UniProtKB identifiers that were imported into FlyMine and processed as per interaction lists. We also analyzed lists of negative control line proteins to establish nonspecific interactions between proteins and affinity resins.
RESULTS
Tag Selection
It was important to select tags that do not alter protein stability, structure or function. Our method of parallel affinity purification of individually tagged endogenous proteins from their native environment requires the incorporation of multiple affinity tags into the bait protein, and we therefore assessed several alternative tags for use in Drosophila. The original Morin protein trap vectors (14) incorporated a 6xHis tag adjacent to the GFP reporter, but attempts to use the His tag for purification were largely unsuccessful, in agreement with findings by other workers (23). We performed pilot experiments with two variants of the His tag, HN6 and HAT, in Drosophila S2 cells and found that the HN6 tag appeared to cause protein aggregation, whereas we failed to purify HAT-tagged proteins (supplemental Figs. S4a and S4b). The use of calmodulin binding protein was also rejected because of low yields and purity (data not shown). Other workers also report that calmodulin binding protein-tagged isolations tend to be of lower purity than either of the two tags we finally used (23). In addition, the calmodulin binding protein tag is reported to interact with endogenous Calmodulin, which can affect either the cellular localization or biological function of the tagged proteins or of Calmodulin itself (24–25). In contrast, we found that StrepII-tagged proteins could be pulled down efficiently (supplemental Figs. S4c–S4e).
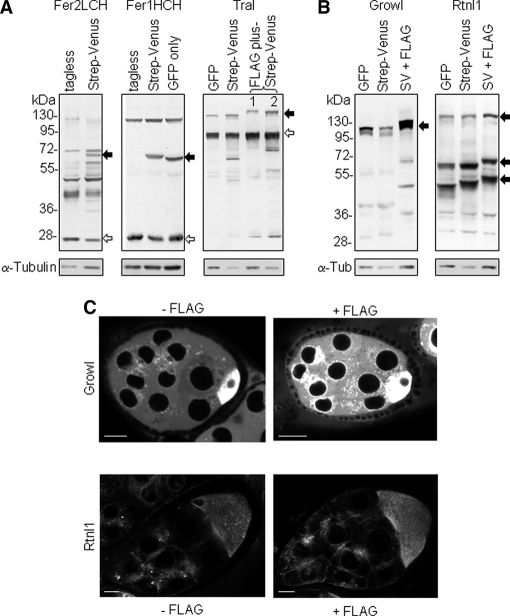
Expression levels for various protein traps in D. melanogaster embryos were compared in order to assess whether variants of the trapping cassette affect expression levels or protein stability. Levels of tagged proteins relative to untagged proteins appeared to vary. For the Ferritins, Fer1HCH and Fer2LCH, levels were ∼70% of the untagged protein (Fig. 2A, left) whereas for Trailer hitch (Tral), a protein involved in membrane trafficking and RNA localization, as little as 19% is tagged according to densitometry analysis of immunoblots (Fig. 2A, right). The proportion of tagged versus untagged protein depends on the efficiency of splicing of the artificial exon, which may be influenced by the insertion site of the exon in the gene, and on the stability of the tagged protein, which will vary according to the position of the tag within the host protein and the domain structure of the protein.
Fig. 2.
Effects of tags on protein levels. A, Protein extracts from heterozygous flies with insertions in Fer1LCH, Fer2HCH and Trailer hitch probed with Fer1LCH, Fer2HCH and Trailer hitch antibodies respectively to show the abundance of the tagged proteins (closed arrows) compared with the untagged proteins (open arrows). For a perfectly spliced and stable protein, the two band intensities would be expected to be equal. For Fer1HCH and Fer2LCH, tagged protein levels were 72 and 73% of the untagged, whereas for Trailer Hitch it was 19%. For Fer1HCH and Trailer hitch more than one fly line was available containing variants of the tag inserted; the figures are averaged from these lines. All these genes are predicted to encode single transcripts and protein products. Antibodies for Fer1LCH and Fer2HCH cross react with other proteins and extracts from untagged flies are run in adjacent lanes for comparison. B, Protein extracts from flies containing traps in growl and Rtnl1 (arrows indicate multiple isoforms) probed with anti-GFP to compare tagged protein levels with GFP, StrepII-tagged Venus YFP (SV), or FLAG-StrepII-tagged Venus YFP proteins. C, Confocal images comparing StrepII-tagged-venus proteins with (right) or without (left) FLAG. The addition of the FLAG tag does not reduce tagged protein levels. For these comparisons, the trap construct is inserted into the same intron and thus in the same position within the protein. Scale bars = 20 μm.
