SUMMARY
The decision by a cell to enter a round of growth and division must be intimately coordinated with nutrient availability and its metabolic state. These metabolic and nutritional requirements, and the mechanisms by which they induce cell growth and proliferation, remain poorly understood. Herein, we report that acetyl-CoA is the downstream metabolite of carbon sources that represents a critical metabolic signal for growth and proliferation. Upon entry into growth, intracellular acetyl-CoA levels increase substantially and consequently induce the Gcn5p/SAGA-catalyzed acetylation of histones at genes important for growth, thereby enabling their rapid transcription and commitment to growth. Thus, acetyl-CoA functions as a carbon-source rheostat that signals the initiation of the cellular growth program by promoting the acetylation of histones specifically at growth genes.
INTRODUCTION
How cell growth and proliferation are coordinated with metabolism and the metabolic state of a cell remains an important unresolved question. When starved for carbon sources or nutrients such as sulfate or phosphate, budding yeast cells utilize dedicated strategies to halt cell division to promote survival (Boer et al., 2008; Gray et al., 2004). Upon receipt of appropriate nutritional or metabolic cues, such as the presence of glucose, they resume proliferation (Dechant and Peter, 2008; Zaman et al., 2008). These considerations indicate that yeast cells must attain a sufficiently favorable metabolic state to initiate a round of growth and division (Dechant and Peter, 2008; Gray et al., 2004; Zaman et al., 2008).
We previously characterized the robust oscillations in oxygen consumption exhibited by budding yeast during continuous, glucose-limited growth, termed yeast metabolic cycles (YMC), which depict the life of a yeast cell population under a slow-growth environment (Tu et al., 2005; Tu and McKnight, 2006; Tu et al., 2007). During such cycles, the highly synchronized cells continuously transition between three metabolic phases (Figure 1), termed OX (oxidative), RB (reductive, building), and RC (reductive, charging) (Tu et al., 2005; Tu and McKnight, 2006). The OX phase represents the peak of mitochondrial respiration and is associated with the rapid induction of genes involved in growth. These include the vast majority of ribosomal, translation, rRNA processing, tRNA processing, and amino acid biosynthesis genes (Tu et al., 2005). Cell division occurs during the RB phase when the rate of oxygen consumption begins to decrease, which is accompanied by the induction of DNA replication and cell cycle genes (Rowicka et al., 2007; Tu et al., 2005). In the RC phase, many genes associated with stress, starvation, and survival-associated responses (e.g., heat shock proteins, stress resistance, vacuole, ubiquitin/proteasome) are activated prior to the next OX phase. Consequently, a variety of fundamental cellular and metabolic processes are precisely orchestrated about these metabolic cycles (Tu et al., 2005).
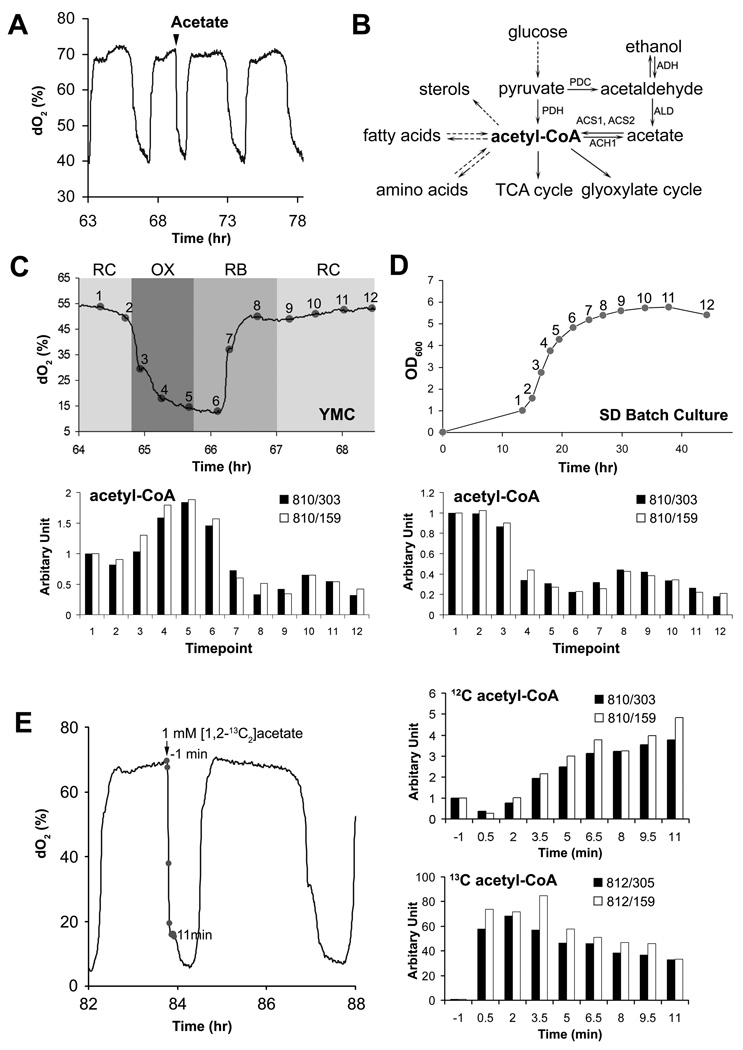
Figure 1. Acetyl-CoA is a Metabolite of Carbon Sources that Induces Entry into Growth.
(A) Entry into the OX, growth phase of the YMC can be induced by addition of glucose as well as products of glycolytic metabolism. Acetate (1 mM) was added to cycling cells in the RC phase and immediately triggered entry into OX phase, which is characterized by a burst of mitochondrial respiration and transcription of growth genes (Tu et al., 2005). In addition to acetate, ethanol, acetaldehyde, and lactate were also able to induce entry into growth.
(B) The yeast pathways that synthesize and consume acetyl-CoA.
(C) Upregulation of acetyl-CoA production upon entry into the OX growth phase. Metabolites were extracted from cells harvested at the indicated 12 time points across one cycle. Acetyl-CoA levels were measured by LC-MS/MS using multiple reaction monitoring (MRM) and quantitating two specific daughter fragments for acetyl-CoA (303, 159 Da) as described previously (Tu et al., 2007). Data were normalized against the first time point.
(D) Acetyl-CoA levels correlate with growth rate in batch culture. Metabolites were extracted from an equivalent number of cells collected at the specified time points during batch culture growth in SD minimal media. Note that acetyl-CoA levels are significantly higher in exponential phase compared to stationary phase.
(E) Acetate-induced entry into growth is accompanied by a significant increase in intracellular acetyl-CoA. Metabolites were extracted from cells collected at 1.5 minute intervals following addition of 13C-labeled acetate to cells in RC phase. 12C and 13C acetyl-CoA were quantitated by LC-MS/MS. 13C acetate was quickly converted to 13C acetyl-CoA, and 12C acetyl-CoA from endogenous, unlabeled carbon sources also increased in response to the stimulus.
