The authors show that macular-specific increases in mtDNA damage, heteroplasmic mutations, and diminished repair are associated with aging and with severity of age-related macular degeneration (AMD). Additionally, they link the accumulation of mtDNA damage to a decline in expression of the DNA repair enzyme OGG1 and speculate that this decline contributes to inefficient DNA repair capacity, especially in eyes with more advanced AMD.
Abstract
Purpose.
Mitochondrial DNA (mtDNA) damage may be associated with age-related diseases, such as age-related macular degeneration (AMD). The present study was designed to test whether the frequency of mtDNA damage, heteroplasmic mtDNA mutations, and repair capacity correlate with progression of AMD.
Methods.
Macular and peripheral RPE cells were isolated and cultured from human donor eyes with and without AMD. The stages of AMD were graded according to the Minnesota Grading System. Confluent primary RPE cells were used to test the frequency of endogenous mtDNA damage by quantitative PCR. Mutation detection kits were used to detect heteroplasmic mtDNA mutation. To test the mtDNA repair capacity, cultured RPE cells were allowed to recover for 3 and 6 hours after exposure to H2O2, and repair was assessed by quantitative PCR. The levels of human OGG1 protein, which is associated with mtDNA repair, were analyzed by Western blot.
Results.
This study showed that mtDNA damage increased with aging and that more lesions occurred in RPE cells from the macular region than the periphery. Furthermore, mtDNA repair capacity decreased with aging, with less mtDNA repair capacity in the macular region compared with the periphery in samples from aged subjects. Most interestingly, the mtDNA damage was positively correlated with the grading level of AMD, whereas repair capacity was negatively correlated. In addition, more mitochondrial heteroplasmic mutations were detected in eyes with AMD.
Conclusions.
These data show macula-specific increases in mtDNA damage, heteroplasmic mutations, and diminished repair that are associated with aging and AMD severity.
There is increasing evidence to support a role for mitochondrial damage and dysfunction in aging and chronic neurodegenerative disorders, including age-related macular degeneration (AMD).1,2 Studies on the aging retina have reported significant age-related decreases in the number and size of mitochondrial in human RPE cells,3 accumulation of mitochondrial DNA deletions,4 and increased mitochondrial DNA damage and downregulation of DNA repair enzymes in aged rodent neural retina and RPE/choroid.5,6 The potential consequences of these detrimental mitochondrial changes include increased generation of reactive oxygen species and decreased metabolic activity, which impede the cells' optimal bioenergetics.1 It is only within the past decade that the potential role for mitochondrial damage and dysfunction in AMD has been realized. mtDNA haplotypes have been identified that are associated with either increased or decreased prevalence of AMD,7 and persons with AMD have high levels of large mtDNA deletions/rearrangements in the retina, unreported and amino acid-changing SNPs in the coding genome, and more SNPs per person in the noncoding MT-Dloop region.8 Studies have focused largely on the RPE given its critical role in photoreceptor maintenance, its atrophy in dry AMD, and the view that the RPE is the site of the primary defect in AMD. Karunadharma et al.9 recently reported that with aging mtDNA damage was observed only in the common deletion region of the mitochondrial genome, whereas, in contrast, with AMD the mtDNA lesions increased significantly in all regions of the mitochondrial genome beyond levels found in age-matched controls. These changes are also associated with a significant decrease in the number and area of mitochondria and the loss of cristae and matrix density in AMD compared with age-matched controls3 and alterations in proteins associated with mitochondrial translation, import of nuclear-encoded proteins, and adenosine triphosphate synthase activity in RPE from AMD patients.10
The cause of mtDNA damage is likely to be endogenous ROS generated either from mitochondria themselves or from photosensitizers within the retina.1,2,11 mtDNA is particularly vulnerable to oxidative damage because of its proximity to the inner mitochondrial membrane, its lack of histones, and its poor repair capacity. The net result impairs the function of mtDNA-encoded subunits of the electron transport chain12 leading to increased generation of ROS, which, in turn, leads to further mtDNA damage in a self-perpetuating, destructive cycle culminating in compromised mitochondrial redox function.13,14 Interestingly, knockdown of mitochondrial superoxide dismutase results in increased superoxide radicals and pathophysiological lesions in mice with a phenotype similar to that observed in AMD.15
Recent studies have begun to elucidate mtDNA repair mechanisms. DNA base damage caused by ROS can be repaired by two general pathways: base excision repair (BER) and nucleotide excision repair.16,17 BER is the main repair pathway in mitochondria of mammalian cells.18,19 The capacity of the BER pathway to elicit DNA repair decreases with age.20,21 Although a decline in both activity and expression of several key mitochondrial DNA repair enzymes, including 8-oxoguanine-DNA glycosylate 1 (OGG1), has been observed with aging in animal retinas,5 the association of OGG1 with AMD pathogenesis is unclear.
In this study, we report that the RPE in AMD is associated with increased mtDNA damage and heteroplasmic DNA mutations that are inversely correlated with decreased mtDNA repair. The results also showed that RPE cells from the macula have greater mtDNA damage and diminished repair capacity than cells from the peripheral region. Taken together, our study suggests that mtDNA damage and reduced repair capacity, especially in macular RPE cells, are associated with aging and AMD pathogenesis.