To assess the affect of introducing multiple tags, we compared StrepII-tagged and FLAG-StrepII-tagged insertions with the original GFP-tagged insertions isolated in the Morin screen. In the cases tested the tag did not abolish protein expression or affect stability (Figs. 2B and 2C). This also demonstrates that acidic tags, such as FLAG, can be tolerated in vivo. Based on this evidence, the final combination of affinity tags we found to be most effective for both imaging and complex purifications was a triple tag comprising FLAG, StrepTagII and YFP. Details of the construct used are provided in supplemental Fig. S5.
Because tandem affinity purification tagging is a popular strategy, we compared protein yields after performing pulldown experiments in parallel and tandem in several trap lines. In our hands we found that bait stability and recovery as well as recovery of potential interaction partners was facilitated by performing parallel purifications with the FLAG and Strep tags instead of serial purification. An example is shown in supplemental Fig. S6. We therefore elected to perform affinity purifications in parallel and focus on proteins common to both purifications: this approach greatly improves throughput and also minimizes the degradation that can result from prolonged binding and additional washing steps needed for tandem strategies. The optimized workflow we developed for using these tags for iPAC is summarized in Fig. 1.
Identification of the Bait
To demonstrate the efficiency and reproducibility of our iPAC approach, six proteins with membrane, cytoplasmic or extracellular localizations, and molecular weights ranging from 22 to 2446 kDa, were studied in detail (Flapwing (FLW), Shaggy (SGG), Reticulon-like 1 (RTNL1), Fasciclin 2 (FAS2), Semaphorin 1a (SEMA-1A), and Dumpy (DP)): Fig. 3 and supplemental Table S2. Following affinity purification, MS analysis positively identified at least 2 peptides with all baits and up to 60% sequence coverage. In the examples shown, an average of 14 peptides were identified per protein from FLAG purifications and nine peptides from StrepII purifications, similar to the eight per protein identified by Brunner and colleagues (26) using a shotgun approach. Importantly, the coverage results in these cases suggest that there is little degradation or cleavage during the purification of even large proteins such as DP. Mapping the observed peptides allowed an investigation of protein cleavage or degradation as truncated proteins are unlikely to interact normally. For example, in the case of SGG, MS showed no peptides mapping to the C terminus (Fig. 3A). Alternatively these C-terminal peptides may have modifications that precluded their identification in MASCOT database searching. The additional data showing peptides liberated by the fused YFP tag confirms the presence of the bait and also serves as a standard across multiple samples, which can give an approximation of tagged protein abundance based on spectral counts (supplemental Table S2).
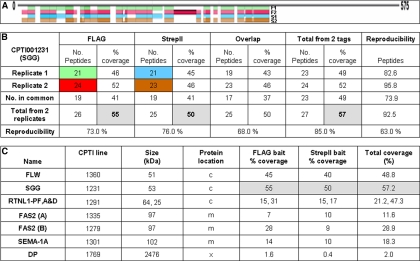
Fig. 3.
Identification, coverage and reproducibility of the bait protein in parallel purifications. A, Peptide coverage for FLAG and StrepII purified baits (SGG) from two replicates and (B) reproducibility of the Mascot processed MS identification data for SGG. Peptide reproducibility % calculated from (No. peptides in common/No. total from 2 reps) × 100. M = oxidation of Met residue. C, Summary of the combined replicate bait data for the six different proteins analyzed. More detailed analysis is in supplemental Table S2 online and supplemental Fig. S7 online. MS identified the presence of three different isoforms for Rtnl1: PA and PD (both 25 kDa) and PF (64 kDa) and the % sequence coverage are shown respectively. Two different protein trap lines were analyzed for Fas2 and the data listed separately as (A) and (B).