Cells in the RC phase of the YMC exhibit several characteristics of stationary phase and quiescent cells (Allen et al., 2006; Shi et al., 2010). During this temporal window, cells express many genes negatively correlated with increasing growth rate (Brauer et al., 2008; Lu et al., 2009). They also become more dense and accumulate the storage carbohydrates trehalose and glycogen in a manner similar to stationary phase cells (Shi et al., 2010). Thus, during the YMC cells alternate between phases that can be likened to quiescence or G0 (RC), and phases whereupon they enter growth and activate growth genes (OX) in preparation for a round of division (RB). Through comprehensive transcript and metabolite profiling studies, we have previously uncovered the temporal sequence of transcriptional and metabolic outputs as cells exit the quiescent-like (RC) phase and enter the growth (OX) and division (RB) phases (Rowicka et al., 2007; Tu et al., 2005; Tu et al., 2007). Herein, we describe how the YMC system enabled us to discover a key metabolic trigger of cell growth and proliferation.
RESULTS
Carbon Sources Fuel the Production of Acetyl-CoA upon Entry into Growth
What are the metabolic and nutritional cues that induce yeast cells to enter growth? To address this question using the YMC, we observed that the addition of select carbon sources can induce metabolically cycling cells to prematurely exit the RC quiescent-like phase and immediately enter the OX growth phase (Figure 1A). Such "phase advancement" into growth is accompanied by a burst of mitochondrial respiration and the induction of growth genes that are typical of a normal OX phase (Chen et al., 2007; Tu and McKnight, 2009). Shortly thereafter, cells begin the division process and normal metabolic cycles resume. While the addition of glucose and galactose readily triggered cells to enter growth, even the end products of glycolytic metabolism, such as ethanol, acetate, and lactate, effectively induced cells to exit the RC phase and rapidly enter the OX growth phase (Figure 1A).
Because ethanol and acetate must be converted to acetyl-CoA during the course of their metabolism (Figure 1B), we suspected that acetyl-CoA itself might represent a metabolic signal to enter growth (Figure S1). We confirmed previous metabolite investigations of the YMC which revealed that acetyl-CoA is one of the most dynamically oscillating metabolites that peaks during the OX growth phase (Figure 1C) (Tu et al., 2007). Moreover, we determined that rapidly-growing, exponential phase yeast cells contained higher levels of acetyl-CoA compared to slower-growing cells entering stationary phase (Figure 1D). Together, these data demonstrate that numerous carbon sources that can be converted to acetyl-CoA can induce cells to enter growth. They further reveal that a substantial increase in intracellular acetyl-CoA is associated with, and may be required, for entry into growth.
Identification of Proteins within the SAGA Transcriptional Coactivator that are Acetylated upon Entry into Growth
There are several possibilities by which acetyl-CoA could be functioning as a metabolic signal for growth. Acetyl-CoA is not only a central intermediate in the oxidation of glucose to produce ATP, but also a precursor for the biosynthesis of numerous metabolites required to build a new cell, such as lipids and sterols (Figure 1B). Therefore, metabolites downstream of acetyl-CoA could be signaling growth. Acetyl-CoA is also the acetyl donor for acetylation reactions within the cell. We hypothesized the existence of proteins that might become acetylated only upon the increase in intracellular acetyl-CoA that accompanies entry into growth. The acetylation of such a protein might then enable it to perform some function required for growth or proliferation.
To test this possibility, we utilized an unbiased proteomic approach in search of proteins that might become acetylated only upon entry into the OX growth phase of the YMC in tune with the increase in intracellular acetyl-CoA levels. We spiked cells in the quiescent-like RC phase of the YMC with 13C acetate, which induced the cycling cell population to phase advance and rapidly enter the OX growth phase (Figure 1E). Several minutes later at a time when ~50% of the acetyl-CoA was still the 13C labeled form, we harvested cells, immunoprecipitated acetylated peptides, and prepared them for LC-MS/MS analysis to identify candidates that contained either a 13C acetyl-lysine or unlabeled 12C acetyl-lysine. Some of the proteins identified in this manner might be newly acetylated upon entry into the OX growth phase.
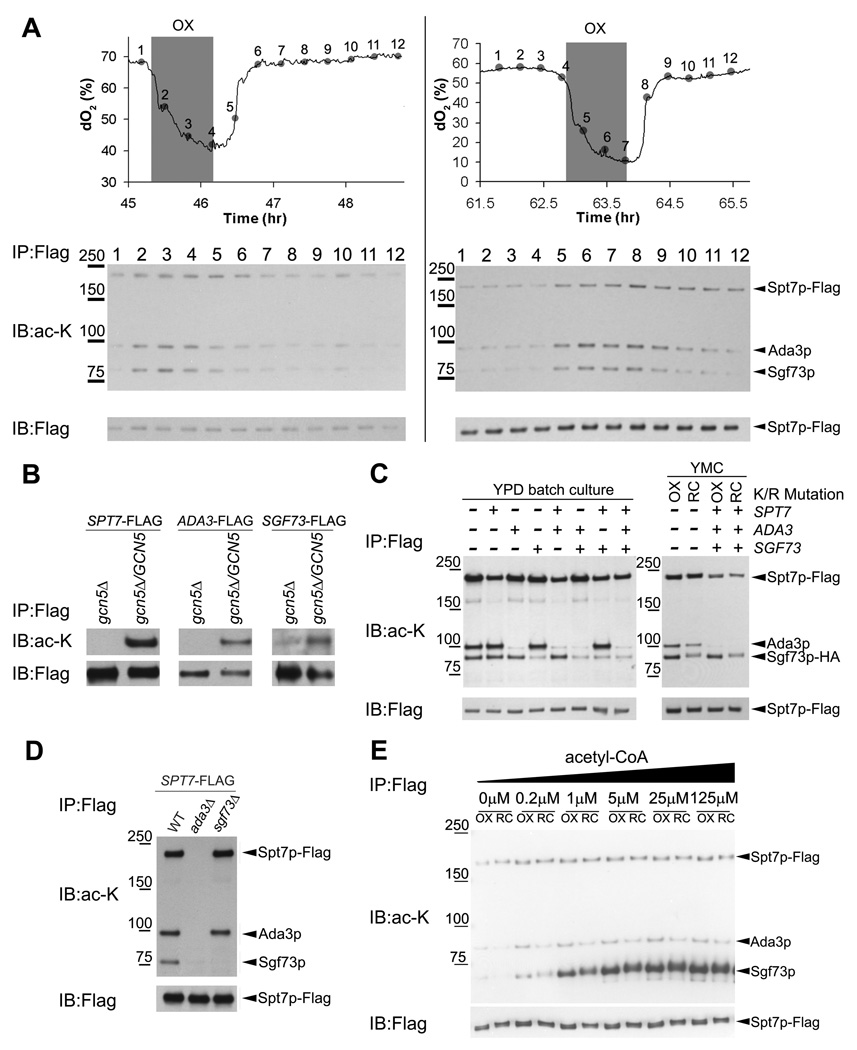
One of the proteins identified in this screen was Spt7p, a component of the SAGA (Spt-Ada-Gcn5-Acetyltransferase) transcriptional coactivator complex (Grant et al., 1997). To test if the acetylation of Spt7p was dynamic and correlated with the fluctuation in acetyl-CoA levels across the YMC, we epitope-tagged Spt7p and immunoprecipitated SAGA across the YMC to assess its acetylation state (Figure 2A). We confirmed that Spt7p was acetylated and discovered that two other proteins (Ada3p, Sgf73p) within the SAGA complex were dynamically acetylated during the OX phase precisely when intracellular acetyl-CoA levels increased (Figure 2A). The cyclic acetylation of Ada3p and Sgf73p (the yeast homolog of Ataxin-7) was more dynamic than that of Spt7p, and the steady state abundance of SAGA remained largely unchanged as a function of the YMC (Figure 2A, Figure S2A). In support of these data, previous studies also detected acetylation of yeast Spt7p and Sgf73p, and human Ada3p within SAGA (Gamper et al., 2009; Mischerikow et al., 2009).