Materials and Methods
Isolation of RPE Cells from Human Donor Eyes
Human donor eyes, including young (20–30 years), aged (70–90 years), and AMD patients (70–90 years) were obtained from the National Disease Research Interchange NDRI (Philadelphia, PA), the Lions Eye Bank of Oregon (Portland, OR), and the Minnesota Lions Eye Bank (Saint Paul, MN), in accordance with the provisions of the Declaration of Helsinki for research involving human tissues. Eyes were received in a moist chamber on wet ice. The average time between donor demise and enucleation of eyes and receipt was approximately 4 hours (4.03 ± 1.2) and 35 hours (34.4 ± 5.61), respectively. Globes were dissected under sterile conditions. Anterior segment, vitreous, and neural retina were carefully removed without damaging the underlying RPE cells. The eyecup was rinsed gently with chilled PBS to remove any debris. Fundus photographs (with and without neural retina) were taken under a dissecting microscope (SMZ1500; Nikon, Inc., Tokyo, Japan) equipped with a digital camera (Coolpix 995; Nikon) with a ruby sphere 1 mm in diameter on the center of the disc in AMD eyes for calibration. Fundus and posterior cup images were examined to detect any RPE abnormalities. Eyes with findings consistent with AMD were graded according to the Minnesota Grading System (MGS).22 An 8-mm sterile trephine was used to remove a disc of the RPE cell layer, Bruch's membrane, and choroid from the macular and peripheral area. RPE cells were loosened after trypsin digestion (30 minutes at 37°C) in 1 mL Dulbecco's modified Eagle's medium (DMEM). Cells were then collected in a 15-mL sterile conical tube and spun at 1000 rpm in a centrifuge at 4°C for 5 minutes. The medium was carefully removed, and the pellet was resuspended in fresh cell growth medium. Purity of the cultures was confirmed using cytokeratin 18 antibody staining, which showed >97% RPE cells. Cells from different regions and different donors were treated as individual samples in all the following experiments.
RPE Cell Culture and Hydrogen Peroxide Treatment
Dissected RPE cells were cultured for up to three to four passages in 35-mm dishes with DMEM (Invitrogen Corp., Carlsbad, CA) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin in a 37°C humidified incubator with 5% CO2. Concentrated H2O2 (30%; Sigma, St. Louis, MO) was diluted into Dulbecco's PBS, pH 7.4. Confluent RPE cells were exposed to different concentrations of H2O2 (100–200 μM) for 1 hour in serum-free, phenol red-free minimum essential medium (MEM). We confirmed that 1 hour of serum deprivation had no significant effect on the viability of RPE cells (data not shown).
Measurement of mtDNA Lesion and Repair
DNA was extracted using a genomic-tip kit (Qiagen, Inc., Valencia, CA) and was stored at −80°C. mtDNA lesions were quantified using a well-validated, sensitive, quantitative polymerase chain reaction (QPCR) assay.12,13,23 The QPCR assay is based on the principle that any lesion on a DNA template, such as strand break, base modification, or apurinic site, will block the progress of thermostable polymerase (rTth DNA polymerase), resulting in decreased amplification of the target sequence. Therefore, only those DNA templates without any DNA damage will be amplified. Sample quality was tested by QPCR of a 222-base pair fragment in the mtDNA and an 84-base pair fragment in the β-globin genes (mtDNA primers) (14619FOR, 5′-CCC CAC AAA CCC CAT TAC TAA ACC CA-3′; 14841REV, 5′-TTT CAT CAT GCG GAG ATG TTG GAT GG-3′) and β-globin primers (48550FOR, 5′-CGA GTA AGA GAC CAT TGT GGC AG-3′; 48634REV, 5′-GCT GTT CTG TCA ATA AAT TTC CTT C-3′), with the expectation that equal template concentrations yielded similar QPCR product (short) concentrations. QPCRs were performed using a PCR kit (GeneAmp XLPCR; Perkin-Elmer, Waltham, MA). Reaction conditions were used as described previously for the 16 ± 2 kb mtDNA product and the 13 ± 4 kb β-globin product12,24 (sense primer, 5′-TGG GAT TAC ACG TGT GAA CCA ACC-3′; antisense primer, 5′-GCT CTA CCC TCT CCT CTA CCG RCC-3′). The same conditions were used for both the 12 ± 2-kb β-polymerase product (sense, 5′-CCT GGA GTA GGA ACA AAAATT GCT G-3′; antisense, 5′-CAT GTC ACC ACT GGA CTCTGC AC-3′) and the 10 ± 4 kb β-globin product. To determine relative amplification, β-globin DNA lesion frequency was calculated as the amplification of the sample from donor eyes relative to the amplification of ARPE-19 cell lines (Ad/Ao).
To determine mtDNA repair, RPE cells from different donors were treated with 200 μM H2O2 for 1 hour. For recovery, normal cell medium was added to cultures, and RPE cells were collected at 0, 3, and 6 hours. DNA lesion frequencies were calculated as described.
Rapid Identification of Heteroplasmic Mitochondrial DNA Mutations
The experiment was carried out according to the published protocol.25 In brief, PCR amplification of the entire human mitochondrial genome in 17 overlapping fragments and identification of mtDNA mismatch by digestion of the 17 amplicons with nuclease (Surveyor; Transgenomic, Inc., Omaha, NE), which is a new mismatch-specific endonuclease that cuts both strands of a DNA heteroduplex on the 3′ side of the mismatch site. Therefore, mismatch-specific cleavage products were seen as two new faster migrating bands in the nuclease (Surveyor; Transgenomic, Inc.)–digested samples. Each of the two new faster migrating bands represented a heteroplasmic mutation. Given that nuclease (Surveyor; Transgenomic, Inc.) cleaves DNA with high specificity at sites of base-substitution mismatch, however, it cannot distinguish novel mutations and polymorphisms. Three- to four-passage primary cultured RPE cells from aged and AMD donors were harvested, and genomic DNA was extracted. PCR amplification of the entire human mtDNA was accomplished using 17 primer sets as previously described.25 Reactions were performed in a final volume of 50 μL, using polymerase (Optimase; Transgenomic, Inc.), as follows: one cycle at 94°C for 4 minutes, 35 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 4 minutes, and finally one cycle at 72°C for 5 minutes. After amplification, 15 μL of each PCR product was digested in a final volume of 60 μL according to the manufacturer's instructions (Surveyor mutation detection kit for WAVE HS System; Transgenomic, Inc.). A total of 20 μL of digested and undigested products was analyzed on a 1.5% agarose gel.