Several tagged proteins are represented by more than one D. melanogaster line in our collection and we used such examples to determine whether the location of the tag within the protein affects the ability to purify the protein and its binding partners. Although in the cases we have analyzed we have not observed a major effect of the location of the trap on bait recovery (and in the case of hth on the recovery of the known interactor exd) (supplemental Fig. S7, supplemental Table S2). However there were unique interactors to each line that were not reported previously and these may be isoform specific. Each trap line would have to be tested as there may be cases where the position of the tag does indeed influence interactions, location and/or function. As several tagged proteins are represented by multiple lines in our collection, parallel purifications from the different lines may overcome this possible drawback.
We were also able to distinguish different isoforms in several cases. For example, unique peptides were identified in the variable N-terminal portions of three isoforms of RTNL1; -PA, -PD and -PF (supplemental Fig. S8). Thus a variety of sizes and types of proteins can be pulled down using both FLAG and StrepII purification procedures, and it appears that our method of tagging does not favor proteins of a particular size range or subcellular localization.
Reproducibility Test
To test the reproducibility of bait and interacting protein identification we replicated the analysis for several lines by performing duplicate purifications from a single protein extraction and analyzing the resulting FLAG and StrepII eluates with identical MS settings. For all the lines tested, the replicates gave similar data in terms of peptides, coverage and number of potential interacting proteins (Fig. 3B). Reproducibility was also high for detection of bait and partners from biological replicates performed months apart (supplemental Table S2).
MS Data Analysis and Exclusion Lists
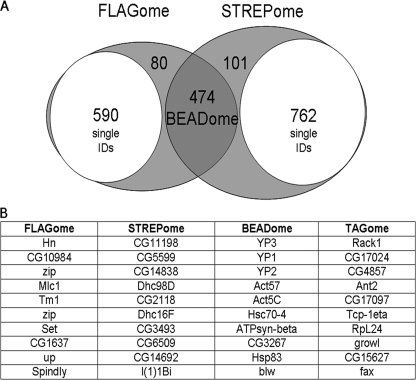
MASCOT results were imported the into the ProteinCenter™ software package (see methods) to compare the FLAG and StrepII interaction data sets, compare Mascot protein probability scores of proteins identified in controls and test samples, and subtract nonspecific proteins that were pulled down in nontagged lines and no-protein systematic controls (supplemental Fig. S2). The software was also used to compare proteins from multiple pull-downs and highlight contaminants specific to each or both resins. Proteins consistently identified with FLAG resins only (FLAGome), StrepII resins only (STREPome) and those binding to both resins (BEADome) are listed in full (supplemental Tables S3a–S3c online) and Fig. 4 lists the top ten hits.
Fig. 4.
Analysis of negative control data. A, Multiple parallel affinity purifications of 25 nontagged lines (negative controls) and the use of ProteinCenter™ identified contaminants specific to FLAG and StrepII resins (FLAGome and STREPome respectively) and those common to both (BEADome). After identifying and removing the single protein hits (subsets) for each affinity method there is 72% overlap. B, The top ten hits for the FLAGome, STREPome and BEADome are listed in the table and the complete listings are presented in supplemental Tables S3A–S3C online. The top ten interacting proteins common to multiple baits are listed as the TRAPome and the complete list is presented in supplemental Table S5 online.
We implemented MS exclusion lists to suppress common contaminants identified above in order to improve coverage of the bait and binding partners. We were conservative when generating the exclusion list to avoid excluding too many m/z values that could represent peptide ions generated from other proteins, thus we used the 20 most abundant Actin peptide m/z values, 10 yolk protein peptide m/z values, two m/z values from two StrepII binding contaminants (CG1516 and CG2118) and one m/z value from a FLAG binding contaminant (CG31974). For example, using the 20 carefully selected m/z values, the sequence coverage for the six Actins fell considerably from 81% to an average of 19% (supplemental Fig. S1). Introducing exclusion lists improved identification of the bait proteins in subsequent replicates, as seen for SGG and SEMA-1A, with the latter resulting in the identification of 50% additional bait peptides in the second StrepII pulldown experiment (PD#15) compared with the initial MS analysis (PD#13) of this low abundance protein (supplemental Table S2 online).