Figure 2. Several Components of the SAGA Transcriptional Coactivator are Acetylated by Gcn5p upon Entry into Growth.
(A) SAGA was immunoprecipitated from a SPT7-FLAG strain across 12 time points of the YMC and protein acetylation was detected by Western blot using an anti-acetyl-lysine antibody. Spt7p, Ada3p, and Sgf73p were found to be dynamically acetylated over the YMC with the highest levels in the OX growth phase, when acetyl-CoA levels increase substantially (Figure 1C). Data from two independent metabolic cycle experiments are shown.
(B) Acetylation of SAGA components is dependent on Gcn5p. Strains expressing C-terminally FLAG-tagged versions of either SPT7, ADA3, or SGF73 in a haploid gcn5Δ background were constructed. Proteins were immunoprecipitated from cells grown to log phase using a FLAG antibody and then acetylation state was detected by Western blot. After mating to a wild type GCN5 haploid, acetylation of each of the three proteins was restored in the resulting diploid.
(C) SAGA components are acetylated at many lysine residues. Mutants were constructed in a SPT7-FLAG, SGF73-HA strain, according to sites identified by LC-MS/MS and by a previous study (Mischerikow et al., 2009). K603, 607 on Spt7p, K183 on Ada3p, and K171, 199, 211, 224, 288, 300 on Sgf73p were mutated to arginine separately or in combination as indicated. The resulting consequences on acetylation of SAGA was determined by immunoprecipitating SAGA from either a log phase YPD batch culture (left), or OX or RC phase of the YMC (right), followed by Western analysis.
(D) The acetylation of SAGA components is dependent on Ada3p. The acetylation status of SAGA components was determined as described above in the indicated strains.
(E) Sgf73p is hyperacetylated upon incubation of SAGA with acetyl-CoA in vitro. Immunopurified SAGA complex from the OX or RC phase of the YMC was incubated with the indicated concentrations of acetyl-CoA for 30 min at 30°C. The acetylation state of SAGA components was monitored by Western blot. Note that Sgf73p within SAGA becomes hyperacetylated, suggesting there are many lysine residues that can be acetylated in response to acetyl-CoA.
The Gcn5p Acetyltransferase Directs the Acetylation of SAGA Components and Histones in Tune with the Increase in Acetyl-CoA upon Entry into Growth
The Gcn5p acetyltransferase is a component of SAGA (Brownell et al., 1996; Grant et al., 1997), suggesting it might be responsible for acetylation of proteins within the complex. Indeed, we confirmed that Spt7p, Sgf73p, and Ada3p were no longer acetylated in a gcn5Δ mutant (Figure 2B), despite still being present within a SAGA complex lacking Gcn5p (Wu and Winston, 2002) (Figure S2B). Multiple lysines within these proteins were confirmed to be sites of acetylation by mutation to arginine (Figure 2C). Moreover, in the absence of Ada3p, the acetylation of both Spt7p and Sgf73p was completely eliminated (Figure 2D).
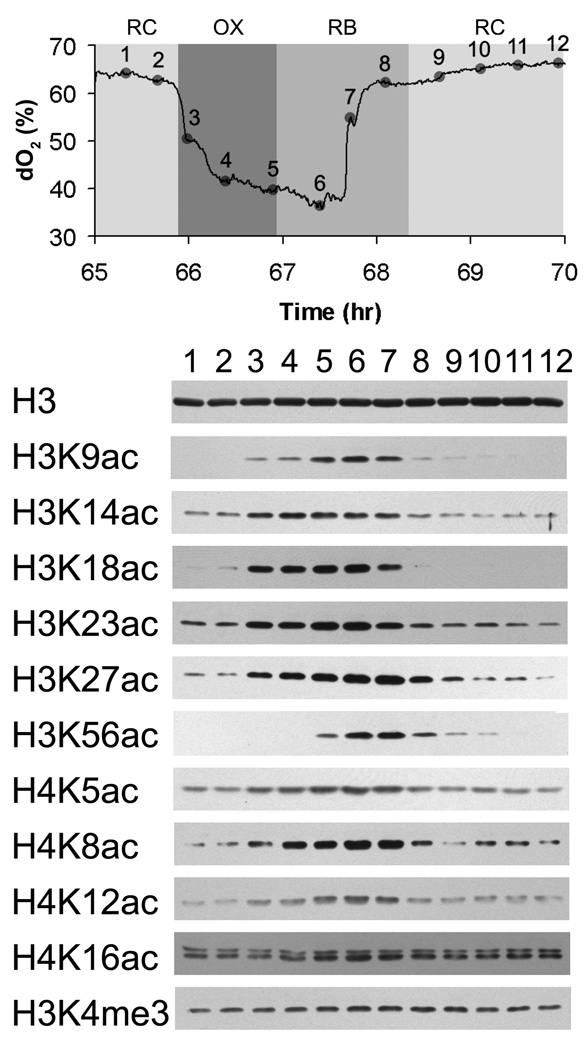
Since SAGA functions as a histone acetyltransferase (Grant et al., 1997; Lee and Workman, 2007; Suka et al., 2001), we next tested whether the acetylation of particular sites on histones might also occur in tune with the increase in acetyl-CoA concentration upon entry into the OX growth phase. Strikingly, we observed that the acetylation of many sites on histone H3 (K9, K14, K18, K23, K27, K56) and histone H4 (K5, K8, K12) was highly periodic over the YMC. All of these sites, with the exception of H3K56 and H4K16, were acetylated beginning in the OX growth phase, which is precisely when acetyl-CoA levels within the cell increase substantially (Figure 3). H3K56 acetylation occurred at later time points during the temporal window of the YMC when cell division occurs (Figure 3) (Tu et al., 2005). These data confirm that H3K56 acetylation occurs during S phase and as a function of the cell cycle (Masumoto et al., 2005; Xu et al., 2005). Interestingly, H3K4 trimethylation, which is a mark associated with active transcription (Pokholok et al., 2005), did not fluctuate significantly at the bulk level (Figure 3). As SAGA has been implicated in the acetylation of many of these sites on H3 that increase in OX phase (Grant et al., 1997; Grant et al., 1999), these data reveal that Gcn5p-containing SAGA might specifically acetylate H3 only following the burst of acetyl-CoA production that is concomitant with entry into growth.
Figure 3. Induction of Histone Acetylation upon Entry into Growth.
Protein extracts were prepared across 12 time points of one metabolic cycle for bulk analysis of histone modifications. Western blots were performed using highly specific antibodies recognizing the indicated modifications on histone H3 or H4. Acetylation of H3K9, H3K14, H3K18, H3K23, H3K27, and H4K5, H4K8, H4K12, increased substantially in tune with the increase in acetyl-CoA that accompanies entry into the OX growth phase. Acetylation of H3K56 increased at later time points in RB phase during the time of cell division. H3K4 trimethylation remained largely unchanged as a function of the YMC as assayed at the bulk level.