Immunoblot for Expression Levels of OGG1 Enzyme
Briefly, RPE cells from various donors were homogenized in PBS, pH 7.2, containing protease inhibitor cocktail (78410 Halt cocktail; Pierce, Rockford, IL). Protein was quantified by Bradford protein assay, and Western blot analysis was carried out as previously described.26 A molecular imaging machine was used to scan the bands. Molecular imaging and analysis software (UltraLum, Inc., Claremont, CA) were used to quantity and analyze the values.
Statistical Analysis
Each experiment was repeated in triplicate. Values are mean ± SEM. Statistical significance was determined using paired Student[b]'s t-test. Statistical analysis was performed (SAS version 12.0; SAS Inc., Cary, NC). Comparison among the groups was made by P value determination using one-way ANOVA followed by Tukey multiple-comparisons test (GraphPad InStat 3 software; La Jolla, CA). P < 0.05 was considered statistically significant.
Results
Comparison of Freshly Isolated Native RPE Cells and Primary RPE Cultures for mtDNA Damage Analysis
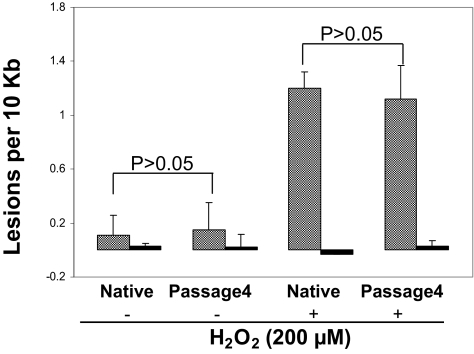
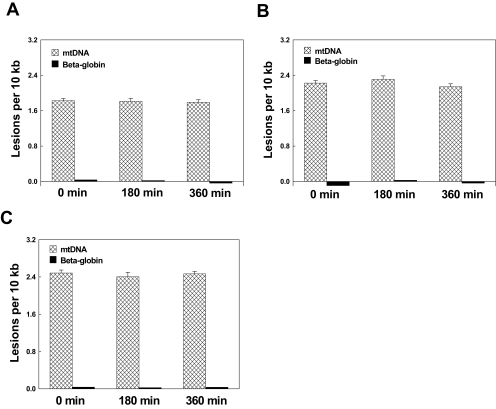
Given that numbers of freshly isolated native RPE cells from the macular region of a single eye were insufficient to undertake mtDNA damage analysis, primary cultured RPE cells were used in this study. To determine up to what passage the cultured RPE cells behaved similarly to native RPE cells, endogenous mtDNA damage in isolated macular RPE cells from four AMD donor eyes and different passages of primary cultured macular RPE cells from the same eyes were analyzed by quantitative PCR.24 The results showed no significant differences (P > 0.1) between the fourth passage of primary cultured RPE cells and native cells (Fig. 1). We also examined the difference in vulnerability of mtDNA to 200 μM H2O2 for 1 hour between the fourth passage of primary cultured RPE cells and native cells. There was no significant difference (P > 0.1) in mtDNA damage (Fig. 1) between the fourth-passage cultured RPE cells and freshly isolated native RPE cells. This confirms that fourth-passage RPE have responses comparable to those of native RPE cells from AMD patient donor eyes. However, RPE cells passaged more than four times exhibited less endogenous mtDNA damage and susceptibility to H2O2 than freshly isolated native RPE cells (data not shown). Therefore, all experiments in this study were performed using primary cultured RPE cells at no more than fourth passage.
Figure 1.
mtDNA damage and repair in freshly isolated native RPE cells and fourth-passage primary cultured RPE cells from AMD MGS III donor. mtDNA lesions were determined in fresh macular RPE cells (n = 4) or from fourth-passage cultures of AMD III (n = 4) donor eyes. Cells were treated with (n = 4) or without (n = 4) 200 μM H2O2 for 1 hour. Shaded bar: mean ± SEM of mtDNA lesions per 10 kb; dark bar: DNA damage of β-globin as the QPCR control.
mtDNA Damage and Repair Capacity Are Age and Topographically Dependent
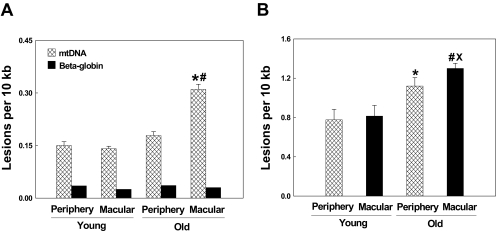
To study the association of mtDNA damage with aging and topography (peripheral and macular regions), native RPE cells were isolated from macular and peripheral regions in young and aged donor eyes. We used 10 eyes from 20- to 30-year-old healthy donors as the young group and 27 eyes from 70- to 90-year-old donors as the aged group for these experiments. The retinas from 70- to 90-year-old donors were graded using the MGS (only MGS I samples were used in this experiment). mtDNA was isolated from the third passage of primary cultured RPE cells. As seen in Figure 2A, RPE cells from young periphery and macula compared with aged periphery and macula showed 0.15 ± 0.08 and 0.14 ± 0.04 and 0.17 ± 0.018 and 0.45 ± 0.025 lesions per 10 kb mtDNA, respectively. Statistical analysis showed that there were no significant differences in mtDNA damage between the periphery and macula in young donor eyes. However, a significant increase in mtDNA lesions was found in the macula compared with the periphery in aged donors (P < 0.001). In addition, there was a significant increase in mtDNA damage in macular regions of aged donors relative to young donors (P < 0.001). These findings indicate that mtDNA damage is increased with aging and more lesions occur within the macular region relative to the periphery in aged, but not young, donors.