Analysis of Interacting Proteins
The ProteinCenter statistics tool was used to subtract the negative control data sets from the replicate FLAG and StrepII data sets and to generate a list of overlapping proteins for analysis. Different baits showed differing overlap and where low overlap was observed known interactors were found multiple times with only one affinity method demonstrating the need for replicates and a variety of affinity tags. Protein interaction lists were therefore generated for all proteins and confidence scores were assigned (supplemental Fig. S3) based on the frequency of appearance in our experiments. Proteins were then ranked as described in the methods section. An example of an annotated interaction list is presented in Table I (top 20 interactors only) and full interaction lists for the selected proteins are presented in supplemental Tables S4a–S4f online. These lists appear in IntAct with Accession #IM-11716.
Table I. Interaction lists and confidence.
(A) Interacting proteins for FLW bait with FlyMine validation. n = number of times observed, cs = confidence score, PP = Protein Probability score (from MASCOT), d = putative direct interaction, i = indirect interaction, F_1/2 = FLAG experiment 1 or 2, S = Strep, Y2H = yeast 2-hybrid, FP = fragment pooling approach, PS = phenotypic suppression, PE = phenotypic enhancement, PD = pulldown, AC = Affinity Capture, CoIP = anti-tag coimmunoprecipitation. Confidence scores derived from Supplementary Fig. S3. Upper row in bold identifies the bait, bold grey highlighted are direct interactions or proteins validated by two independent methods and grey highlighted are those validated by one method. Italicised are potential contaminants. *plus ID codes 12134149, 12750335, 14605208, 15575970, 15710747, 17007873, 9230081.
| Uniprot ID | Gene name | Description | n | cs | PP mean | F1 | F2 | S1 | S2 | Experimental evidence | d/i | PubMed ID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q8T0U6 | flw | Serine/threonine protein phosphatase | 4 | 1.00 | 601.8 | 1 | 1 | 1 | 1 | |||
| Q9VUX6 | Mbs | CG32156-PA | 4 | 1.00 | 337.5 | 1 | 1 | 1 | 1 | Y2H-FP, FB | d | 15710747 |
| Q9VBD6 | CG5471 | CG5471-PA | 3 | 0.80 | 97.3 | 1 | 0 | 1 | 1 | Y2H | i | 14605208 |
| Q8IM86 | unc-13 | CG2999-PB | 3 | 0.80 | 76.0 | 0 | 1 | 1 | 1 | Y2H | i | 14605208 |
| P28668 | Aats-glupro | Bifunctional aminoacyl-tRNA synthetase | 3 | 0.80 | 57.7 | 0 | 1 | 1 | 1 | |||
| Q8SXP0 | CG8709 | GH19076p | 3 | 0.80 | 46.7 | 0 | 1 | 1 | 1 | Y2H | i | 14605208 |
| Q94883 | Dref | CG5838-PA | 3 | 0.80 | 35.7 | 0 | 1 | 1 | 1 | |||
| P05661–19 | Mhc | Myosin Heavy Chain-PF | 2 | 0.50 | 244.0 | 1 | 0 | 0 | 1 | Y2H-FP | i | 15710747 |
| P12982 | Pp1–87B | Serine/threonine-protein phosphatase α-2 isoform | 2 | 0.50 | 127.5 | 1 | 0 | 1 | 0 | CoIP, PE, PS, Y2H, Y2H-FP | d | 11514446, 11729158* |
| Q9VMT8 | CG33715 | Muscle specific protein | 2 | 0.50 | 124.0 | 0 | 1 | 0 | 1 | |||
| Q9VLT5 | poe | Protein purity of essence | 2 | 0.50 | 105.0 | 0 | 1 | 0 | 1 | |||
| Q9VB44 | CG3339 | CG3339-PA | 2 | 0.50 | 102.5 | 0 | 1 | 0 | 1 | |||
| Q27331 | Vha68–2 | Vacuolar ATP synthase catalytic subunit A isoform 2 | 2 | 0.50 | 97.0 | 0 | 1 | 0 | 1 | |||
| Q9VBE2 | CG5468 | CG5468-PA | 2 | 0.50 | 90.5 | 0 | 1 | 1 | 0 | Y2H | i | 14605208 |