SAGA Acetylates Histones Specifically at those Genes Important for Growth upon Entry into Growth
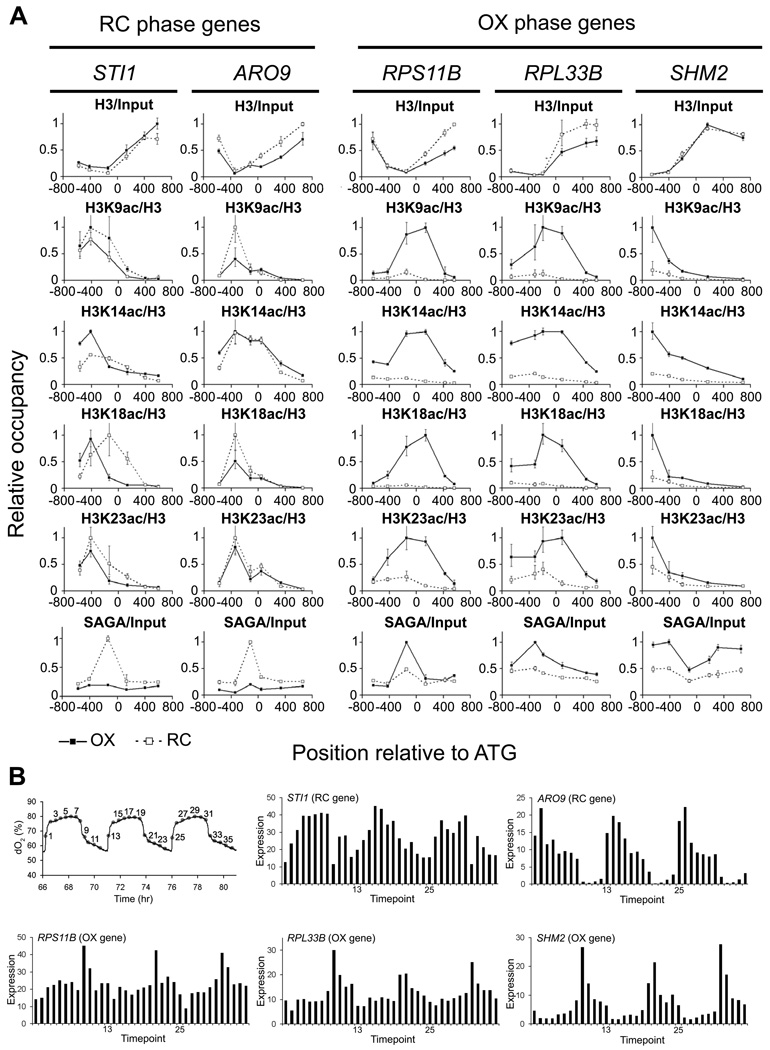
We next tested whether SAGA might be recruited to acetylate H3 specifically at OX phase growth genes to enable their transcription upon entry into growth. Interestingly, many genes previously reported to be regulated by Gcn5p or SAGA are genes involved in growth (e.g., ribosomal) that comprise the OX phase expression program, while others are linked to stress-associated functions (Huisinga and Pugh, 2004; Robert et al., 2004). We chose several SAGA target genes that are induced during the RC phase (STI1, ARO9) and several targets that are induced during the OX phase of the YMC (RPS11B, RPL33B, SHM2) to determine whether they might be regulated by SAGA-catalyzed histone acetylation during the YMC (Huisinga and Pugh, 2004; Robert et al., 2004). We performed chromatin immunoprecipitation (ChIP) analysis to quantitate SAGA binding and histone acetylation at these target genes in either OX or RC phase (Figure 4). We confirmed that SAGA preferentially bound OX phase targets during OX phase, and RC phase targets during RC phase (Figure 4A). H3K9, H3K14, H3K18, H3K23, and H3K27 were significantly acetylated at the OX phase SAGA targets specifically during OX phase, perhaps as expected. In striking contrast, for RC phase SAGA targets, there was no significant increase in H3 acetylation in either RC phase or OX phase (Figure 4A). These data suggest that acetyl-CoA enables SAGA to acetylate histones specifically at OX phase growth genes to facilitate enhanced transcription. In support of this idea, the transcripts of OX phase genes peak very sharply in the midst of the respiratory burst and H3 acetylation has been shown to correlate with transcription rate (Figure 4B) (Kurdistani et al., 2004; Pokholok et al., 2005; Tu et al., 2005).
Figure 4. SAGA and Acetylated Histones are Present at Growth Genes upon Entry into Growth.
(A) Chromatin immunoprecipitation was performed to quantitate SAGA binding and histone acetylation at previously identified SAGA target genes that peak either during RC phase (STI1, ARO9) or OX phase (RPS11B, RPL33B, SHM2). SPT7-FLAG cells were crosslinked for ChIP analysis in early OX phase corresponding to time point 3 and in early RC phase corresponding to time point 9 (Figure 3). Histones at these OX phase genes are significantly acetylated upon gene activation, in contrast to RC phase genes. Several acetylated sites on H4 were also present at ribosomal genes during OX phase (Figure S3). Error bars denote standard deviation for triplicate experiments.
(B) YMC expression profiles of SAGA target genes from RC and OX Phase used for ChIP analysis. Expression data are from 36 time points over 3 consecutive metabolic cycles as reported previously (Tu et al., 2005).
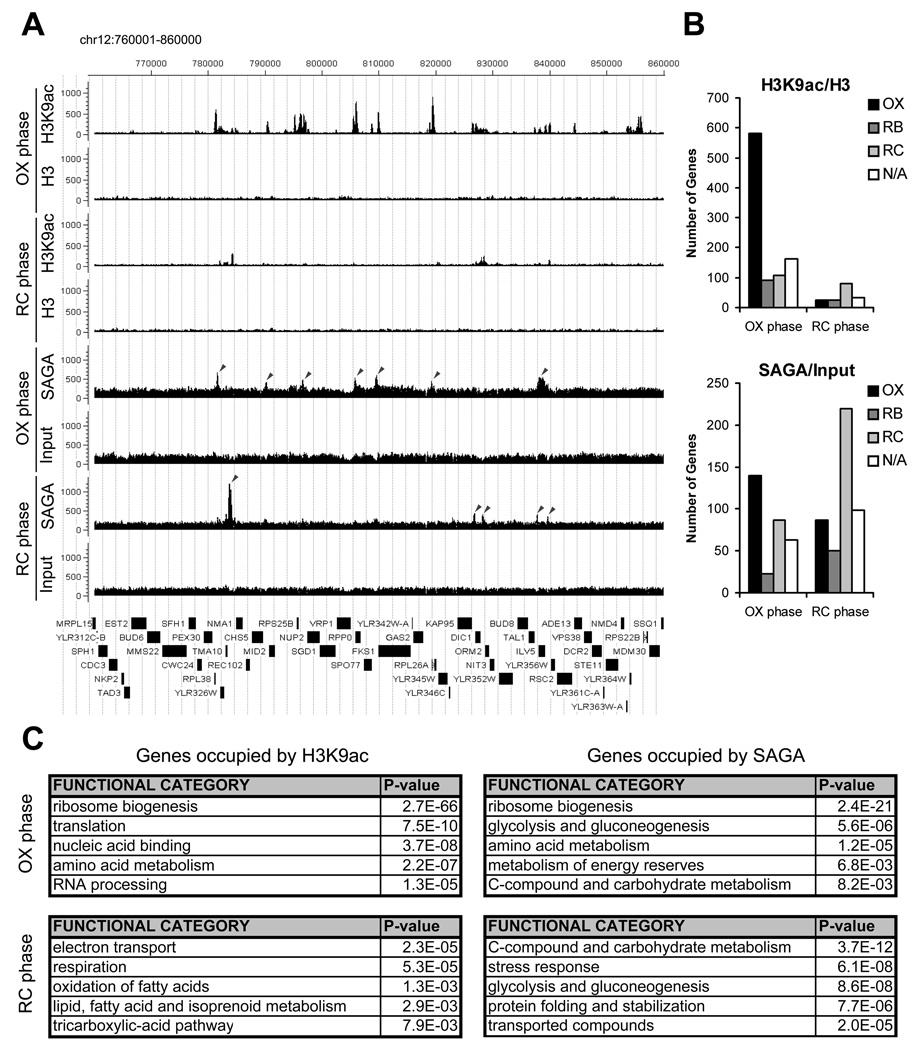
Having established that induction of histone acetylation occurs at several growth genes upon entry into growth, we next performed ChIP-seq analysis to assess the genome-wide locations of H3K9 acetylation, a highly cyclic OX phase histone modification catalyzed by SAGA (Grant et al., 1999; Zhang et al., 1998) (Figure 5). Substantially more genomic regions containing acetylated-H3K9 were observed in OX phase compared to RC phase (Figure 5A, 5B), in agreement with Western analysis (Figure 3). Strikingly, H3K9 acetylation was present almost exclusively at growth genes (e.g., ribosome biogenesis, translation, amino acid metabolism), specifically during OX phase (Figure 5B, 5C and Figure S4). Many chromosomal locations containing H3K9 acetylation correlated well with SAGA binding, especially at ribosomal genes (Figure 5A, 5C). In contrast, many fewer genomic regions containing H3K9 acetylation were detected in RC phase (Figure 5B), despite the fact that >43% of all periodic YMC genes are expressed in RC phase (Tu et al., 2005). Moreover, the majority of regions bound by SAGA in the RC phase were not associated with a corresponding increase in H3K9 acetylation (Figure 5A). Collectively, these data strongly suggest that histone acetylation induced by acetyl-CoA is rate-limiting primarily for the transcription of OX phase growth genes, whereas RC phase genes are much less dependent on histone acetylation for activation.