Figure 2.
Comparison of mtDNA damage in peripheral and macular RPE cells from young and aged (MGS I) eyes. (A) mtDNA lesions in RPE cells from young peripheral (n = 10) and macular (n = 10) regions and aged peripheral (n = 27) and macular (n = 27) regions were detected using QPCR assay. β-Globin was used as a negative control. Results are mean ± SEM. *P < 0.001, old macular versus young macular; #P < 0.001, old macular versus aged peripheral. (B) mtDNA lesions in RPE cells from young peripheral (n = 10) and macular regions (n = 10) and normal aged peripheral (n = 27) and macular (n = 27) regions caused by 200 μM H2O2 exposure for 1 hour. Results are mean ± SEM. *P < 0.001, aged peripheral versus young peripheral; #P < 0.001, aged macular versus young macular; ×P < 0.01, old macular versus aged peripheral.
To determine mtDNA vulnerability to oxidative stress, third-passage primary cultured RPE cells from young periphery and macula and aged periphery and macula (MGS I) were exposed to 200 μM H2O2 for 1 hour. To calculate the mtDNA damage caused by H2O2, the number of mtDNA lesions before exposure to H2O2 was subtracted from the number of mtDNA lesions after exposure to H2O2. As shown in Figure 2B, mtDNA lesions caused by H2O2 exposure to RPE cells in young peripheral and macular (n = 10) and aged peripheral and macular (n = 27) cultures were 0.78 ± 0.10, 0.81 ± 0.11, 1.12 ± 0.13, and 1.3 ± 0.02, respectively. Statistical analyses indicate no significant differences in mtDNA damage between young peripheral and macular RPE cells but a significant difference (P < 0.01) between aged peripheral and macular cells. In addition, there was a significant difference between aged macula and young macula (P < 0.001). Although there was no difference in the number of endogenous mtDNA lesions in peripheral RPE cells from young and aged donors, there was a significant difference (P < 0.001) in mtDNA lesions after treatment with H2O2. These results indicate that aged peripheral RPE cells are more vulnerable to oxidative stress than younger ones.
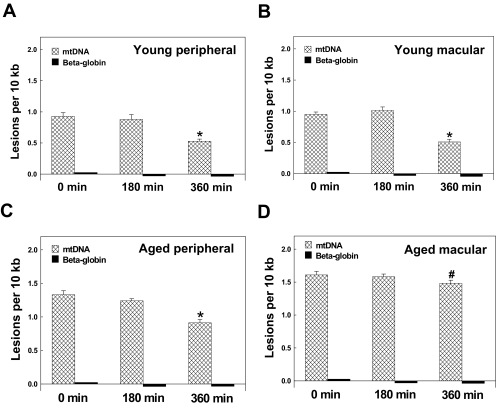
To test the hypothesis that the efficiency of mtDNA repair may be age-dependent and topographically dependent, we investigated the repair capacity of mtDNA lesions in young peripheral and macular as well as aged peripheral and macular (MGS I) samples. Briefly, RPE cells were treated for 1 hour with 200 μM H2O2 and transferred to fresh medium, and mtDNA lesions were measured at 0-, 3-, and 6-hour time points during this recovery period. At 6 hours after switching to fresh medium from H2O2, mtDNA lesion frequency was significantly decreased in young peripheral (Fig. 3A, n = 10; from 0.927 ± 0.06 to 0.527 ± 0.03; P < 0.001), young macular (Fig. 3B, n = 10; from 0.95 ± 0.04 to 0.51 ± 0.04; P < 0.001), and aged peripheral (Fig. 3C, n = 10; from 1.33 ± 0.06 to 0.91 ± 0.05; P < 0.001) samples compared with the 0-hour time point (Figs. 3A-C). There was a lesser degree of mtDNA lesion repair in aged macular regions (n = 10; 1.61 ± 0.03) at 6 hours compared with the 0-hour time point (Fig. 3D; 1.51 ± 0.03; P < 0.01). The lesions calculated as percentages of control in young peripheral, young macular, aged peripheral, and aged macular regions were 43.0%, 46.2%, 31.6%, and 12.4%, respectively. This result suggested that the repair capacity of mtDNA lesions in the macular region decreases with aging. There was no significant repair of mtDNA at the 3-hour time point in all groups, possibly because of inadequate time to repair.
Figure 3.
Time-course analysis of mtDNA damage repair after oxidative stress in young macular and peripheral and aged macular and peripheral RPE cells (MGS I). RPE cells were harvested at 0, 3, and 6 hours after recovery to determine the mtDNA lesions after 1-hour exposure to 200 μM H2O2. The results are mean ± SEM. *P < 0.001 versus 0 hour; #P < 0.05 versus 0 h. (A) mtDNA in RPE cells from young peripheral regions (n = 10). (B) mtDNA in RPE cells from young macular regions (n = 10). (C) mtDNA in RPE cells from aged peripheral regions (n = 10). (D) mtDNA in macular RPE cells (n = 10) from aged macular regions.