| Q9VNE0 | CG2926 | CG2926-PA | 2 | 0.50 | 80.5 | 0 | 1 | 0 | 1 | Y2H-FP | i | 15710747 |
| Q9W2Z4 | CG2990 | CG2990-PA | 2 | 0.50 | 77.5 | 0 | 1 | 0 | 1 | |||
| Q9W2Z3 | CG2989 | CG2989-PA | 2 | 0.50 | 76.0 | 0 | 1 | 0 | 1 | |||
| Q9W053 | zormin | CG33484-PA | 2 | 0.50 | 72.5 | 0 | 1 | 0 | 1 | Y2H, Y2H-FP | i | 14605208, 15710747 |
| Q8I8U7 | Nipped-A | Transcription-associated protein 1 | 2 | 0.50 | 72.5 | 0 | 1 | 0 | 1 | Y2H FP, AC-western | i | 15710747, 12697829 |
| Q9VZQ3 | kst | CG12008-PA | 2 | 0.50 | 70.5 | 0 | 1 | 0 | 1 | Y2H, Y2H-FP | i | 14605208, 15710747 |
| (B) Summary of interacting proteins from the other selected baits and validation analysis using FlyMine. Lists of interacting proteins for the remaining proteins are in supplementary Tables S4a-f.[1]. *after removal of negative controls (W-), ¶ based on FlyMine and DroID, + after removal of single IDs. No's in parentheses indicate proteins found >10% of contaminants lists. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bait | No. of hits* |
Published Interactors¶ |
Validated Interactors+ |
||||||||
| Total | Score≥ 0.75 | Score>0.2-<0.75 | Score≤ 0.2 | Total (inc. genetic) | Affinity method | % | Direct (with bait) | Direct (all) | Orthologs (direct bait) | Indirect (with bait) | |
| FLW | 460 | 7(2) | 49(11) | 405 | 55 | 1 | 44 | 8 | 81 | 2 | 37 |
| SGG | 843 | 12(1) | 83(7) | 748 | 31 | 1 | 48 | 0 | 18 | 5 | 0 |
| RTNL1 | 166 | 1 | 3(1) | 162 | 10 | 0 | 50 | 0 | 1 | 0 | 7 |
| FAS2(A) | 209 | 1(1) | 4(1) | 204 | 16 | 2 | 64 | 1 | 30 | 0 | 9 |
| FAS2(B) | 583 | 11(7) | 46(13) | 526 | 16 | 2 | 69 | 1 | 124 | 0 | 19 |
| SEMA-1A | 129 | 3(3) | 26(10) | 100 | 7 | 0 | 56 | 0 | 12 | 0 | 6 |
| DP | 28 | 0 | 0 | 28(16) | 2 | 0 | 12 | 0 | 1 | 0 | 0 |
| PKA-R2 | 63 | 1 | 15 | 47 | 8 | 0 | 50 | 2 | 11 | 6 | 1 |
| fl(2)d | 29 | 0 | 5 | 24 | 20 | 3 | 68 | 2 | 2 | 0 | 2 |
| ATPsyn-β | 91 | 6 | 12 | 73 | 25 | 0 | 81 | 1 | 12 | 20 | 12 |
| CAM | 84 | 0 | 2 | 82 | 32 | 0 | 38 | 0 | 6 | 1 | 9 |
| GS2 | 78 | 1 | 18 | 59 | 1 | 0 | 71 | 0 | 15 | 12 | 1 |
| NOTCH | 315 | 0 | 6 | 309 | 183 | 1 | 64 | 3 | 54 | 7 | 63 |
| BEL | 43 | 0 | 2 | 41 | 0 | 0 | 57 | 0 | 6 | 15 | 0 |
After analyzing several lists, it became apparent that several proteins were present in multiple lists but not in all negative control lists. These included ribosomal proteins, which may indicate interactions between the bait and protein synthesis machinery, heat shock and cognate proteins that interact with the bait as part of the protein folding machinery, and structural proteins such as Myosins. To analyze the frequency of these proteins, interaction lists were merged using ProteinCenter and the proteins ranked according to frequency of appearance in interaction lists and the average mascot protein probability score: we termed this the TRAPome (supplemental Table S5). We are reluctant to exclude these proteins at the ProteinCenter processing level, because they may genuinely participate in many complexes. We report the ten most abundant proteins in the TRAPome as well as the tag specific contaminants and derivation of the BEADome in Fig. 4. We also show the growth of the BEADomes and TRAPome as more experiments are added and at present these have not yet reached saturation (supplemental Fig. S9).