Figure 5. Genome-wide Analysis Reveals that Histone H3 at Growth Genes becomes Acetylated upon Entry into Growth.
(A) ChIP-seq analysis was performed to reveal genomic regions occupied by H3K9ac and SAGA at a time point in either early OX or early RC phase, as in Figure 4. Sequencing data were analyzed and displayed using CisGenome (Ji et al., 2008). The arrows mark peaks associated with the following genes (maximal YMC phase of expression in parentheses): SAGA OX phase - RPL38 (OX), MID2 (OX), RPS25B (OX), RPP0 (OX), FKS1 (OX), RPL26A (OX), ILV5 (OX); SAGA RC phase - TMA10 (RC), KAP95 (OX), DIC1 (RC), TAL1 (N/A), ILV5 (OX). The temporal expression profiles of these genes across the YMC were highly consistent with these ChIP-seq data (Figure S4). Note that H3K9ac peaks are much more frequent and significant in OX phase compared to RC phase. Many SAGA binding sites in OX phase corresponded directly to significant regions of H3K9 acetylation, while many SAGA binding sites in RC phase did not.
(B) YMC phase distribution of genes occupied by H3K9 acetylation or SAGA in either early OX or RC phase. Many more genomic regions of H3K9 acetylation were present in OX phase (n=942) compared to RC phase (n=161) at the same level of statistical significance. H3K9 acetylation was present almost exclusively at OX phase growth genes, specifically during OX phase (p-value < 9.8 × 10−140). As determined previously by microarray analysis, there are ~1,016 OX, 975 RB, and 1,508 RC phase genes in the YMC (Tu et al., 2005).
(C) Functional categories of genes occupied by H3K9 acetylation or SAGA as determined by MIPS (Munich Information Center for Protein Sequences) functional classification. The genes called by each ChIP-seq experiment were used to identify overrepresented gene functions (the top 100 genes are listed in Tables S1–S4). Note that genes involved in ribosome biogenesis were found to be the most significant group of H3K9ac and SAGA targets. Genes involved in translation, amino acid metabolism, and rRNA processing which are also critical for growth, are also highly overrepresented.
Acetyl-CoA Drives Cell Growth through Gcn5p and SAGA
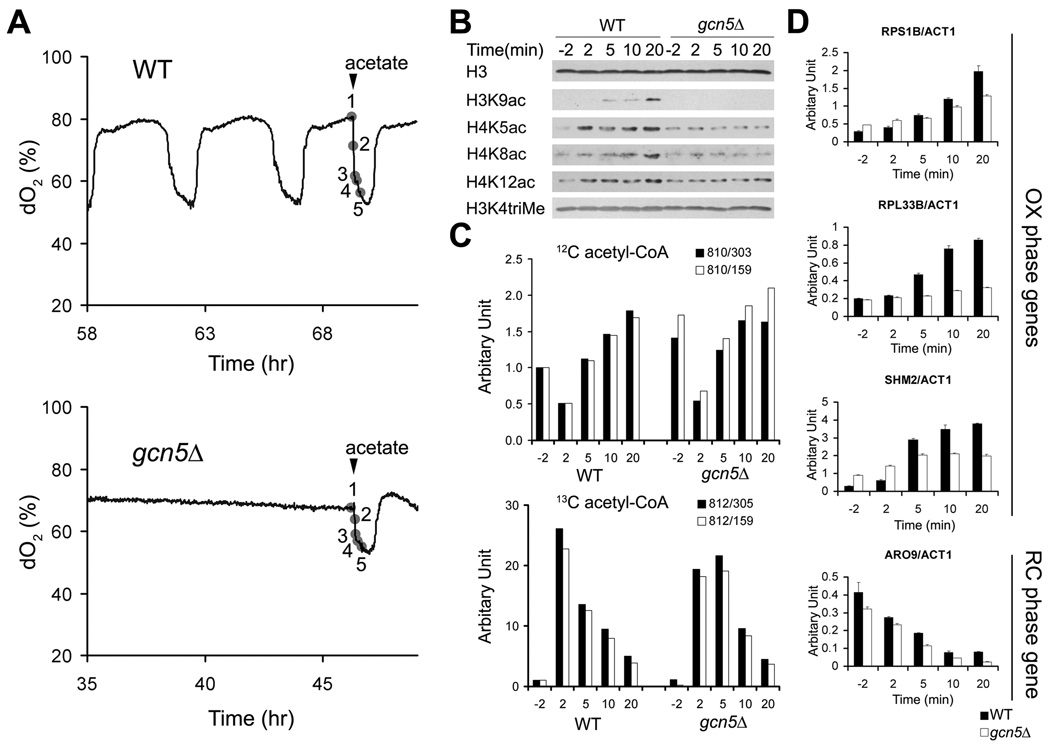
Multiple lines of genetic evidence substantiate our findings that acetyl-CoA sets in motion a series of acetylation events to enable cellular commitment to growth. Bulk histone acetylation was previously found to be compromised in yeast containing mutations in acetyl-CoA synthetase enzymes that control acetyl-CoA flux (Takahashi et al., 2006). Furthermore, a synthetic genetic interaction between a temperature sensitive mutation in acetyl-CoA synthetase (acs2-ts) and gcn5Δ has been reported (Takahashi et al., 2006). Notably, we observed that a gcn5Δ mutant exhibited slow growth and did not undergo the synchronized bursts of respiratory growth that are a hallmark of metabolic cycles, in contrast to mutants lacking other acetyltransferases (Figures 6A, 6C and Figures S5C, S5D). A catalytically inactive Gcn5p point mutant (E173Q) also exhibited a severe cycling phenotype (Figures S5A, S5B) (Trievel et al., 1999). Moreover, an acs1Δ mutant, which lacks the acetyl-CoA synthetase expressed under glucose-poor conditions (van den Berg et al., 1996), did not undergo metabolic cycles (Figure S5E).