mtDNA Damage and Repair Capacity Are Associated with AMD Severity
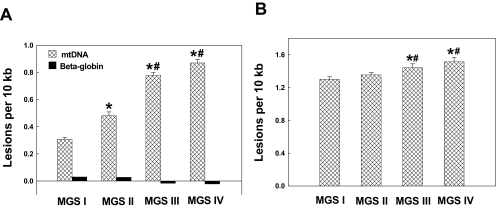
To study the association of mtDNA damage with AMD level, we isolated macular RPE cells from 60- to 90-year-old donors—27 eyes with MGS I, 12 eyes with MGS II, 12 eyes with MGS III, and 12 eyes with MGS IV. mtDNA was isolated from the third-passage cultured macular RPE cells. As seen in Figure 4A, RPE cells from MGS I, MGS II, MGS III, and MGS IV showed 0.31 ± 0.02, 0.48 ± 0.03, 0.78 ± 002, and 0.87 ± 0.03 lesions per 10 kb mtDNA, respectively. Statistical analysis showed a significant increase in mtDNA lesions in macular RPE cells from MGS IV compared with MGS III, MGS III compared with MGS II, and MGS II compared with MGS I. These findings indicate that mtDNA damage in RPE cells is associated with AMD and AMD severity.
Figure 4.
mtDNA damage in macular RPE cells from aged and AMD eyes. (A) mtDNA lesions in RPE cells from MGS I (n = 27), MGS II (n = 12), MGS III (n = 12), and MGS IV (n = 12) donor eyes were detected using QPCR. β-Globin was used as a control. Results are mean ± SEM. *P < 0.001 versus MGS I; #P < 0.01 versus earlier levels. (B) mtDNA lesions induced by 200 μM H2O2 exposure for 1 hour in RPE cells from MGS I, II, III, and IV donor eyes. *P < 0.001 versus MGS I; #P < 0.05 versus MGS II.
To determine mtDNA vulnerability to oxidative stress, third-passage primary cultured macular RPE cells from MGS I (n = 27), MGS II (n = 12), MGS III (n = 12), and MGS IV (n = 12) macular regions were exposed to 200 μM H2O2 for 1 hour. To calculate the mtDNA damage caused by H2O2, the number of mtDNA lesions before exposure to H2O2 was subtracted from the number of mtDNA lesions after exposure to H2O2. As seen in Figure 4B, mtDNA lesions caused by H2O2 in RPE cells from MGS I, MGS II, MGS III, and MGS IV macular regions showed 1.3 ± 0.02, 1.35 ± 0.01, 1.44 ± 0.03, and 1.51 ± 0.03 lesions per 10 kb mtDNA, respectively. Statistical analysis showed no significant mtDNA lesion caused by H2O2 between MGS I and II. There was also no difference between MGS III and IV. Significantly more mtDNA lesions were caused by H2O2 in RPE cells from MGS III than from MGS I and II and from MGS IV compared with MGS I and II. These findings indicate that mtDNA vulnerability to H2O2 in RPE cells is associated with AMD.
To determine the mtDNA repair capability in macular RPE cells in different stages of AMD, we investigated the repair capacity of mtDNA lesions in macular region RPE cells from different MGS levels. Briefly, RPE cells were treated for 1 hour with 200 μM H2O2 and then allowed to recover in fresh new medium; mtDNA lesions were measured at 0-, 3-, and 6-hour time points. At 6 hours, after switching to fresh medium from H2O2, some mtDNA lesion repair was observed in aged macular regions (MGS I) compared with the 0-hour time point (Fig. 3D). There was no significant mtDNA repair seen in macular RPE cells from MGS II, III, and IV at the 6-hour compared with the 0-hour time point; MGS II (Fig. 5A, n = 10, from 1.83 ± 0.07 to 1.79 ± 0.03; P > 0.05), MGS III (Fig. 5B, n = 10; from 2.22 ± 0.06 to 2.14 ± 0.03; P > 0.05), MGS IV (Fig. 5C, n = 10; from 2.48 ± 0.03 to 2.47 ± 0.03; P > 0.05). These results indicate that the repair capacity of mtDNA lesions in the macular regions is decreased in AMD.
Figure 5.
Time-course analysis of mtDNA damage repair after oxidative stress in macular RPE cells from MGS II to MGS IV eyes. RPE cells were harvested at 0-, 3-, and 6-hour recovery time points to determine mtDNA lesions after exposure to 200 μM H2O2 for 1 hour to assess the repair capability. β-Globin was used as the control. Results are mean ± SEM. (A) Total mtDNA lesion in macular RPE cells from MGS II (n = 10). (B) Total mtDNA lesions in macular RPE cells from MGS III (n = 10). (C) Total mtDNA lesions in macular RPE cells from MGS IV (n = 10).