Interaction Validation
We used FlyMine to compare our lists of interacting proteins identified in our parallel affinity purifications with published data for putative direct interactors (binary: typically from Y2H screens) and indirect interactors (Y2H interactions through an intermediate protein, and data from genetic and affinity purification screens) and have summarized and highlighted these in Table I and supplemental Tables S4a–S4f online respectively. From these data we were able to identify putative direct binders of the bait, and interactors of these, derived from prey-prey validations generating bait-(prey-preyn)n. Up to 81% of the proteins in each interaction list have previously reported interactions, mostly indirect, with other proteins within the list. To assess the significance of this validation rate, we generated mock interaction lists containing an equivalent number of randomly selected proteins, and observed that 7% of these had reported “interactions” (direct and indirect) with other proteins in the list. Because this is much lower than what we observe for the real protein interaction data, we conclude that we have successfully enriched for genuine interacting proteins. As a further comparison, we used the negative control lists of abundant, sticky proteins that bind to the affinity columns in the absence of a tagged protein, and observed that 90% of these proteins have reported interactions with each other in the literature. This suggests that the public databases are contaminated with spurious interactions among these sticky proteins, justifying their exclusion from our analysis.
Many candidate interactions are not supported by published data from Drosophila, and we therefore broadened our validation screen by using FlyMine to identify direct interactions among orthologs of the Drosophila proteins in eight other species (see methods). Reported interactions between orthologous proteins supported much of our Drosophila data and many of our additional interactors, depending on the bait, that were not identified in Drosophila databases, further validating the quality of our interaction data.
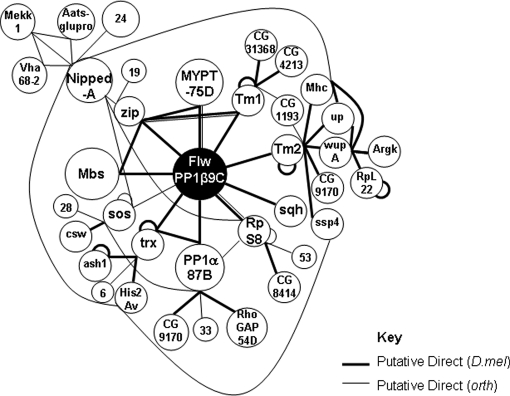
Published network maps of our selected proteins are limited and where available, show little overlap depending on the screens, most of which are Y2H. Based on our stringent methodology and validation we have attempted to generate more comprehensive networks to map the proteins we have identified. Fig. 5 shows an interaction network for FLW indicating all putative binary interactions for bait and prey and their binary interactions with other prey in the list (bait-[prey-preyn]n), using full interaction lists that include lower confidence data. Because of the multiple queries in FlyMine (see Methods) we are able to link many more proteins than those that are highlighted on the interaction lists (supplemental Table S4a) as these static lists can show only proteins involved in putative bait-prey or prey-prey interactions and don't distinguish individual pairs, or hubs or any dynamic information. In addition, proteins we found that have interactions in orthologues have also been mapped. For simplicity, indirect interactions have been excluded but are shown in supplemental Table S4a.
Fig. 5.
Interaction network map for FLW, a protein phosphatase that targets Mbs. All putative direct bait-prey interacting proteins immediately surround the bait, FLW, indicated in black. Additional direct interactions from within the entire list are mapped if linked to the bait. Direct ortholog interactions are represented by lighter lines. Indirect interactions have been omitted for simplicity apart from where the “via” proteins are also direct. The size of the circle is proportional to the confidence score of the interaction.
DISCUSSION
Here we report the development of a pipeline for reproducibly isolating endogenously tagged proteins of all types along with their specific interaction partners. Because of the adoption of stringent thresholds at the affinity purification and MS stage, our approach has generated a catalogue of high confidence interactions with low false discovery rates. Considerable optimization was performed in terms of tag choice, extraction, purification and MS analysis, to ensure reliable identification of interacting proteins for a range of different bait proteins. Our analysis indicates that this high-throughput method is sensitive and reproducible, and can be applied to a wide range of proteins. Although our starting material is D. melanogaster embryo extracts, we believe the method is more widely applicable and also suitable for other organisms, especially metazoans where some protein abundances are lower.