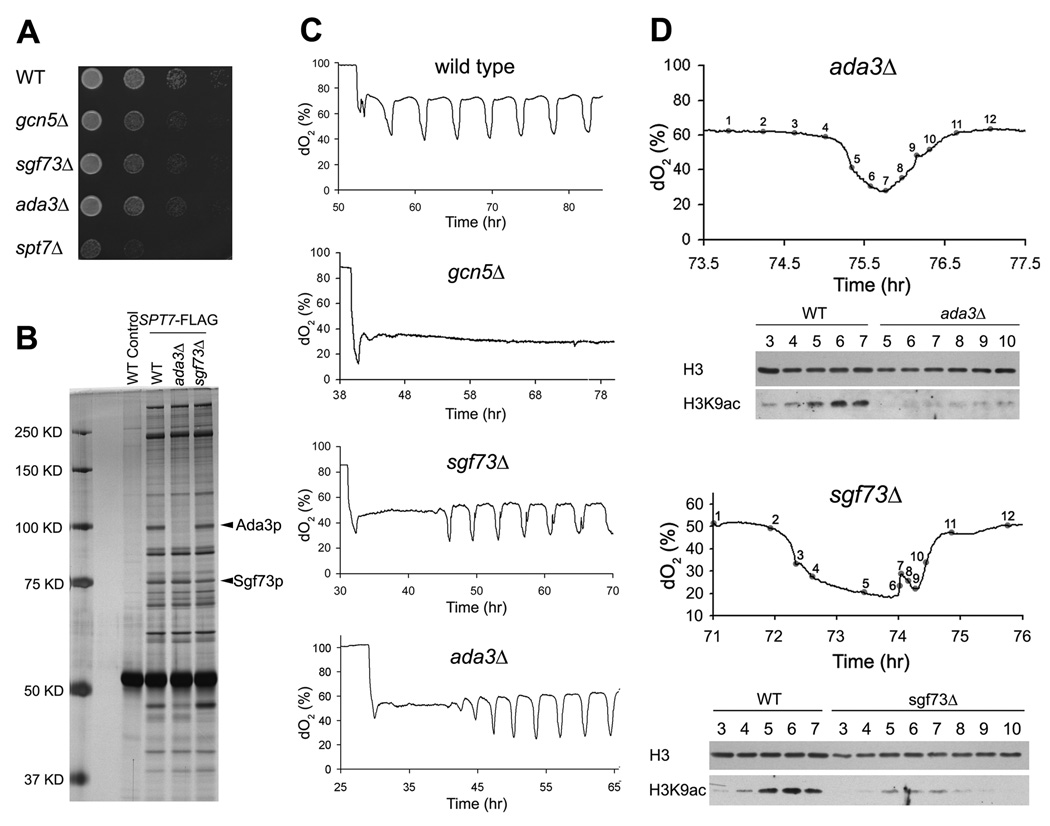
Figure 6. Deletion Mutants of the Acetylated SAGA Components Result in Slow Growth, Reduced H3 Acetylation and Abnormal Metabolic Cycles.
(A) Serial dilutions of the indicated strains were grown on SD minimal media at 30°C.
(B) Composition of the SAGA complex in WT, ada3Δ or sgf73Δ strains.
(C) YMC traces of the indicated strains. Note that the ada3Δ or sgf73Δ mutants experience a long delay (>10 h) before they start cycling.
(D) ada3Δ and sgf73Δ strains have significantly reduced H3K9 acetylation at OX, RB phase time points compared to WT. Note that ada3Δ exhibited a shorter OX phase compared to WT, while sgf73Δ exhibited an abnormal RB phase during which cells exhibit a second burst of oxygen consumption.
How might the acetylation of Sgf73p, Ada3p, and Spt7p by Gcn5p affect the SAGA complex and its ability to acetylate histones at growth genes? To test whether the composition of the SAGA complex might change due to acetylation, we immunoprecipitated SAGA across the YMC and observed no obvious changes in its protein composition (Figure S2B). To test whether the acetylation of SAGA enhances its activity as a histone acetyltransferase, we immunopurified SAGA from either the OX or RC phase and did not detect differences in its ability to acetylate free histones or core nucleosomes in vitro (data not shown). However, we observed that SAGA isolated in RC phase autoacetylates in vitro to a level equivalent to the OX phase, acetylated form, thus making it difficult to assess the acetylation activity of hypoacetylated SAGA (Figure 2E). Moreover, incubation of SAGA with acetyl-CoA resulted in hyperacetylation of Sgf73p, while the acetylation state of other proteins within SAGA remained largely unchanged (Figure 2E). These data suggest that Sgf73p in particular can be acetylated at many sites and that its acetylation might be particularly sensitive to acetyl-CoA fluctuations. Interestingly, the sgf73Δ mutant exhibited slow growth, defects in H3 acetylation, and abnormal metabolic cycles (Figures 6A–6D). Although Sgf73p is also required for histone deubiquitylation catalyzed by SAGA (Kohler et al., 2008), this activity does not appear critical for entry into growth, as the ubp8Δ mutant exhibited normal growth and more normal metabolic cycles (Figure S5F). In addition, the ada3Δ mutant, which completely eliminates acetylation of Spt7p and Sgf73p (Figure 2D), also exhibited slow growth, defects in H3 acetylation, and abnormal cycles (Figures 6A–6D).
Given that SAGA components can be acetylated at many sites (Figures 2C and 2E), we then attempted to address the functional role of the acetylation of SAGA components by constructing a diploid strain expressing one copy of wild type Gcn5p and one copy of a catalytically inactive Gcn5p (E173Q) mutant, each marked with its own epitope tag. Interestingly, we observed that the catalytically inactive Gcn5p was present within a SAGA complex that did not appear to be acetylated, while as expected wild type Gcn5p was present within a SAGA complex that is acetylated (Figure S6A). This diploid strain expressing two different alleles of Gcn5p was able to undergo normal metabolic cycles, and so we tested whether the acetylation of SAGA might influence its ability to be recruited to particular genes. We observed that while both acetylated and deacetylated SAGA were present at an RC phase gene during RC phase, the acetylated form of SAGA (as pulled down using WT Gcn5p) exhibited increased binding to a ribosomal gene during the OX growth phase (Figure S6A). These data suggest that the Gcn5p-catalyzed acetylation of SAGA components might function to aid its recruitment to certain classes of genes, in particular the OX phase growth genes. In this experiment, we cannot exclude the possibility that Gcn5p-dependent acetylation of a chromosome-bound target might be required for the stabilization of SAGA on chromatin. Moreover, it is possible that some of the acetylation sites within SAGA could be a coincidental consequence of having an acetyltransferase enzyme situated within a complex of proteins that is activated in response to a burst of acetyl-CoA production. Nonetheless, previous studies on the role of Ada3p and the observation that the acetylation of SAGA components Spt7p and Sgf73p is dependent on Ada3p (Figure 2D) suggest the possibility that acetylation of SAGA could function to stimulate its nucleosomal acetyltransferase activity and perhaps its activation or recruitment to growth genes (Balasubramanian et al., 2002; Wu et al., 1999).