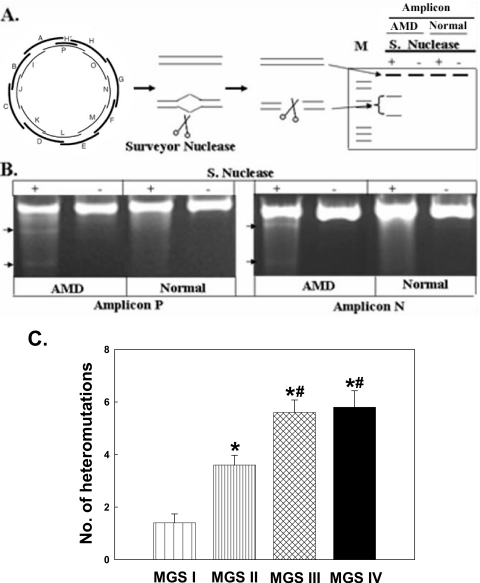
Mitochondrial Genome Heteroplasmic Mutations Are Associated with AMD Severity
Because of the polyploid nature of the mitochondrial genome, mutant mtDNA molecules may coexist with wild-type mtDNA, a condition termed heteroplasmy. In general, mtDNA mutations do not cause mitochondrial dysfunction until the damage reaches a critical threshold. The numbers of whole mitochondrial genome heteroplasmic mutations in macular RPE cells from MGS levels I, II, III, and IV were determined. Figure 6B shows an example of a level III AMD donor sample that has a heteroplasmic mutation in both amplicon P and amplicon N. Our analysis revealed that approximately one heteroplasmic mutation was detected per RPE sample from young periphery and young macula and from aged periphery and aged macula. There was no difference in heteroplasmic mutation frequency between these groups (data not shown). However, there was a statistically significant increase in the number of mtDNA heteroplasmic mutations in macular RPE cells at the MGS level II (3.6 ± 0.3) compared with MGS I (1.4 ± 0.3) and between MGS III (5.6 ± 0.4) and MGS IV (5.8 ± 0.6) compared with MGS II (Fig. 6C). These results show that mtDNA heteroplasmic mutations are positively associated with AMD severity.
Figure 6.
The association of mitochondrial genome heteroplasmic mutation and AMD severity in RPE cells. (A) Flow chart of heteroplasmic mtDNA mutation identification with nuclease. mtDNA was amplified with 17 sets of primers. PCR products were screened with nuclease that cleaved DNA with high specificity at sites of base substitution mismatch. (B) PCR amplification of amplicons P and N. Bottom arrow: heteroplasmic mutation digested with nuclease to produce two bands in AMD, not shown in normal control. (C) Numbers of heteroplasmic mutations in the whole mitochondrial genome with all sets of primers in macular RPE cells from MGS I (n = 10), MGS II (n = 10), MGS III (n = 10), and MGS IV (n = 10) donor eyes. Results are mean ± SEM. *P < 0.001 versus MGS I; #P < 0.001 versus MGS II.
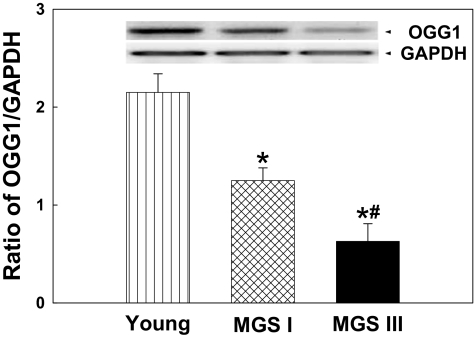
Lower Levels of hOGG1 Expression in Aged and AMD Macular RPE Cells
OGG1 is among the major enzymes responsible for mtDNA repair.1,27,28 To study the association of OGG1 with aging and AMD, the fourth passages of primary cultured macular RPE cells from young, aged healthy (MGS I), and AMD (MGS III) eyes were collected. OGG1 expression level was determined by Western blot and was expressed as a ratio of OGG1 to the internal control protein GAPDH. As shown in Figure 7, the expression level of OGG1 was approximately twofold higher in young macular RPE cells compared with aged macular RPE cells. More interestingly, the expression level of OGG1 decreased by almost half in AMD macular RPE cells compared with aged macular RPE cells. These results suggest that reduced levels of this DNA repair enzyme in aging and in AMD RPE cells may partially contribute to the accumulation of mtDNA lesions.
Figure 7.
Levels of hOGG1 expression in macular RPE cells. (A) Expression of hOGG1 in macular RPE cells from young, aged (MGS I), and AMD (MGS III) eyes determined by Western blot analysis. (B) Quantitation by densitometry was performed for the levels of hOGG1 expression in young, aged, and AMD macular RPE cells. Results are mean ± SEM. n = 5 for each group. *P < 0.001 versus young; #P < 0.01 versus MGS I.
Discussion
AMD is a complex disease involving a combination of environmental and genetic risk factors.29,30 Human donor eyes affected by AMD contain increased levels of protein adducts resulting from the oxidative modification of carbohydrates and lipids31,32 and higher levels of antioxidant enzymes than healthy eyes.33,34 Mitochondrial proteomic studies of RPE cells identified multiple proteins that are changed in AMD.10,35 In this study we used primary RPE cell lines isolated from fresh human donor eyes as we postulated that a propensity for mtDNA damage would be retained over several passages given the poor capacity for mtDNA repair. Our experimental results (Fig. 1) confirmed a similar effect from oxidative stress on mtDNA damage between primary RPE cell cultures and freshly isolated native RPE cells from human donor eyes.
In the present study, we measured the relative amplification of the mtDNA genome from macular and peripheral RPE cells in young, aged, and AMD donor eyes using a sensitive and validated QPCR method. Briefly, a damaged DNA template contains base modifications, strand breaks, and apurinic sites that block the progression of DNA polymerase, resulting in decreased target DNA amplification.23 We detected higher mtDNA damage in AMD eyes than in aged normal eyes. These findings are consistent with previous studies,9 as is our observation of higher mtDNA lesion frequencies in macular RPE cells in aged eyes compared with young eyes24 and RPE and retina from donor eyes with AMD.3,9,36 Multiple lines of evidence suggest that mtDNA is vulnerable to oxidative damage because of multiple factors. First, constant exposure to ROS makes it inherently unstable. Second, its proximity to the electron transport chain makes it an easy target for electrophile and oxidant attack. Third, mtDNA lacks protective histones and nonhistone proteins.2 RPE from eyes with AMD consistently showed more mtDNA lesions, positively correlating with increasing phenotypic severity. Our studies found that there was also an increase in the number of heteromutations in macular RPE from AMD donor eyes compared with age-matched controls (Fig. 5B). In a healthy cell, all mtDNA is identical, a condition called homoplasty. However, in various disease conditions, mutated mtDNA may coexist with normal DNA, leading to the heteroplasmic state seen in AMD sample groups. Because mtDNA is intron-less and has a high transcription rate, oxidatively mediated mtDNA mutations likely result in abnormal protein accumulation, leading to RPE cell dysfunction or death. Further studies may be needed to find the threshold level of mtDNA heteroplasmy that results in the AMD phenotype.