Because of the scale of our study, we were able to identify the contaminants that bind nonspecifically to each affinity resin, to all resins, and the promiscuous Drosophila proteins that bind to many baits or to the tags themselves. These “BEADomes” will provide a valuable resource for identifying nonspecific contaminants in similar affinity purification experiments using D. melanogaster extracts. Similar “BEADomes” have been reported for affinity purifications from human tissue culture cells (27), and it is striking that these contain many orthologs of proteins in the D. melanogaster BEADome, including cytoskeletal and ribosomal proteins. The prevalence of ribosomes and protein chaperones in these lists may indicate that affinity tags delay the folding of the proteins to which they are attached, causing their retention at the ribosome exit site and prolonged association with chaperones.
Using software packages we were able to remove contaminants in silico, confirm the presence of known interactors, validate the genuine interacting partners, and output the data in a format that is easily deposited in public repositories. We used this procedure to analyze the interaction partners of protein baits that had been labeled by the introduction of affinity tags into the middle of the protein by the transposon-mediated insertion of an artificial exon. This method has the advantage that it can efficiently tag many proteins in a multicellular organism without the labor intensive generation of large numbers of constructs or homologous recombination. It has the drawback, however, that the insertion of the affinity tags into protein may disrupt its folding, stability or function. Nevertheless, we identified known interaction partners of several baits. Although the effect on the insertion will obviously vary depending on the nature of the tagged protein and the site of the insertion, this approach provides a straightforward way to introduce affinity tags into a large number of proteins in vivo.
Although there are groups using proteomics approaches in Drosophila, such as the Peptide Atlas project (26, 28), there are few large scale published proteomics interaction data sets with which to compare our results, and most interaction databases provide no confidence scores. Our comprehensive interaction lists have experimental-based confidence scores and by comparing our data with publicly available data, most of which is derived from Y2H studies, we can also validate a significant number of interactors as well as identify proteins that are common interactors irrespective of the bait. Any high throughput approaches generate many false positives, predicted to be as high as 90% in some instances. It is likely that many of the putative interactors we have identified, particularly those with low confidence scores are also spurious. However, when one of these interactors is identified in our affinity purification approach and is also found in Y2H screens this dramatically raises the likelihood that this is a bona fida interaction that occurs in vivo.
An example used in our study is FLW, a myosin phosphatase. We found all components of its complex and its target substrates according to Vereshchagina (29) and were able to produce an interaction network involving many regulatory and structural proteins, confirming data from genetic studies. Other proteins identified in our FLW iPAC screen have similar GO annotations as the FLW interacting partners indicating that the majority are structural proteins or kinases and phosphatases involved in the regulation of the cytoskeletal components. In contrast, we expected to see very few proteins interacting with the extracellular matrix protein DP. Indeed our interaction list is short and none are verified, with the majority (59%) found in multiple interaction lists (the TRAPome).
Although this paper only presents a small number of examples to demonstrate the reliability of our approach, our protein trap screen is ongoing and will generate many more tagged proteins. Not only does this project have the power to purify and identify the components of complexes, but it also allows the analysis of protein localization and expression dynamics in vivo. The number of proteins that can be tagged by this transposon-based protein trap approach is limited by the number of proteins that will tolerate an artificial insert and by the insertional bias of the transposon (30). Nevertheless, we predict that we can tag hundreds of proteins in this way, and thereby provide high quality interaction data for a significant fraction of the D. melanogaster proteome. The fly-trap lines studied to date have been deposited into the Kyoto stock center (http://kyotofly.kit.jp/stocks/documents/CPTI.html) and the interaction data has been uploaded into FlAnnotator (www.flyprot.org).
Acknowledgments
We would like to thank Serena Tolin for assistance in the high-throughput purifications, Julie Howard and Svenja Hester for performing some LC-MS/MS. We thank J. Drummond, E. Drummond, H. Spriggs, and T. Morley for assistance with the fly work and A. Carr for assistance with FlyMine templates.
Footnotes
* This project is funded by the Welcome Trust.
 This article contains supplemental Figs. S1 to S9 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S9 and Tables S1 to S5.
1 The abbreviations used are:
- Y2H
- yeast two-hybrid
- iPAC
- interactomes by parallel affinity capture
- m/z
- mass/charge ratio
- YFP
- yellow fluorescent protein
- MS
- Mass Spectrometry.