Lastly, we determined that the acetate-induced increase in H3 acetylation and growth gene expression was severely reduced in a gcn5Δ mutant, indicating that the effects of acetyl-CoA on histone acetylation and cell growth are mediated primarily through Gcn5p (Figures 7A–7D). Moreover, the increase in bulk levels of H4 acetylation in response to acetyl-CoA is also dependent on Gcn5p (Figure 7B), suggesting the activity of the NuA4 acetyltranferase complex can be influenced by SAGA in some instances. Together, these data show that both Gcn5p and acetyl-CoA are at the core of the metabolic cycling response and provide compelling evidence that a burst of acetyl-CoA production triggers a series of acetylation events that culminates in the expression of growth genes and commitment to cell growth and division.
Figure 7. Acetyl-CoA-induced Histone Acetylation and Growth Gene Expression are Driven by Gcn5p and SAGA.
(A) Acetate was added to WT or gcn5Δ cells during continuous growth to provide a burst of acetyl-CoA. Samples were harvested at the 5 indicated time points (2 min before, and 2 min, 5 min, 10 min, 20 min after acetate dosing).
(B) H3K9 acetylation as well as H4 acetylation increase in response to acetate addition in WT cells but not in gcn5Δ cells.
(C) 13C acetate is converted to 13C acetyl-CoA and stimulates 12C acetyl-CoA production in both WT and gcn5Δ cells.
(D) gcn5Δ cells exhibit much slower induction of growth genes in response to acetate addition. Transcript levels of representative genes highly expressed in the OX growth phase (RPS1B, RPL33B, SHM2) or RC quiescent-like phase (ARO9) were measured by real-time PCR and normalized against actin (ACT1) transcript levels. Note that despite the defect in growth gene induction, transcript levels of the RC phase gene ARO9 still decreased with similar kinetics in the gcn5Δ mutant. Error bars denote standard deviation for triplicate experiments.
DISCUSSION
In this study, we have shown how cellular metabolism is directly linked to the regulation of growth genes and hence, entry into growth and the cell cycle. By regulating the acetylation of select proteins important for growth in tune with acetyl-CoA levels, a yeast cell appears capable of coordinating the decision to enter growth with the production of this central metabolite of carbon sources that is indicative of its metabolic and nutritional state. The observation that acetyl-CoA levels tend to be constitutively high under nutrient-rich growth conditions commonly used in the laboratory (Figure 1D) might explain why this fundamental mode of regulation has been previously overlooked. In the YMC, we have slowed the growth of a highly synchronized yeast cell population through the use of continuous, glucose-limited conditions, thereby facilitating the observation of the molecular and metabolic events that accompany entry into growth (Laxman and Tu, 2010).
Notably, not all acetylated proteins are dynamically acetylated across the YMC in tune with metabolic state (Figure S6B and data not shown). How then does the Gcn5p-containing SAGA complex acetylate its substrates in response to increases in acetyl-CoA concentration? Based on previous in vitro Kd (~8.5 µM) and Km (~2.5 µM) measurements for Gcn5p (Berndsen and Denu, 2008; Langer et al., 2002), our estimates of intracellular acetyl-CoA concentration across the YMC (~3 to 30 µM) indicate it is conceivable that the activity of SAGA could be regulated in vivo by physiological fluctuations of acetyl-CoA (Figure S7A). Accordingly, the observation that a catalytically inactive point mutant of Gcn5p fails to undergo the bursts of growth that are a hallmark of the YMC is consistent with a critical role for Gcn5p in driving cell growth (Figure 6C and Figures S5A, S5B). Moreover, we observed that the A190T mutant, which increases its affinity for acetyl-CoA by ~10-fold from 8.5 µM to 0.56 µM (Langer et al., 2002), exhibits a temperature sensitive growth defect on acetate as the carbon source, suggesting that Gcn5p might be finely tuned to sense and respond to a higher threshold of acetyl-CoA levels in vivo (Figure S7B). However, the kinetic parameters for Gcn5p are comparable to those measured for other acetyltransferases such as Esa1p (Berndsen and Denu, 2008). In contrast to this observation, the increase in H4 acetylation catalyzed by the Esa1p–containing NuA4 complex in response to acetyl-CoA is dependent on Gcn5p (Figure 7B), indicating the vivo behavior of acetyltransferases in relation to changes in acetyl-CoA substrate levels might not be immediately predictable from estimates of their kinetic parameters alone. Regardless, multiple lines of evidence strongly indicate that Gcn5p-containing SAGA distinctively acetylates substrates in vivo in response to the burst of acetyl-CoA production that accompanies entry into growth. As such, acetyl-CoA triggers a series of acetylation events within SAGA and at histone tails surrounding growth genes, thereby enabling rapid gene activation and consequent commitment to growth. Future studies will be required to resolve the molecular changes to the SAGA complex following acetylation and how they affect its function and recruitment to particular classes of genes, as well as the basis for its ability to acetylate substrates in tune with increases in intracellular acetyl-CoA levels.
In closing, our data have revealed one of the elusive mechanisms by which carbon sources trigger growth and raise the possibility that the products of glycolytic metabolism (e.g., ethanol, acetate, lactate) may be important activators of growth within microbial communities and cancerous tumors (Sonveaux et al., 2008). In addition, genetic alterations that cause a net accumulation of intracellular acetyl-CoA may enhance cellular proliferative capacity. As protein acetylation has also been linked to metabolism in several previous studies (Choudhary et al., 2009; Friis et al., 2009; Takahashi et al., 2006; Wang et al., 2010; Wellen et al., 2009; Zhao et al., 2010), we predict that a similar control of gene expression and protein activities with respect to intracellular acetyl-CoA levels might represent a widely conserved mechanism by which cell growth and proliferation are coordinated with metabolic state.
EXPERIMENTAL PROCEDURES
Yeast Strains and Methods
All yeast strains were constructed in the CEN.PK background (van Dijken et al., 2000). Gene deletion strains were constructed by replacing the target gene with a drug cassette by homologous recombination (Longtine et al., 1998). C-terminal tagging of genes was also performed by homologous recombination followed by drug selection. Site-directed mutagenesis was performed by PCR in which primers containing the mutation were used to generate two PCR fragments that contain ~45 bp overlapping regions containing the mutation. Fusion PCR or isothermal assembly (Gibson et al., 2009) was then performed to fuse the fragments together and the product was used for transformation. The introduced mutations and the full-length gene sequences were verified by DNA sequencing.