We observed that young RPE cells have a high capacity to repair mitochondrial DNA (Fig. 6). By contrast, AMD eyes showed poor repair efficiency with minimal amelioration in mtDNA damage levels throughout the study period. We also noted impaired repair efficiency in RPE samples from aged donor eyes compared with samples from young donors. An explanation for this may be related to our findings of decreased expression of the DNA repair enzyme OGG1 in samples with AMD (approximately threefold) compared with age-matched controls. Similar reductions in OGG1 expression was seen in samples from aged eyes (twofold) compared with samples from the young group.
In mitochondria, the primary antioxidants, their chaperones, antioxidant enzymes, and specific protein repair systems work in concert to protect against ROS-induced damage.37 mtDNA lesions accumulate progressively with age in various tissues,4 and higher levels of 8-oxo-dG in mtDNA with increasing age confirm the vulnerability of mtDNA.38 Such accumulations are perhaps caused by defective repair of oxidative damage, as shown in our study and by others.39 The substrate-specific repair enzyme 8-oxoG DNA glycosylase 1 has been localized to the mitochondria in human cells.40 Our experiments demonstrate that the level of OGG1 expression is reduced in aged RPE cells, especially in those from the macular region, and the expression is further reduced in eyes with a more severe AMD phenotype. We speculate that impairment of the DNA repair enzymes in mitochondria of RPE cells increases vulnerability to oxidative stress, leading to the accumulation of mtDNA mutations and diminished or defective mitochondrial proteins and may ultimately contribute to cell dysfunction. Evidence supporting our hypothesis comes directly from a previous study,41 in which targeting hOGG1 to the mitochondria of oligodendrocytes enhanced mtDNA repair efficiency and protected against caspase-9–dependent apoptosis after oxidative stress.
In conclusion, our studies demonstrate that RPE cells from aged and AMD donor eyes have increased mtDNA damage. Additionally, mtDNA damage in macular RPE cells was positively correlated with greater phenotypic AMD severity. Not only did macular RPE cells from AMD eyes have higher frequencies of mtDNA lesions, more mtDNA genomic heteroplasmic mutations were detected than in their age-matched controls. Finally, we link the accumulation of mtDNA damage to a decline in expression of the DNA repair enzyme OGG1 and speculate that this decline contributes to some extent to inefficient DNA repair capacity, especially in more advanced AMD levels. These data strongly support our hypothesis of the mitochondria-based ROS model for the development of AMD.1,13 Increased mtDNA damage in aged RPE cells, exacerbated by a decline in its repair capacity, lead to further accumulation of mtDNA lesions. As damage accumulates, mitochondrial redox function decreases, leading to a vicious circle of ROS production and mitochondrial damage. This, in turn, leads to diminished energy production. When the level of energy production drops, it triggers future dysfunction and apoptosis of RPE, possibly an early pathologic event of AMD. Our studies suggest that preventing mtDNA damage and enhancing mtDNA repair capacity may point toward novel therapeutic strategies for AMD.
Acknowledgments
The authors thank Gangduo Wang for helping with data analysis.
Footnotes
Supported by National Institutes of Health Grants EY12850 and R01EY019688 and by Research to Prevent Blindness, Inc.
Disclosure: H. Lin, None; H. Xu, None; F.-Q. Liang, None; H. Liang, None; P. Gupta, None; A.N. Havey, None; M.E. Boulton, None; B.F. Godley, None
References
- 1. Jarrett SG, Lewin AS, Boulton ME. The importance of mitochondria in age-related and inherited eye disorders. Ophthalmic Res. 2010;44:179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarrett SG, Lin H, Godley BF, Boulton ME. Mitochondrial DNA damage and its potential role in retinal degeneration. Prog Retin Eye Res. 2008;27:596–607 [DOI] [PubMed] [Google Scholar]
- 3. Feher J, Kovacs I, Artico M, Cavallotti C, Papale A, Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol Aging. 2006;27:983–993 [DOI] [PubMed] [Google Scholar]
- 4. Barreau E, Brossas JY, Courtois Y, Treton JA. Accumulation of mitochondrial DNA deletions in human retina during aging. Invest Ophthalmol Vis Sci. 1996;37:384–391 [PubMed] [Google Scholar]
- 5. Wang AL, Lukas TJ, Yuan M, Neufeld AH. Increased mitochondrial DNA damage and down-regulation of DNA repair enzymes in aged rodent retinal pigment epithelium and choroid. Mol Vis. 2008;14:644–651 [PMC free article] [PubMed] [Google Scholar]
- 6. Wang AL, Lukas TJ, Yuan M, Neufeld AH. Age-related increase in mitochondrial DNA damage and loss of DNA repair capacity in the neural retina. Neurobiol Aging. 2010;31:2002–2010 [DOI] [PubMed] [Google Scholar]
- 7. Udar N, Atilano SR, Memarzadeh M, et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:2966–2974 [DOI] [PubMed] [Google Scholar]
- 8. Kenney MC, Atilano SR, Boyer D, et al. Characterization of retinal and blood mitochondrial DNA from age-related macular degeneration patients. Invest Ophthalmol Vis Sci. 2010;51:4289–4297 [DOI] [PubMed] [Google Scholar]