REFERENCES
- 1. Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 2. Parrish J. R., Gulyas K. D., Finley R. L., Jr. (2006) Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 17, 387–393 [DOI] [PubMed] [Google Scholar]
- 3. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabasi A. L., Tavernier J., Hill D. E., Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322(5898): 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bork P., Jensen L. J., von Mering C., Ramani A. K., Lee I., Marcotte E. M. (2004) Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 14, 292–299 [DOI] [PubMed] [Google Scholar]
- 5. Vidalain P. O., Boxem M., Ge H., Li S., Vidal M. (2004) Increasing specificity in high-throughput yeast two-hybrid experiments. Methods 32, 363–370 [DOI] [PubMed] [Google Scholar]
- 6. von Mering C., Jensen L. J., Kuhn M., Chaffron S., Doerks T., Krüger B., Snel B., Bork P. (2007) STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 35, D358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang I. F. (2006) Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics 6, 6158–6166 [DOI] [PubMed] [Google Scholar]
- 8. Brizzard B. L., Chubet R. G., Vizard D. L. (1994) Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. BioTechniques 16, 730–735 [PubMed] [Google Scholar]
- 9. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 10. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 11. Bader G. D., Hogue C. W. (2002) Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 20, 991–997 [DOI] [PubMed] [Google Scholar]
- 12. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A. S., Venkatesan K., Rual J. F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., Vidal M. (2009) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J. A., Goud B., de Gunzburg J., Johannes L., Junier M. P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rossé C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morin X., Daneman R., Zavortink M., Chia W. (2001) A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 15050–15055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A. D., Nystul T. G., Ohlstein B., Allen A., Wilhelm J. E., Murphy T. D., Levis R. W., Matunis E., Srivali N., Hoskins R. A., Spradling A. C. (2007) The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., Misra S., Rubin G. M. (1999) The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital drosophila genes. Genetics 153, 135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cary L. C., Goebel M., Corsaro B. G., Wang H. G., Rosen E., Fraser M. J. (1989) Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172, 156–169 [DOI] [PubMed] [Google Scholar]
- 18. Horn C., Offen N., Nystedt S., Häcker U., Wimmer E. A. (2003) piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics 163, 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Missirlis F., Holmberg S., Georgieva T., Dunkov B. C., Rouault T. A., Law J. H. (2006) Characterization of mitochondrial ferritin in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 5893–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilhelm J. E., Buszczak M., Sayles S. (2005) Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9, 675–685 [DOI] [PubMed] [Google Scholar]
- 21. Veraksa A., Bauer A., Artavanis-Tsakonas S. (2005) Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Developmental Dynamics 232, 827–834 [DOI] [PubMed] [Google Scholar]
- 22. Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., Guillier F., Janssens H., Ji W. Y., Mclaren P., North P., Rana D., Riley T., Sullivan J., Watkins X., Woodbridge M., Lilley K., Russell S., Ashburner M., Mizuguchi K., Micklem G. (2007) FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007; 8(7):R129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lichty J. J., Malecki J. L., Agnew H. D., Michelson-Horowitz D. J., Tan S. (2005) Comparison of affinity tags for protein purification. Protein Expression and Purification 41, 98–105 [DOI] [PubMed] [Google Scholar]
- 24. Knuesel M., Wan Y., Xiao Z., Holinger E., Lowe N., Wang W., Liu X. (2003) Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell Proteomics 2, 1225–1233 [DOI] [PubMed] [Google Scholar]
- 25. Przewloka M. R., Zhang W., Costa P., Archambault V., D'Avino P. P., Lilley K. S., Laue E. D., McAinsh A. D., Glover D. M. (2007) Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One 2, e478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Heck A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 27. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vereshchagina N., Bennett D., Szöor B., Kirchner J., Gross S., Vissi E., White-Cooper H., Alphey L. (2004) The essential role of PP1 beta in Drosophila is to regulate nonmuscle myosin. Mol. Biol. Cell 15, 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aleksic J., Lazic R., Müller I., Russell S. R., Adryan B. (2009) Biases in Drosophila melanogaster protein trap screens. BMC Genomics 10, 249 [DOI] [PMC free article] [PubMed] [Google Scholar]