Yeast Cell Lysis and Immunoprecipitation
Typically ~100 OD yeast cells were lysed in 1 ml lysis buffer (100 mM Tris-Cl pH 7.5, 100 mM NaCl, 50 mM NaF, 1 mM EDTA, 1 mM EGTA, 0.1% Tween-20, 10% glycerol, 50 mM sodium butyrate, 50 mM nicotinamide, 5 µM trichostatin A, 1 mM PMSF, 10 µM leupeptin, 5 µM pepstatin A, Roche protease inhibitor cocktail, 14 mM β-mercaptoethanol) by bead beating and immunoprecipitation was performed by incubating the lysate with 15 µl magnetic beads (Invitrogen) conjugated to 2 µg Flag M2 antibody (Sigma). After washing 3 times in the same buffer, the beads were boiled. For the in vitro SAGA autoacetylation experiment (Figure 2E), the SAGA complex was eluted off the beads with Flag peptide.
Identification of Proteins Acetylated during the OX Growth Phase of the YMC
Identification of acetylated proteins by affinity purification of the acetylated peptides was performed largely according to a previously published protocol (Kim et al., 2006). Details are provided in the Supplemental Experimental Procedures. Candidate genes were FLAG-tagged at the C-terminus. Immunoprecipitation of the tagged protein from cells grown to log phase in YPGE (YP + glycerol, ethanol) culture was performed followed by Western blot using an antibody against acetyl-lysine (Immunechem #ICP0380) to confirm acetylation of the candidate protein. To determine whether the candidate protein was dynamically acetylated, cells were collected over 12 time points of the YMC and analyzed similarly. Not all proteins were dynamically acetylated across the YMC (Figure S6).
Histone Western Blots
Western blots to detect histone modifications at the bulk level were performed using a protocol adapted from previously published studies (Keogh et al., 2006). Western blots were performed using commercially available antibodies that are highly specific for the indicated modifications (Suka et al., 2001) listed in the Supplemental Experimental Procedures.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed according to a well-established protocol with minor modifications (Ezhkova and Tansey, 2006). Lysates were sonicated using Bioruptor. For ChIP with histone antibodies, 50 µl whole cell extract (WCE) was diluted 1:10 and used for each ChIP. For SAGA ChIP, 500 µl WCE and 2 µg Flag M2 antibody was used. Real-time PCR was performed in triplicate using SYBR green reagents from Applied Biosystems. PCR primers were designed by Primer Express software from Applied Biosystems to target every ~250 bp region from −750 bp before to 750 bp after the start codon of the selected gene.
ChIP-Seq
Library construction was performed using a protocol from the following website: http://bioinfo.mbb.yale.edu/array/Solexa_LibraryPrep_20080229ge.pdf Sequencing was performed on an Illumina GA IIX supervised by the UTSW Microarray Core Facility. H3K9ac/H3 data was analyzed using CLC Genomics Workbench software. Peaks were called with a maximum FDR = 1% and maximum p-value for Wilcoxon test = 1.0E-4. SAGA/Input data was assembled to the reference genome by Bowtie and analyzed by CisGenome as described in its online tutorial (Ji et al., 2008). Peaks with a fold-enrichment value max|FC| above 2 (>4-fold change) were selected. Peaks were annotated with the gene whose transcription start site (TSS) is closest to the peak region. The CisGenome browser was used to visualize ChIP-Seq peaks (Ji et al., 2008).
In vitro HAT Assay
SAGA was immunoprecipitated from OX and RC phase of the YMC as described above. Beads were washed twice with HAT buffer (50 mM Tris•Cl pH 8.0, 1 mM DTT, 5% glycerol, 1 mM PMSF, 0.03% NP-40) and incubated with HeLa core histones or long oligonucleosomes in the presence of 3H-acetyl-CoA in HAT buffer at 30°C for 1 hr. The reaction was spotted onto phosphocellulose filter paper, dried and washed twice with 50 mM NaHCO3-Na2CO3 buffer (pH 9.2), and rinsed with acetone. The dried filter paper was placed into vials for liquid scintillation counting to quantitate 3H signal.
Metabolic Cycles and Metabolite Analysis
Chemostat growth, metabolite extraction, and LC-MS/MS analysis of cellular metabolites were performed as described previously (Tu et al., 2005; Tu et al., 2007).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jerry Workman, Joe Reese, Cheng-Ming Chiang for generously sharing antibodies, the UTSW Protein Chemistry Technology Core Facility and the Scripps Center for Mass Spectrometry for protein identification and post-translational modification analysis, the UTSW Microarray Core for Illumina sequencing, Zheng Kuang, Mark Borromeo for help with ChIP-Seq analysis, Steve McKnight, Sung Chan Kim, Michael Springer, Noah Dephoure, Sean Taverna, Ralph Deberardinis, Kosaku Uyeda, and members of the Tu Lab for helpful discussions. This work was supported by the UTSW Endowed Scholars Program (B.P.T.), a Burroughs Wellcome Fund Career Award in Biomedical Sciences (B.P.T.), a Welch Foundation Research Grant I-1697 (B.P.T.), a Research Excellence Award from the W. M. Keck Foundation (B.P.T.), award R01GM094314 from the National Institute of General Medical Sciences (B.P.T.), a David and Lucile Packard Foundation Fellowship (B.P.T.), a Damon Runyon-Rachleff Innovation Award (B.P.T.), and a Frank and Sara McKnight Graduate Fellowship (L.C.). ChIP-Seq data have been deposited at GEO (accession number GSE28734).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci U S A. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Dechant R, Peter M. Nutrient signals driving cell growth. Curr Opin Cell Biol. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP. Chromatin immunoprecipitation to study protein-DNA interactions in budding yeast. Methods Mol Biol. 2006;313:225–244. doi: 10.1385/1-59259-958-3:225. [DOI] [PubMed] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37:3969–3980. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Kim J, Roeder RG. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol. 2009;29:266–280. doi: 10.1128/MCB.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. "Sleeping beauty": quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Langer MR, Fry CJ, Peterson CL, Denu JM. Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J Biol Chem. 2002;277:27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- Laxman S, Tu BP. Systems approaches for the study of metabolic cycles in yeast. Curr Opin Genet Dev. 2010;20:599–604. doi: 10.1016/j.gde.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lu C, Brauer MJ, Botstein D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol Biol Cell. 2009;20:891–903. doi: 10.1091/mbc.E08-08-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Mischerikow N, Spedale G, Altelaar AF, Timmers HT, Pijnappel WW, Heck AJ. In-depth profiling of post-translational modifications on the related transcription factor complexes TFIID and SAGA. J Proteome Res. 2009;8:5020–5030. doi: 10.1021/pr900449e. [DOI] [PubMed] [Google Scholar]
- Parrella E, Longo VD. The chronological life span of Saccharomyces cerevisiae to study mitochondrial dysfunction and disease. Methods. 2008;46:256–262. doi: 10.1016/j.ymeth.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowicka M, Kudlicki A, Tu BP, Otwinowski Z. High-resolution timing of cell cycle-regulated gene expression. Proc Natl Acad Sci U S A. 2007;104:16892–16897. doi: 10.1073/pnas.0706022104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Sutter BM, Ye X, Tu BP. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell. 2010;21:1982–1990. doi: 10.1091/mbc.E10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. Evidence of carbon monoxide-mediated phase advancement of the yeast metabolic cycle. Proc Natl Acad Sci U S A. 2009;106:14293–14296. doi: 10.1073/pnas.0907786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol. 2000;26:706–714. doi: 10.1016/s0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Newcomb L, Heideman W. Regulation of gene expression by glucose in Saccharomyces cerevisiae: a role for ADA2 and ADA3/NGG1. J Bacteriol. 1999;181:4755–4760. doi: 10.1128/jb.181.16.4755-4760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.