- 9. Karunadharma PP, Nordgaard CL, Olsen TW, Ferrington DA. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51:5470–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:2848–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Godley BF, Shamsi FA, Liang FQ, Jarrett SG, Davies S, Boulton M. Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J Biol Chem. 2005;280:21061–21066 [DOI] [PubMed] [Google Scholar]
- 12. Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang FQ, Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res. 2003;76:397–403 [DOI] [PubMed] [Google Scholar]
- 14. Santos JH, Hunakova L, Chen Y, Bortner C, Van Houten B. Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death. J Biol Chem. 2003;278:1728–1734 [DOI] [PubMed] [Google Scholar]
- 15. Justilien V, Pang JJ, Renganathan K, et al. SOD2 knockdown mouse model of early AMD. Invest Ophthalmol Vis Sci. 2007;48:4407–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dianov GL, Souza-Pinto N, Nyaga SG, Thybo T, Stevnsner T, Bohr VA. Base excision repair in nuclear and mitochondrial DNA. Prog Nucl Acid Res Mol Biol. 2001;68:285–297 [DOI] [PubMed] [Google Scholar]
- 17. Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol Aging. 2006;27:1129–1136 [DOI] [PubMed] [Google Scholar]
- 18. LeDoux SP, Druzhyna NM, Hollensworth SB, Harrison JF, Wilson GL. Mitochondrial DNA repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience. 2007;145:1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst). 2008;7:605–616 [DOI] [PubMed] [Google Scholar]
- 20. Chen D, Cao G, Hastings T, et al. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J Neurochem. 2002;81:1273–1284 [DOI] [PubMed] [Google Scholar]
- 21. Shen GP, Galick H, Inoue M, Wallace SS. Decline of nuclear and mitochondrial oxidative base excision repair activity in late passage human diploid fibroblasts. DNA Repair (Amst). 2003;2:673–693 [DOI] [PubMed] [Google Scholar]
- 22. Olsen TW, Feng X. The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45:4484–4490 [DOI] [PubMed] [Google Scholar]
- 23. Ballinger SW, Bouder TG, Davis GS, Judice SA, Nicklas JA, Albertini RJ. Mitochondrial genome damage associated with cigarette smoking. Cancer Res. 1996;56:5692–5697 [PubMed] [Google Scholar]
- 24. Ballinger SW, Van Houten B, Jin GF, Conklin CA, Godley BF. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;68:765–772 [DOI] [PubMed] [Google Scholar]
- 25. Bannwarth S, Procaccio V, Paquis-Flucklinger V. Rapid identification of unknown heteroplasmic mutations across the entire human mitochondrial genome with mismatch-specific Surveyor Nuclease. Nat Protoc. 2006;1:2037–2047 [DOI] [PubMed] [Google Scholar]
- 26. Cai J, Jiang WG, Grant MB, Boulton M. Pigment epithelium-derived factor inhibits angiogenesis via regulated intracellular proteolysis of vascular endothelial growth factor receptor 1. J Biol Chem. 2006;281:3604–3613 [DOI] [PubMed] [Google Scholar]
- 27. de Souza-Pinto NC, Eide L, Hogue BA, et al. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–5381 [PubMed] [Google Scholar]
- 28. Singh KK, Sigala B, Sikder HA, Schwimmer C. Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Res. 2001;29:1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration—emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W, Dean DC, Kaplan HJ. Age-related macular degeneration. Discov Med. 9:13–15 [PubMed] [Google Scholar]
- 31. Crabb JW, Miyagi M, Gu X, et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:14682–14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howes KA, Liu Y, Dunaief JL, et al. Receptor for advanced glycation end products and age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45:3713–3720 [DOI] [PubMed] [Google Scholar]
- 33. Decanini A, Nordgaard CL, Feng X, Ferrington DA, Olsen TW. Changes in select redox proteins of the retinal pigment epithelium in age-related macular degeneration. Am J Ophthalmol. 2007;143:607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frank RN, Amin RH, Puklin JE. Antioxidant enzymes in the macular retinal pigment epithelium of eyes with neovascular age-related macular degeneration. Am J Ophthalmol. 1999;127:694–709 [DOI] [PubMed] [Google Scholar]
- 35. Nordgaard CL, Berg KM, Kapphahn RJ, et al. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:815–822 [DOI] [PubMed] [Google Scholar]
- 36. Gramajo AL, Zacharias LC, Neekhra A, et al. Mitochondrial DNA damage induced by 7-ketocholesterol in human retinal pigment epithelial cells in vitro. Invest Ophthalmol Vis Sci. 51:1164–1170 [DOI] [PubMed] [Google Scholar]
- 37. Brennan LA, Kantorow M. Mitochondrial function and redox control in the aging eye: role of MsrA and other repair systems in cataract and macular degenerations. Exp Eye Res. 2009;88:195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miceli MV, Jazwinski SM. Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2005;46:1765–1773 [DOI] [PubMed] [Google Scholar]
- 39. Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev. 2008;129:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cortina MS, Gordon WC, Lukiw WJ, Bazan NG. Oxidative stress-induced retinal damage up-regulates DNA polymerase gamma and 8-oxoguanine-DNA-glycosylase in photoreceptor synaptic mitochondria. Exp Eye Res. 2005;81:742–750 [DOI] [PubMed] [Google Scholar]
- 41. Druzhyna NM, Hollensworth SB, Kelley MR, Wilson GL, Ledoux SP. Targeting human 8-oxoguanine glycosylase to mitochondria of oligodendrocytes protects against menadione-induced oxidative stress. Glia. 2003;42:370–378 [DOI] [PubMed] [Google Scholar]